ABSTRACT
The phenomenon of protein superstructural polymorphism has become the subject of increased research activity. Besides the relevance to explain the existence of multiple prion strains, such activity is partly driven by the recent finding that in many age-related neurodegenerative diseases highly ordered self-associated forms of peptides and proteins might be the structural basis of prion-like processes and strains giving rise to different disease phenotypes.
Biophysical studies of prion strains have been hindered by a lack of tools to characterize inherently noncrystalline, heterogeneous and insoluble proteins. A description of the pressure response of prion quaternary structures might change this picture. This is because applying pressure induces quaternary structural changes of PrP, such as misfolding and self-assembly. From the thermodynamics of these processes, structural features in terms of associated volume changes can then be deduced. We suggest that conformation-enciphered prion strains can be distinguished in terms of voids in the interfaces of the constituting PrP protomers and thus in their volumetric properties.
The assembly of complex protein superstructures, such as oligomers and amyloid fibrils, from simpler misfolded proteins or peptides is a hallmark of various, seemingly unrelated disorders.Citation1 These include prion diseases, Alzheimer's, synucleinopathies, and type II diabetes, all of which are progressive disorders with associated high morbidity and mortality.
In prion diseases, the prion protein (PrPC) misfolds and assembles into various self-propagating quaternary structures with predominantly β sheet secondary structure, designated PrPSc (Sc for scrapie, a prion disease of small ruminants).Citation2 PrP assemblies formed in vitro can show significant variability in their structures and biological effects,Citation3-8 yet the structural basis of PrPSc in vivo has remained elusive so far. It is generally accepted that the extreme structural plasticity of brain-derived PrPSc occurs through conformational changes within protomers or through rearrangements of the subunits relative to each other. Besides underlying the generally observed amyloid polymorphism, these changes appear sufficiently different and stable to allow the existence of biologically distinct prion strains within the same host-species.Citation9-16 Within the context of the protein-only hypothesis, a strain is therefore viewed as a protein superstructural identity that confers distinctive symptomology, as well as a precise pathobiology and infectious character to PrPSc. Moreover, such PrP superstructures can be maintained indefinitely, under defined conditions, by repeated passaging in laboratory animals.
Another layer of complexity in describing PrP structural features comes from recent findings suggesting that beyond the inter-strain variability, a prion strain itself may be composed of an ensemble of superstructures. Indeed, PrPSc might exist as an ensemble of nonidentical but related molecular forms in a dynamic equilibrium (i.e. that can interconvert).Citation17 Such a structural network or quasispecies,Citation18,19 might be maintained in vivo under the selection pressure imposed by complex interactions with other macromolecular partners. In this regard, a number of molecules have been proposed to be involved in the in vivo PrP misfolding.Citation20-22 We have recently demonstrated that both endogenous Shadoo, a protein that resembles the flexibly disordered N-terminal domain of PrPC, and acetylcholinesterase can affect PrP structural dynamics and therefore modify the physicochemical properties of PrPSc.Citation23,24 From a thermodynamic point of view, a particular environment leads predictably to a dominant PrPSc morphotype preferentially stabilized/propagated, together with less populated superstructures. The “strain shift” or “strain mutation” phenomenon that is observed in prion diseases upon passage to another host,Citation25 could therefore be readily explained by a preferential selection of alternative dominant superstructures, probably due to changes in the PrP primary sequence (between species) or to a fine-tuned, tissue-specific interactome (within and between species).
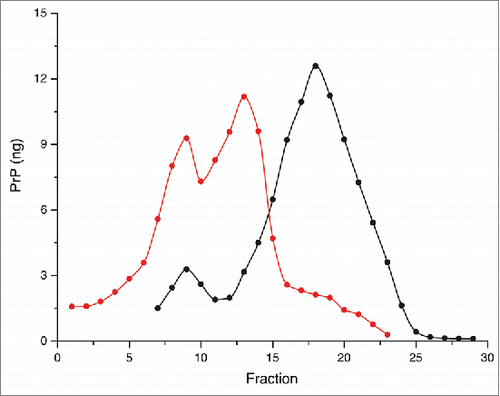
While a close correlation between diverse prion strain phenotypes and PrPSc conformational stability (against thermal or chaotropic denaturation), as well as relative susceptibility to proteolytic cleavage is widely recognized,Citation5,26-32 less is known about the underlying physical and structural basis characterizing such assemblies. There is limited evidence showing the diversity and complexity of PrP assemblies within an isolated strain. Our recent results obtained applying sedimentation fractionation techniques to a panel of prion strains are consistent with the essential role of the hydrodynamic properties (hydrodynamic size) of PrPSc in prion conversion efficiency and duration of diseaseCitation33 Another key parameter distinguishing various PrPSc superstructures, and differing between PrPSc and PrPC is the volume (i.e density).Citation34 Of particular interest is that PrPSc is found in fractions of lower density than PrPC after isopycnic density gradient centrifugation (equilibrium separation) ( and ref. Citation33). This observation is consistent with the presence of packing defects that might arise during the assembly of the PrP protomers to form the PrPSc assemblies. Remarkably, the sedimentation profile of several distinct prion strains from different species reveals at least 2 PrPSc populations with different density properties ( and ref. Citation33), supporting the existence of prion superstructural heterogeneity.
