ABSTRACT
In bacterial plasmids, Rep proteins initiate DNA replication by undergoing a structural transformation coupled to dimer dissociation. Amyloidogenesis of the ‘winged-helix’ N-terminal domain of RepA (WH1) is triggered in vitro upon binding to plasmid-specific DNA sequences, and occurs at the bacterial nucleoid in vivo. Amyloid fibers are made of distorted RepA-WH1 monomers that assemble as single or double intertwined tubular protofilaments. RepA-WH1 causes in E. coli an amyloid proteinopathy, which is transmissible from mother to daughter cells, but not infectious, and enables conformational imprinting in vitro and in vivo; i.e. RepA-WH1 is a ‘prionoid’. Microfluidics allow the assessment of the intracellular dynamics of RepA-WH1: bacterial lineages maintain two types (strains-like) of RepA-WH1 amyloids, either multiple compact cytotoxic particles or a single aggregate with the appearance of a fluidized hydrogel that it is mildly detrimental to growth. The Hsp70 chaperone DnaK governs the phase transition between both types of RepA-WH1 aggregates in vivo, thus modulating the vertical propagation of the prionoid. Engineering chimeras between the Sup35p/[PSI+] prion and RepA-WH1 generates [REP-PSI+], a synthetic prion exhibiting strong and weak phenotypic variants in yeast. These recent findings on a synthetic, self-contained bacterial prionoid illuminate central issues of protein amyloidogenesis.
Introduction
Along the last few years, the field of functional amyloids, i.e., amyloid assemblies with a physiological role and no proteinopathic side effects, has attracted much attention.Citation1,2 Bacterial functional amyloids are invariantly extracellular, acting as scaffolding devices in building biofilms, three-dimensional networks that serve to colonize solid surfaces and confer to bacteria persistence against chemotherapy with antibiotics.Citation3 Amyloids also serve as inactive storage deposits of the antimicrobial microcins.Citation4 In eukaryotic cells, functional amyloids are either epigenetic determinants of non-Mendelian transmissible characters, as in the case of yeast prions,Citation5 or scaffolding modules, as for mammalian Pmel17 in melanocytes;Citation6 or even regulators of long-term synaptic potentiation, as proteins of the CPEB family from mollusks to mice.Citation7 Among functional amyloids, the ability of the Escherichia coli curli system (CsgABC/CsgDEFG) to export nearly any protein tagged to a CsgA-derived secretion signal has enabled the extracellular screening of the amyloidogenic potential of whole proteomes and also of anti-amyloidogenic compounds.Citation8,9
The lack of natural intracellular amyloid proteinopathies in bacteria and the fact that expressing any amyloidosis-related human protein in E. coli leads to the formation of inclusion bodies, as many other heterologous mammalian proteins do, initially provoked the dismissal of bacteria as model organisms for studies on protein amyloidoses. Furthermore, although inclusion bodies exhibit amyloid features, they do not hamper bacterial viability in a significant way.Citation10 Interestingly, aggregation is a natural resource in some bacterial proteins in which irreversible structural changes are used to transit across distinct, mutually exclusive, functional states.Citation1 This is the case of the replication protein RepA of the Pseudomonas plasmid pPS10,Citation11 which undergoes a transition through three association states, each of them linked to a defined function: from stable soluble dimers (transcriptional repressors of repA gene expression), RepA dissociates into metastable monomers (acting as plasmid DNA replication initiators), which then aggregate as oligomers that inhibit new rounds of DNA replication (by holding together two plasmid molecules through their replication origins).Citation12-15 In all these cases, binding to distinct DNA sequences in the plasmid triggers allosteric conformational changes that affect the structure of the N-terminal dimerization winged-helix domain (WH1).Citation16 While tracking the molecular basis for the functional aggregation of RepA, in 2007 we reported that its isolated WH1 domain, in its mutant variant A31V, was able to assemble into amyloid fibers, built on the core amyloidogenic stretch L26VLCAVSLI34 and upon a conformational change promoted by transient WH1 binding to a short plasmid-specific dsDNA sequence.Citation17 We proved that, similarly to its action enabling DNA replication initiation through RepA monomerization,Citation13,16 dsDNA acted as an allosteric effector on RepA-WH1 amyloidogenesis by enhancing its assembly into amyloid fibers.Citation17 Furthermore, we showed that a small organic molecule (S4-indigo) was able to inhibit such a process by competing with DNA for the binding to RepA-WH1 in vitro.Citation18 Aiming to characterize RepA-WH1 amyloidogenesis in vivo, we discovered that expressing fusions of the bacterial protein to a fluorescent protein tracer (mCherry/mRFP), thus replacing the natural C-terminal WH2 domain in RepA,Citation12,13 generated in E. coli a synthetic amyloid proteinopathy that severely reduced bacterial proliferation and finally led to cell death.Citation19 These in vivo generated aggregates templated the amyloid conformation on soluble RepA-WH1 molecules in vitro,Citation19 as they also did in a mutant variant-specific way in vivo,Citation20 a signature for prion and prion-like proteins. These findings were already discussed in another publication in this journal dated in 2011,Citation21 and thus will not be treated extensively again here. Instead, we will now review novel results recently reported on the RepA-WH1 prionoid and provide additional insight on the topic.