Figure 1. Solubilized brain material from uninfected (black) or 263K (red) infected hamsters was fractionated by sedimentation at the equilibrium. The collected fractions (numbered from top to bottom of the gradient) were analyzed for PrP content by immunoblot. The amount (in ng) of PrP per fraction was reported on the graph by using a standard curve of recombinant PrP. For methodological details see ref. Citation34.
It is therefore likely that thermodynamically and conformationally unique PrP superstructures differ in their hydration and packing, 2 properties that modify directly the volume of the system. In line with this view, a recent study using 2 PrP amyloid fibrils formed from the recombinant protein demonstrated that stability differences might be governed by distinct packing arrangements.Citation35 Hence, we believe that the bedrock principle of PrPSc polymorphism must be largely determined by the interfaces of the constituting PrP protomers. These interfaces should exhibit significantly different packing defects between distinct superstructures, yet conserving their basic structural arrangement of cross β-sheet. Nevertheless, the physical nature of such protein boundaries, the influence of inter-subunit cavities in PrP conformational changes, and its functional link to prion replication and pathogenesis remain to be determined.
It was recently demonstrated that eliminating the cavity volume in a protein acts as the principal determinant for its destabilization by pressure.Citation36 In contrast to temperature, or the use of exogenous chemical agents, pressure is a fundamental thermodynamic parameter that affects only the volume of the system under study. Pressure changes the Gibbs free energy of a reversible process in proportion to its volume variation, favoring processes accompanied by a volume decrease. Because of its unique potential, the influence of pressure on the energy landscapes for protein folding and aggregation has not escaped the consideration of the scientific community.Citation37,38 The pressure response of a protein mainly consists in the unfolding of its 3 dimensional structure and, in the case of a protein assembly, in its dissociation.
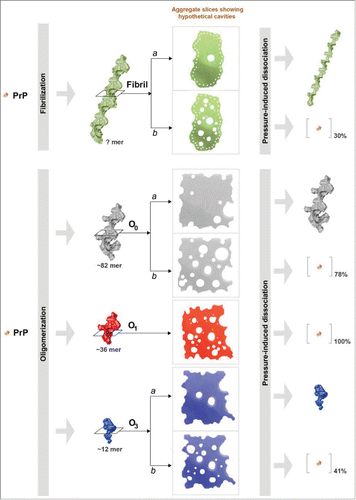
We and other groups have used pressure as an external perturbation for shifting the dynamic balance between native and unfolded/misfolded PrP states.Citation39-44 These studies evidenced that the conformational changes mimicking the PrP structural conversion have important impacts in the PrP volumetric properties. Therefore, PrP structural variations are characteristically associated with changes in hydration and cavities. We previously found that the application of pressure to the native PrP state effectively populates aggregation-prone intermediates and, depending on the interplay of different parameters, such as temperature, pH, incubation time, distinct thermodynamically stable assemblies were obtained.Citation42,43 The combined effects of pressure and temperature also lead to a decrease in the infectivity titer in processed meat contaminated by prions.Citation45-47 Although not proven, these irreversible effects are certainly due to a pressure-induced destabilization of PrPSc. This would mean that PrP assemblies (i.e., amyloid fibrils or oligomers) dissociate under pressure—at least to some extent. The impact of pressure on various PrP quaternary structures was recently studied in our group. We demonstrated that a pressure treatment leads to increasing destabilization of the assembled state, relative to the monomeric form. We therefore characterized the kinetic and energetic behavior of the pressure-induced dissociation of mature amyloid PrP fibrilsCitation48 and of 3 distinct misfolded PrP oligomers of different sizesCitation49 that were generated in vitro from recombinant PrP. shows a schematic representation of the effects of pressure on these various quaternary structures. The pressure-induced dissociation of fibrils was not complete, indicating structural heterogeneity of the starting amyloid fibrils. We considered a population of initial PrP fibrils with few packing defects (denoted as “a” in the scheme). These fibrils underwent a conformational rearrangement under pressure that led to a physicochemically different fibril, characterized by an irreversible loss of several amyloid specific features, including loss of thioflavin T binding. In contrast, a population of fibrils characterized by a low degree of packing (denoted as “b”), disaggregates into native monomers. In agreement with these results, different PrP oligomers exhibit different pressure-sensitivities. This can be explained by strongly varying void volumes in their quaternary structure. As for amyloid fibrils, we also brought evidence of 2 different degrees of barostability within a particular oligomer type (specifically for O0 and O3 oligomers), reflecting that 2 populations of PrP superstructures coexist in these samples of initial PrP fibrils.
Figure 2. Schematic representation of the effects of high pressure on mature PrP amyloid fibrils and 3 distinct β-sheet-rich PrP oligomers. This diagram illustrates the proposed model of pressure-induced structural changes, which depends on the strongly varying cavities present in their quaternary structure. The amount (% on the entire protein sample) of resolubilized PrP rescued from the superstructure after the pressure treatment is also shown.
These results open the opportunity to detect small differences in the conformational stability of quaternary structures associated with a particular strain, and between strains. Many important questions and extra-views lay ahead. Expanding this study to a large collection of brain samples from scrapie-infected animals might therefore be useful, not only to unravel the predicted wealth of PrPSc superstructures, but also to induce a shift of the dominant superstructures. If minor PrPSc structures that coexist during disease propagation have less solvent excluded volumes, these otherwise dismissed forms will be selected and enriched under high pressure, and could be subsequently amplified by standard techniques, such as including protein misfolding cyclic amplification (PMCA),Citation50,51 the amyloid seeding assay (ASA),Citation52 or quaking-induced conversion (QUIC).Citation53,54
The latest results reviewed here show that defects in atomic packing may deserve consideration as a new factor that influences the PrPSc assembly puzzle. We conclude that pressure provides an efficient tool to ascertain the highly diversified spatial arrangement between PrP protomers. Its use will provide information not only on mammalian prion self-perpetuating structural states, but also on other complex protein superstructures associated with a range of debilitating medical disorders, as well as on many newly discovered beneficial prions and prion-like proteins that are turning out to be versatile components in various physiological processes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
REFERENCES
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol 2014; 15:384-96; PMID:24854788; http://dx.doi.org/10.1038/nrm3810
- Prusiner SB. Molecular biology of prion diseases. Science 1991; 252:1515-22; PMID:1675487; http://dx.doi.org/10.1126/science.1675487
- Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol 2010; 119:177-87; PMID:20052481; http://dx.doi.org/10.1007/s00401-009-0633-x
- Legname G, Baskakov IV, Nguyen HO, Riesner D, Cohen FE, DeArmond SJ, Prusiner SB. Synthetic mammalian prions. Science 2004; 305:673-6; PMID:15286374; http://dx.doi.org/10.1126/science.1100195
- Colby DW, Giles K, Legname G, Wille H, Baskakov IV, DeArmond SJ, Prusiner SB. Design and construction of diverse mammalian prion strains. Proc Natl Acad Sci U S A 2009; 106:20417-22; PMID:19915150; http://dx.doi.org/10.1073/pnas.0910350106
- Colby DW, Wain R, Baskakov IV, Legname G, Palmer CG, Nguyen HO, Lemus A, Cohen FE, DeArmond SJ, Prusiner SB. Protease-sensitive synthetic prions. PLoS Pathog 2010; 6:e1000736; PMID:20107515; http://dx.doi.org/10.1371/journal.ppat.1000736
- Raymond GJ, Race B, Hollister JR, Offerdahl DK, Moore RA, Kodali R, Raymond LD, Hughson AG, Rosenke R, Long D, et al. Isolation of novel synthetic prion strains by amplification in transgenic mice coexpressing wild-type and anchorless prion proteins. J Virol 2012; 86:11763-78; PMID:22915801; http://dx.doi.org/10.1128/JVI.01353-12
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132-5; PMID:20110469; http://dx.doi.org/10.1126/science.1183748