Structural Polymorphism in RepA-WH1: from Amyloid Nano-Springs and Tubes to Chimeric Yeast Prions
Structural polymorphism, i.e. the alternative ways in which a particular macromolecule can arrange its tertiary and/or quaternary structures, is an intrinsic feature of amyloid assemblies.Citation22 Polymorphism has been described from the subtleties of the packing of protein side chains and strands in cross-β assemblies to the pitch of twisted amyloid fibers.Citation23,24 Moreover, the ways and strengths in which phenotypes associated to a particular amyloidogenic protein show up, either in disease or within a physiological function, bear an intimate relation with polymorphism.Citation25,26
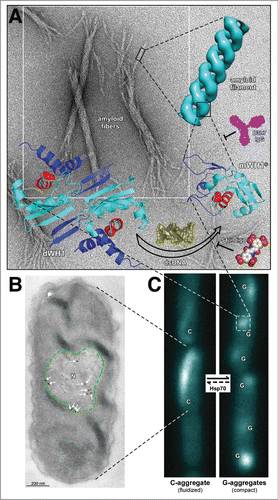
Recent electron microscopy (EM) and atomic force microscopy (AFM) studies on the amyloid fibers, assembled in vitro by templating on soluble RepA-WH1 molecules the amyloid conformation present in the RepA-WH1-mCherry aggregates purified from bacteria, revealed that the characteristic ≈25 nm-wide amyloid fibers are actually composed of several coiled filaments, each of them with ≈4 nm width ().Citation27 In turn, these filaments consist either in a single or a double thread of RepA-WH1 molecules that, with the limitations imposed by the low resolution of the EM reconstruction, are built by distorted monomers. This was inferred from the loose fit of a model based on the crystal structure of a replication-competent monomeric WH1 domain into the EM volume, as well as from circular dichroism (CD) spectra showing an increase in β-sheet structure upon RepA-WH1 assembly, as expected in any amyloidogenesis.Citation27 Polymorphism is thus manifested at three levels of increasing complexity: the number of threads that constitute the amyloid filaments (either one or two); the number of filaments per fiber bundle (mode value, 6); and variations in the pitch (on average, 64 nm) of the superhelix that results from twisting several filaments into the mature fibers. Disregarding if the assembly of the fibers was triggered by dsDNA or by templating with purified RepA-WH1-mCherry seeds, and by either leaving the samples to stand in the fridge or agitating them at higher temperatures, which respectively retards or accelerates amyloidogenesis, the fibers generated exhibit the aforementioned polymorphism.Citation27 Interestingly, the overall architecture of the twisted RepA-WH1 amyloid filaments can be described as hollow springs (single) or tubules (double filaments) with an axial cavity with a diameter around 2.5 nm. The latter is close to the average dimension of a similar cavity in the short tubular proteotoxic oligomers assembled by α-synuclein.Citation28 If this is a mere coincidence or if it points to a common mechanism of cytotoxicity for both proteins (e.g., the assembly of pores at membranes) remains to be determined.