- Bessen RA, Marsh RF. Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J Virol 1994; 68:7859-68; PMID:7966576
- Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 1995; 375:698-700; PMID:7791905; http://dx.doi.org/10.1038/375698a0
- Telling GC, Parchi P, DeArmond SJ, Cortelli P, Montagna P, Gabizon R, Mastrianni J, Lugaresi E, Gambetti P, Prusiner SB. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 1996; 274:2079-82; PMID:8953038; http://dx.doi.org/10.1126/science.274.5295.2079
- Caughey B, Raymond GJ, Bessen RA. Strain-dependent differences in β-sheet conformations of abnormal prion protein. J Biol Chem 1998; 273:32230-5; PMID:9822701; http://dx.doi.org/10.1074/jbc.273.48.32230
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157-65; PMID:9771749; http://dx.doi.org/10.1038/2654
- Parchi P, Giese A, Capellari S, Brown P, Schulz-Schaeffer W, Windl O, Zerr I, Budka H, Kopp N, Piccardo P, et al. Classification of sporadic Creutzfeldt-Jakob disease based on molecular and phenotypic analysis of 300 subjects. Ann Neurol 1999; 46:224-33; PMID:10443888; http://dx.doi.org/10.1002/1531-8249(199908)46:2%3c224::AID-ANA12%3e3.0.CO;2-W
- Parchi P, Zou W, Wang W, Brown P, Capellari S, Ghetti B, Kopp N, Schulz-Schaeffer WJ, Kretzschmar HA, Head MW, et al. Genetic influence on the structural variations of the abnormal prion protein. Proc Natl Acad Sci U S A 2000; 97:10168-72; PMID:10963679; http://dx.doi.org/10.1073/pnas.97.18.10168
- Collinge J, Clarke AR. A general model of prion strains and their pathogenicity. Science 2007; 318:930-6; PMID:17991853; http://dx.doi.org/10.1126/science.1138718
- Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science 2010; 327:869-72; PMID:20044542; http://dx.doi.org/10.1126/science.1183218
- Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften 1971; 58:465-523; PMID:4942363; http://dx.doi.org/10.1007/BF00623322
- Epstein IR, Eigen M. Selection and self-organization of self-reproducing macromolecules under the constraint of constant flux. Biophys Chem 1979; 10:153-60; PMID:486701; http://dx.doi.org/10.1016/0301-4622(79)85035-8
- Deleault NR, Walsh DJ, Piro JR, Wang F, Wang X, Ma J, Rees JR, Supattapone S. Cofactor molecules maintain infectious conformation and restrict strain properties in purified prions. Proc Natl Acad Sci U S A 2012; 109:E1938-46; PMID:22711839; http://dx.doi.org/10.1073/pnas.1206999109
- Ma J. The role of cofactors in prion propagation and infectivity. PLoS Pathog 2012; 8:e1002589; PMID:22511864; http://dx.doi.org/10.1371/journal.ppat.1002589
- Aguzzi A, Heikenwalder M, Polymenidou M. Insights into prion strains and neurotoxicity. Nat Rev Mol Cell Biol 2007; 8:552-61; PMID:17585315; http://dx.doi.org/10.1038/nrm2204
- Ciric D, Richard CA, Moudjou M, Chapuis J, Sibille P, Daude N, Westaway D, Adrover M, Béringue V, Martin D, et al. Interaction between Shadoo and PrP Affects the PrP-Folding Pathway. J Virol 2015; 89:6287-93; PMID:25855735; http://dx.doi.org/10.1128/JVI.03429-14
- Torrent J, Vilchez-Acosta A, Munoz-Torrero D, Trovaslet M, Nachon F, Chatonnet A, Grznarova K, Acquatella-Tran Van Ba I, Le Goffic R, Herzog L, et al. Interaction of prion protein with acetylcholinesterase: potential pathobiological implications in prion diseases. Acta Neuropathol Commun 2015; 3:18; PMID:25853328; http://dx.doi.org/10.1186/s40478-015-0188-0
- Li J, Mahal SP, Demczyk CA, Weissmann C. Mutability of prions. EMBO Rep 2011; 12:1243-50; PMID:21997293; http://dx.doi.org/10.1038/embor.2011.191
- Legname G, Nguyen HO, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S A 2006; 103:19105-10; PMID:17142317; http://dx.doi.org/10.1073/pnas.0608970103
- Legname G, Nguyen HO, Baskakov IV, Cohen FE, Dearmond SJ, Prusiner SB. Strain-specified characteristics of mouse synthetic prions. Proc Natl Acad Sci U S A 2005; 102:2168-73; PMID:15671162; http://dx.doi.org/10.1073/pnas.0409079102
- Sigurdson CJ, Nilsson KP, Hornemann S, Manco G, Polymenidou M, Schwarz P, Leclerc M, Hammarström P, Wüthrich K, Aguzzi A. Prion strain discrimination using luminescent conjugated polymers. Nat Methods 2007; 4:1023-30; PMID:18026110; http://dx.doi.org/10.1038/nmeth1131
- Bett C, Kurt TD, Lucero M, Trejo M, Rozemuller AJ, Kong Q, Nilsson KP, Masliah E, Oldstone MB, Sigurdson CJ. Defining the conformational features of anchorless, poorly neuroinvasive prions. PLoS Pathog 2013; 9:e1003280; PMID:23637596; http://dx.doi.org/10.1371/journal.ppat.1003280
- Ayers JI, Schutt CR, Shikiya RA, Aguzzi A, Kincaid AE, Bartz JC. The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog 2011; 7:e1001317; PMID:21437239; http://dx.doi.org/10.1371/journal.ppat.1001317
- Peretz D, Williamson RA, Legname G, Matsunaga Y, Vergara J, Burton DR, DeArmond SJ, Prusiner SB, Scott MR. A change in the conformation of prions accompanies the emergence of a new prion strain. Neuron 2002; 34:921-32; PMID:12086640; http://dx.doi.org/10.1016/S0896-6273(02)00726-2
- Bett C, Joshi-Barr S, Lucero M, Trejo M, Liberski P, Kelly JW, Masliah E, Sigurdson CJ. Biochemical properties of highly neuroinvasive prion strains. PLoS Pathog 2012; 8:e1002522; PMID:22319450; http://dx.doi.org/10.1371/journal.ppat.1002522
- Tixador P, Herzog L, Reine F, Jaumain E, Chapuis J, Le Dur A, Laude H, Béringue V. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog 2010; 6:e1000859; PMID:20419156; http://dx.doi.org/10.1371/journal.ppat.1000859
- Laferriere F, Tixador P, Moudjou M, Chapuis J, Sibille P, Herzog L, Reine F, Jaumain E, Laude H, Rezaei H, et al. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog 2013; 9:e1003702; PMID:24130496; http://dx.doi.org/10.1371/journal.ppat.1003702
- Cobb NJ, Apostol MI, Chen S, Smirnovas V, Surewicz WK. Conformational stability of mammalian prion protein amyloid fibrils is dictated by a packing polymorphism within the core region. J Biol Chem 2014; 289:2643-50; PMID:24338015; http://dx.doi.org/10.1074/jbc.M113.520718
- Roche J, Caro JA, Norberto DR, Barthe P, Roumestand C, Schlessman JL, Garcia AE, García-Moreno BE, Royer CA. Cavities determine the pressure unfolding of proteins. Proc Natl Acad Sci U S A 2012; 109:6945-50; PMID:22496593; http://dx.doi.org/10.1073/pnas.1200915109
- Silva JL, Foguel D, Royer CA. Pressure provides new insights into protein folding, dynamics and structure. Trends Biochem Sci 2001; 26:612-8; PMID:11590014; http://dx.doi.org/10.1016/S0968-0004(01)01949-1
- Marchal S, Font J, Ribo M, Vilanova M, Phillips RS, Lange R, Torrent J. Asymmetric kinetics of protein structural changes. Acc Chem Res 2009; 42:778-87; PMID:19378977; http://dx.doi.org/10.1021/ar800266r
- Kuwata K, Li H, Yamada H, Legname G, Prusiner SB, Akasaka K, James TL. Locally disordered conformer of the hamster prion protein: a crucial intermediate to PrPSc? Biochemistry 2002; 41:12277-83; PMID:12369815; http://dx.doi.org/10.1021/bi026129y