Figure 1 (See next page). Overview of RepA-WH1 amyloidogenesis, remarking hierarchical assembly in vitro (A) and phase transitions in vivo (B,C). (A) Stable dimers of RepA-WH1 (dWH1) undergo a structural transformation upon transient, low affinity binding to dsDNA, thus resulting in metastable, aggregation-prone monomers (mWH1*).Citation13 The core of the WH domain is colored cyan, whereas segments showing significant conformational changes are in blue. The amyloidogenic peptide L26VLCAVSLI34 is depicted in red, with the side-chain of the hyper-amyloidogenic mutant residue A31V shown as spheres.Citation17 Binding of dsDNA (yellow) to dWH1 disrupts the dimerization interface, thus generating partially unfolded mWH1* monomers which assemble as helical amyloid tubular filaments.Citation17,27 Binding of RepA-WH1 to dsDNA, and thus amyloidogenesis, can be competed by molecules of S4-indigo (spheres),Citation18 whereas the conformation specific antibody B3h7 (magenta) inhibits the assembly of RepA-WH1 oligomers into filaments.Citation38 Filaments further associate laterally and coil to generate the mature amyloid fibers (background EM).Citation17,27 (B) Electron micrograph showing an ultra-thin section through an E. coli cell incubated with the B3h7 antibody (arrows/dots: gold-conjugated secondary anti-mouse antibody), which reveals the preferential location of pre-amyloidogenic RepA-WH1 aggregates at the nucleoid (N; green dashed line).Citation38 A fluidized C-type aggregate hydrogel is also outlined (cyan dotted line).Citation39 (C) E. coli cells growing in microfluidic channels and expressing the prionoid.Citation39 RepA-WH1 amyloid aggregates show two distinct appearances, i.e. single comet-like (C) aggregates behaving as a fluidized hydrogel that readily splits on cell division; or multiple globular (G) cytotoxic aggregates with the compactness characteristic of typical amyloid plaques.Citation39 While the phase transition from the C to the G aggregates occurs spontaneously in vivo, the reverse uphill transition is promoted by a single cell factor: the Hsp70 chaperone DnaK.Citation39
Regarding the lateral association of RepA-WH1 filaments, it is known for other amyloid fibers that divalent anions, in particular SO42−, play a role in bridging them together, side-by-side.Citation28-30 In the crystal structure of a RepA-WH1 dimer (PDB entry 1HKQ),Citation13 two symmetry-related molecules were in contact through a PO43− ion, which established a triple salt bridge between Lys85 in one molecule and Lys62 and His63 in the other. During the crystallization attempts, it became evident that incubation with monovalent anions (such as Cl− or acetate) did not result in crystal growth, whereas SO42− rendered poorer diffracting crystals, although this anion yielded the best fibers. The dependence on multivalent anions for RepA-WH1 crystallization matches their requirement in the lateral association of amyloid filaments to build the mature fibers. This could be interpreted as resulting from the overall electropositive charge of the RepA-WH1 molecule,Citation31 because lowering the ionic strength enhances the dissociation of the fiber ends to the point of releasing their constituent filaments.Citation27
What we have learnt on the structure of RepA-WH1 fibers opens an intriguing possibility: that the kind of assembly that monomers of this protein build upon amyloidogenesis could bear some relation to the regulatory complexes of the full-length RepA protein that inhibit new rounds of amplification in recently replicated plasmids.Citation15 This hypothesis, that needs experimental support, would imply that such regulatory complexes would be a new case of functional amyloid and so the cytotoxic RepA-WH1 prionoid would be the outcome of decoupling amyloidogenesis from its natural linkage to WH2 and plasmid replication,Citation12,13 thus unveiling the proteotoxic potential typical of amyloids.
Aiming to test in vivo the amyloidogenic potential of the L26VLCAVSLI34 stretch in RepA, 1-5 repeats of this sequence were used to replace, following the design of a parallel superpleated β-structure,Citation32 the oligopeptide Q/N-rich repeats at the N-terminal domain yeast protein Sup35p that, in its aggregated state, constitutes the [PSI+] prion.Citation33 As previously proved for a number of other mutant and heterologous sequences,Citation34-36 the resulting chimeric protein enables an epigenetically inheritable prion, named [REP-PSI+],Citation33 capable of reading-through the stop codon UAG, the phenotype characteristic of [PSI+],Citation5 provided that at least three repeats of the bacterial-related sequence were present.Citation33 The simultaneous presence of hydrophobic (RepA-WH1 derived) and polar (from the Sup35p scaffold) sequences in [REP-PSI+] results in a gradation of weak prion phenotypes/variants, as assessed by the pink color of colonies and the generation of very large aggregates, as evident in semi-denaturing detergent agarose gel electrophoresis (SDD-AGE).Citation33 Interestingly, biophysical analyses of these functional prion variants revealed that the presence of the hydrophobic RepA-WH1 repeats within the Sup35p prion domain increased the β-sheet structure while resulting in the formation of oligomers, not fibers as for the wild-type prion. The generation of [REP-PSI+] variants, which are mitotically stable in terms of propagation, illustrates the feasibility of engineering phenotypic traits in prions by fully exploiting the structural polymorphism of amyloids, from oligomers to fibers, as a constructive resource.Citation37
DNA and Hsp70-Modulated Phase Transitions in RepA-WH1 Amyloidogenesis
The effect of dsDNA as an allosteric effector of protein amyloidogenesis in vitro was a seminal discovery in RepA-WH1 research.Citation17 However, the evidence for a similar process in vivo was indirect, i.e., a role of DNA in promoting RepA-WH1 amyloidogenesis in E. coli cells was inferred from an increase in the number and size of protein aggregates when the vector expressing the prionoid carried tandem repeats of the DNA sequence that efficiently promoted amyloidogenesis in vitro.Citation19 The development of a monoclonal antibody (termed B3h7), characterized in vitro as specific for a pre-amyloid oligomeric conformation on-pathway to mature RepA-WH1 fibers (),Citation38 has recently made possible to unveil, through immuno-electron microscopy (iEM) (), that in E. coli cells expressing the prionoid the very first amyloid oligomeric precursors are found at the bacterial nucleoid,Citation38 as expected from the known ability of DNA to promote RepA-WH1 amyloidogenesis in vitroCitation17,18 and possibly from the original function of the WH1 domain when, in the whole RepA, controls plasmid DNA replication.Citation15 It was found, also by iEM, that amyloid particles exceeding ≈100 nm in diameter are placed out of the nucleoid territory,Citation39 as expected if entropic exclusion by the nucleoid were playing a mayor role in the localization of the aggregates. These findings could have implications for the preferential or alternative tropism of amyloid aggregates toward the cytoplasm or the nucleus in some human neurodegenerative proteinopathies, or in the case of proteins such as FUS, whose ability to assemble functional ribonucleoparticles in either location becomes impaired in amyotrophic lateral sclerosis and frontotemporal dementia.Citation40-42
Concerning the intracellular inclusions formed by amyloidogenic proteins either in the cytoplasm or in the nucleus of eukaryotic cells, it is remarkable the recent description of the existence of phase transitions between soluble, hydrogel and fibrilar states, in some cases through intermediates with the appearance of liquid droplets.Citation40-44 Such partitions imply changes in the association state beyond the quaternary structure, generating the so-called quinary structural level, having consequences for the functionality and bioavailability of the macromolecules involved. In many instances, these are proteins bearing domains with amino-acid compositions biased toward low complexities. In the case of Q/N/G-rich domains, phase transitions have been related with the package of mRNA in stress/P-granules and are relevant in amyloidogenesis.Citation40-42 Interestingly, albeit RepA-WH1 lacks such simplified amino-acid composition and has a very stable fold in its native state, when assembled as cytoplasmic aggregates in vivo, exhibits similar phase transitions, the first instance in which this has been observed in bacteria.Citation39 In these experiments, a combination of fluorescence microscopy and microfluidics was used, because this enabled monitoring bacterial proliferation and the dynamics of RepA-WH1 aggregation at the level of multiple single cell lineages, for many generations (up to 200) and under controlled environmental conditions. For studies on amyloidosis, surveying the physiology of bacteria in microfluidic devices surpasses the approaches based on agarose pads in terms of the number of generations achievable before the cessation of growth, the feasibility to track lineages and the ease to inject into, or extract from, the cultures diverse molecules of interest. Two clearly distinct RepA-WH1 aggregates became evident at the microfluidic device (): i) multiple globular, compact cytotoxic (G) particles that inhibit cell division; ii) a single, comet-shaped (C) aggregate that appears to be fluidized, i.e. elongates, contracts and undulates along the cell axis and readily splits between the daughter cells on cytokinesis.Citation39 The outcome is that while bacteria expressing the G-type aggregates show a tendency toward filamentation and cells die after a few generations, those bearing the C-type aggregates keep dividing indefinitely at a reasonable rate. In an isogenic population expressing RepA-WH1, bacteria at distinct microfluidic channels propagate for generations one or the other type of aggregates, although interconversion (more commonly C to G) occurs at low rates.Citation39 We also explored the effect of the Hsp70 chaperone DnaK on RepA-WH1 amyloidogenesis, because it is central in counteracting protein aggregation in bacteria and a role for DnaK in the activation of RepA-type proteins for plasmid replication was recognized for long.Citation11 It is the activity of DnaK, i.e., its expression induced from a regulated promoter vs. its specific inhibition with myricetin, what determines the balance and switching (phase transition) between both kinds of aggregates: DnaK promotes the conversion of most cells to the elongated phenotype, whereas its inactivation results in the population dominated by the globular phenotype.Citation39 iEM studies revealed that DnaK is preferentially located at the aggregate with an elongated and fluidized appearance, which constitutes a territory with a clearly different density toward the electrons compared with that observed for the globular aggregates.Citation39 Therefore this Hsp70 chaperone, most likely through its aggregate remodeling activity, is the factor determining quinary structural phase transitions for RepA-WH1 in the bacterial cytoplasm. On the contrary, the bacterial ClpB (Hsp100) chaperone has no apparent contribution to this effect. Coming to the point, the work carried out with our synthetic bacterial model prionoid suggests that the long sought cell factor(s) that would ameliorate amyloid cytotoxicity in human proteinopathies might be Hsp70 chaperones by promoting the assembly of pro-amyloidogenic proteins as non-fibrilar hydrogels.