- Cordeiro Y, Kraineva J, Ravindra R, Lima LM, Gomes MP, Foguel D, Winter R, Silva JL. Hydration and packing effects on prion folding and β-sheet conversion. High pressure spectroscopy and pressure perturbation calorimetry studies. J Biol Chem 2004; 279:32354-9; PMID:15173173; http://dx.doi.org/10.1074/jbc.M404295200
- Kuwata K, Kamatari YO, Akasaka K, James TL. Slow conformational dynamics in the hamster prion protein. Biochemistry 2004; 43:4439-46; PMID:15078089; http://dx.doi.org/10.1021/bi036123o
- Torrent J, Alvarez-Martinez MT, Harricane MC, Heitz F, Liautard JP, Balny C, Lange R. High pressure induces scrapie-like prion protein misfolding and amyloid fibril formation. Biochemistry 2004; 43:7162-70; PMID:15170353; http://dx.doi.org/10.1021/bi049939d
- Torrent J, Alvarez-Martinez MT, Heitz F, Liautard JP, Balny C, Lange R. Alternative prion structural changes revealed by high pressure. Biochemistry 2003; 42:1318-25; PMID:12564935; http://dx.doi.org/10.1021/bi0269916
- Torrent J, Alvarez-Martinez MT, Liautard JP, Balny C, Lange R. The role of the 132-160 region in prion protein conformational transitions. Protein Sci 2005; 14:956-67; PMID:15772306; http://dx.doi.org/10.1110/ps.04989405
- Brown P, Meyer R, Cardone F, Pocchiari M. Ultra-high-pressure inactivation of prion infectivity in processed meat: a practical method to prevent human infection. Proc Natl Acad Sci U S A 2003; 100:6093-7; PMID:12732724; http://dx.doi.org/10.1073/pnas.1031826100
- Fernandez Garcia A, Heindl P, Voigt H, Buttner M, Wienhold D, Butz P, Stärke J, Tauscher B, Pfaff E. Reduced proteinase K resistance and infectivity of prions after pressure treatment at 60 °C. J Gen Virol 2004; 85:261-4; PMID:14718641; http://dx.doi.org/10.1099/vir.0.19410-0
- Garcia AF, Heindl P, Voigt H, Buttner M, Butz P, Tauber N, Tauscher B, Pfaff E. Dual nature of the infectious prion protein revealed by high pressure. J Biol Chem 2005; 280:9842-7; PMID:15598650; http://dx.doi.org/10.1074/jbc.M410679200
- El Moustaine D, Perrier V, Acquatella-Tran Van Ba I, Meersman F, Ostapchenko VG, Baskakov IV, Lange R, Torrent J. Amyloid features and neuronal toxicity of mature prion fibrils are highly sensitive to high pressure. J Biol Chem 2011; 286:13448-59; PMID:21357423; http://dx.doi.org/10.1074/jbc.M110.192872
- Torrent J, Lange R, Rezaei H. The volumetric diversity of misfolded prion protein oligomers revealed by pressure dissociation. J Biol Chem 2015; 290(33):20417-26; PMID:26126829
- Saa P, Castilla J, Soto C. Presymptomatic detection of prions in blood. Science 2006; 313:92-4; PMID:16825570; http://dx.doi.org/10.1126/science.1129051
- Atarashi R, Moore RA, Sim VL, Hughson AG, Dorward DW, Onwubiko HA, Priola SA, Caughey B. Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nat Methods 2007; 4:645-50; PMID:17643109; http://dx.doi.org/10.1038/nmeth1066
- Colby DW, Zhang Q, Wang S, Groth D, Legname G, Riesner D, Prusiner SB. Prion detection by an amyloid seeding assay. Proc Natl Acad Sci U S A 2007; 104:20914-9; PMID:18096717; http://dx.doi.org/10.1073/pnas.0710152105
- Atarashi R, Wilham JM, Christensen L, Hughson AG, Moore RA, Johnson LM, Onwubiko HA, Priola SA, Caughey B. Simplified ultrasensitive prion detection by recombinant PrP conversion with shaking. Nat Methods 2008; 5:211-2; PMID:18309304; http://dx.doi.org/10.1038/nmeth0308-211
- Orru CD, Wilham JM, Hughson AG, Raymond LD, McNally KL, Bossers A, Ligios C, Caughey B. Human variant Creutzfeldt-Jakob disease and sheep scrapie PrP(res) detection using seeded conversion of recombinant prion protein. Protein Eng Des Sel 2009; 22:515-21; http://dx.doi.org/10.1093/protein/gzp031