Further perspectives
Ongoing research on the bacterial prionoid RepA-WH1 aims to trace a scheme of the molecular basis for amyloid cytotoxicity in such a minimal host as Escherichia coli, with the ambition of providing clues on the undeniably more complex mammalian amyloid proteinopathies. Such endeavor spans from a systems biology perspective, to highlight gene networks and metabolic pathways related to amyloid disease, to bottom-up approaches based on reconstructive, cytomimetic biochemistry performed with lipid vesicles.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
Acknowledgments
We are indebted to our colleagues who originally contributed to the work reviewed here, especially to M.E. Fernández-Tresguerres, A. Serrano and F. Gasset-Rosa, as well as to A. Lindner, O. Llorca, F. Moreno-Herrero and their co-workers.
Funding
Research on RepA-WH1 amyloids at CIB-CSIC is currently financed by Spanish MINECO grants BIO2012-30852 and CSD2009-00088.
REFERENCES
- Otzen D. Functional amyloid: Turning swords into plowshares. Prion 2010; 4:256-64; PMID:20935497; http://dx.doi.org/10.4161/pri.4.4.13676
- Romero D, Kolter R. Functional amyloids in bacteria. Int Microbiol 2014; 17:65-73; PMID:26418850
- DePas WH, Chapman MR. Microbial manipulation of the amyloid fold. Res Microbiol 2012; 163:592-606; PMID:23108148; http://dx.doi.org/10.1016/j.resmic.2012.10.009
- Bieler S, Estrada L, Lagos R, Baeza M, Castilla J, Soto C. Amyloid formation modulates the biological activity of a bacterial protein, J Biol Chem 2005; 280:26880-5; PMID:15917245; http://dx.doi.org/10.1074/jbc.M502031200
- Liebman SW, Chernoff YO. Prions in yeast. Genetics 2012; 191:1041-72; PMID:22879407; http://dx.doi.org/10.1534/genetics.111.137760
- Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, Marks MS. Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol 2003; 161:521-33; PMID:12732614; http://dx.doi.org/10.1083/jcb.200302072
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. The CPEB3 protein is a functional prion that interacts with the actin cytoskeleton. Cell Rep 2015; 11:1772-85; PMID:26074072; http://dx.doi.org/10.1016/j.celrep.2015.04.060
- Van Gerven N, Klein RD, Hultgren SJ, Remaut H. Bacterial amyloid formation: Structural insights into curli biogenesis. Trends Microbiol 2015; 23:693-706; PMID:26439293; http://dx.doi.org/10.1016/j.tim.2015.07.010
- Sivanathan V, Hochschild A. Generating extracellular amyloid aggregates using E coli cells. Genes Dev 2012; 26:2659-67; PMID:23166018; http://dx.doi.org/10.1101/gad.205310.112
- García-Fruitós E, Sabate R, de Groot NS, Villaverde A, Ventura S. Biological role of bacterial inclusion bodies: A model for amyloid aggregation. FEBS J 2011; 278:2419-27; http://dx.doi.org/10.1111/j.1742-4658.2011.08165.x
- Giraldo R, Fernández-Tresguerres ME. Twenty years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids. Plasmid 2004; 52:69-83; PMID:15336485; http://dx.doi.org/10.1016/j.plasmid.2004.06.002
- Giraldo R, Andreu JM, Díaz-Orejas R. Protein domains and conformational changes in the activation of RepA, a DNA replication initiator. EMBO J 1998; 17:4511-26; PMID:9687517; http://dx.doi.org/10.1093/emboj/17.15.4511
- Giraldo R, Fernández-Tornero C, Evans PR, Díaz-Orejas R, Romero A. A conformational switch between transcriptional repression and replication initiation in the RepA dimerization domain. Nat Struct Biol 2003; 10: 565-71; PMID:12766757; http://dx.doi.org/10.1038/nsb937
- Díaz-López T, Dávila-Fajardo C, Blaesing F, Lillo MP, Giraldo R. Early events in the binding of the pPS10 replication protein RepA to single iteron and operator DNA sequences. J Mol Biol 2006; 364:909-20; http://dx.doi.org/10.1016/j.jmb.2006.09.013
- Gasset-Rosa F, Díaz-López T, Lurz R, Prieto A, Fernández-Tresguerres ME, Giraldo R. Negative regulation of pPS10 plasmid replication: Origin pairing by zipping-up DNA-bound RepA monomers. Mol Microbiol 2008; 68:560-72; PMID:18284592; http://dx.doi.org/10.1111/j.1365-2958.2008.06166.x
- Díaz-López T, Lages-Gonzalo M, Serrano-López A, Alfonso C, Rivas G, Giraldo R. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J Biol Chem 2003; 278:18606-16; http://dx.doi.org/10.1074/jbc.M212024200
- Giraldo R. Defined DNA sequences promote the assembly of a bacterial protein into distinct amyloid nanostructures. Proc Natl Acad Sci USA 2007; 104:17388-93; PMID:17959784; http://dx.doi.org/10.1073/pnas.0702006104
- Gasset-Rosa F, Maté MJ, Dávila-Fajardo C, Bravo J, Giraldo R. Binding of sulphonated indigo derivatives to RepA-WH1 inhibits DNA-induced protein amyloidogenesis. Nucl Acids Res 2008; 36:2249-56; PMID:18285361; http://dx.doi.org/10.1093/nar/gkn067
- Fernández-Tresguerres ME, Moreno-Díaz de la Espina S, Gasset-Rosa F, Giraldo R. A DNA-promoted amyloid proteinopathy in Escherichia coli. Mol Microbiol 2010; 77:1456-69; http://dx.doi.org/10.1111/j.1365-2958.2010.07299.x
- Molina-García L, Giraldo R. Aggregation interplay between variants of the RepA-WH1 prionoid in Escherichia coli. J Bacteriol 2014; 196:2536-42; http://dx.doi.org/10.1128/JB.01527-14
- Giraldo R, Moreno-Díaz de la Espina S, Fernández-Tresguerres ME, Gasset-Rosa F. RepA-WH1 prionoid: A synthetic amyloid proteinopathy in a minimalist host. Prion 2011; 5:60-4; PMID:21293179; http://dx.doi.org/10.4161/pri.5.2.14913
- Tycko R. Physical and structural basis for polymorphism in amyloid fibrils. Prot Sci 2014; 23:1528-39; http://dx.doi.org/10.1002/pro.2544
- Diaz-Avalos R, King CY, Wall J, Simon M, Caspar DLD. Strain-specific morphologies of yeast prion amyloid fibrils. Proc Natl acad Sci USA 2005; 102:10165-70; PMID:16006506; http://dx.doi.org/10.1073/pnas.0504599102
- Wiltzius JJW, Landau M, Nelson R, Sawaya MR, Apostol MI, Goldschmidt L, Soriaga AB, Cascio D, Rajashankar K, Eisenberg D. Molecular mechanisms for protein-encoded inheritance. Nat Struct Mol Biol 2009; 16:973-8; PMID:19684598; http://dx.doi.org/10.1038/nsmb.1643
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of β-amyloid fibrils in Alzheimer disease brain tissue. Cell 2013; 154:1257-68; PMID:24034249; http://dx.doi.org/10.1016/j.cell.2013.08.035
- Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, Lindquist S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem Biol 2014; 21:295-305; PMID:24485763; http://dx.doi.org/10.1016/j.chembiol.2013.12.013
- Torreira E, Moreno-del Álamo M, Fuentes-Perez ME, Fernández C, Martín-Benito J, Moreno-Herrero F, Giraldo R, Llorca O. Amyloidogenesis of bacterial prionoid RepA-WH1 recapitulates dimer to monomer transitions of RepA in DNA replication initiation. Structure 2015; 23:183-9; PMID:25543255; http://dx.doi.org/10.1016/j.str.2014.11.007
- Chen SW, Drakulic S, Deas E, Ouberai M, Aprile FA, Arranz R, Ness S, Roodveldt C, Guilliams T, De-Genst EJ, et al. Structural characterization of toxic oligomers that are kinetically trapped during α-synuclein fibril formation. Proc Natl Acad Sci USA 2015; 112:E1994-2003; PMID:25855634; http://dx.doi.org/10.1073/pnas.1421204112
- Campioni S, Mannini B, Lopez-Alonso JP, Shalova IN, Penco A, Mulvihill E, Laurents DV, Relini A, Chiti F. Salt anions promote the conversion of HypF-N into amyloid-like oligomers and modulate the structure of the oligomers and the monomeric precursor state. J Mol Biol 2012; 424:132-49; PMID:23041425; http://dx.doi.org/10.1016/j.jmb.2012.09.023
- Marek PJ, Patsalo V, Green DF, Raleigh DP. Ionic strength effects on amyloid formation by amylin are a complicated interplay among Debye screening, ion selectivity, and Hofmeister effects. Biochemistry 2012; 51:8478-90; PMID:23016872; http://dx.doi.org/10.1021/bi300574r
- Buell AK, Hung P, Salvatella X, Welland ME, Dobson CM, Knowles TP. Electrostatic effects in filamentous protein aggregation. Biophys J 2013; 104:1116-26; PMID:23473495
- Kajava AV, Baxa U, Steven AC. b arcades: recurring motifs in naturally occurring and disease-related amyloid fibrils. FASEB J 2010; 24:1311-9; PMID:20032312
- Gasset-Rosa F, Giraldo R. Engineered bacterial hydrophobic oligopeptide repeats in a synthetic yeast prion. [REP-PSI+]. Front Microbiol 2015; 6:311
- Crist CG, Kakayashiki T, Kurahashi H, Nakamura Y. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells 2003; 8:603-18; PMID:12839621
- Toombs J.A, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. Proc Natl Acad Sci USA 2012; 109:6519-24; PMID:22474356
- Bondarev SA, Shchepachev VV, Kajava A, Zhouravleva GA. Effect of charged residues in the N-domain of Sup35 protein on prion [PSI+] stability and propagation. J Biol Chem 2013; 288:28503-13; PMID:23965990
- Giraldo R. Amyloid assemblies: Protein Legos at a crossroads in bottom-up synthetic biology. Chem Bio Chem 2010; 11:2247-357; PMID:20922739
- Moreno-del Álamo M, Moreno-Díaz de la Espina S, Fernández-Tresguerres ME, Giraldo R. Pre-amyloid oligomers of the proteotoxic RepA-WH1 prionoid assemble at the bacterial nucleoid. Sci Rep 2015; 5:14669; PMID:26423724
- Gasset-Rosa F, Coquel AS, Moreno-del Álamo M, Chen P, Song X, Serrano AM, Fernández-Tresguerres ME, Moreno-Díaz de la Espina S, Lindner AB, Giraldo R. Direct assessment in bacteria of prionoid propagation and phenotype selection by Hsp70 chaperone. Mol Microbiol 2014; 91:1070-87; PMID:24417419
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 2015; 162:1066-77; PMID:26317470
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015; 210:529-39; PMID:26283796
- Murakami T, Qamar S, Lin JQ, Schierle GS, Rees E, Miyashita A, Costa AR, Dodd RB, Chan FT, Michel CH, et al. ALS/FTD Mutation-induced phase transition of FUS liquid droplets and reversible hydrogels into irreversible hydrogels impairs RNP granule function. Neuron 2015; 88:678-90; PMID:26526393
- Escusa-Toret S, Vonk WI, Frydman J. Spatial sequestration of misfolded proteins by a dynamic chaperone pathway enhances cellular fitness during stress. Nat Cell Biol 2013; 15:1231-43; PMID:24036477
- Wallace EW, Kear-Scott JL, Pilipenko EV, Schwartz MH, Laskowski PR, Rojek AE, Katanski CD, Riback JA, Dion MF, Franks AM, et al. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 2015; 162:1286-98; PMID:26359986
