ABSTRACT
Prion-infected cells have been used for analyzing the effect of compounds on the formation of abnormal isoform of prion protein (PrPSc). PrPSc is usually detected using anti-prion protein (PrP) antibodies after the removal of the cellular isoform of prion protein (PrPC) by proteinase K (PK) treatment. However, it is expected that the PK-sensitive PrPSc (PrPSc-sen), which possesses higher infectivity and conversion activity than the PK-resistant PrPSc (PrPSc-res), is also digested through PK treatment. To overcome this problem, we established a novel cell-based ELISA in which PrPSc can be directly detected from cells persistently infected with prions using anti-PrP monoclonal antibody (mAb) 132 that recognizes epitope consisting of mouse PrP amino acids 119–127. The novel cell-based ELISA could distinguish prion-infected cells from prion-uninfected cells without cell lysis and PK treatment. MAb 132 could detect both PrPSc-sen and PrPSc-res even if all PrPSc molecules were not detected. The analytical dynamic range for PrPSc detection was approximately 1 log. The coefficient of variation and signal-to-background ratio were 7%–11% and 2.5–3.3, respectively, demonstrating the reproducibility of this assay. The addition of a cytotoxicity assay immediately before PrPSc detection did not affect the following PrPSc detection. Thus, all the procedures including cell culture, cytotoxicity assay, and PrPSc detection were completed in the same plate. The simplicity and non-requirement for cell lysis or PK treatment are advantages for the high throughput screening of anti-prion compounds.
INTRODUCTION
Prion diseases are a group of neurodegenerative disorders that include scrapie in sheep and goats, bovine spongiform encephalopathy in cattle, chronic wasting disease in deer, and Creutzfeld-Jakob disease (CJD) in human. The preclinical period of the diseases is extremely long; however, after the clinical onset, the diseases are subacutely progressive and inevitably fatal. The diseases are characterized by neuronal vacuolation, astrocytosis, microglial activation and accumulation of an abnormal isoform of prion protein (PrPSc) in the central nervous system (CNS).Citation1 PrPSc is the only known proteinaceous component of prions, the causative agents of the diseases, and the infectivity of prions is thought to be associated with PrPSc oligomers.Citation2,3 PrPSc is generated from a host-encoded cellular isoform of prion protein (PrPC) by post-translational modifications including conformational transformation. Once PrPSc appears, PrPSc is gradually generated and accumulates in CNS. The conversion of PrPC to PrPSc in neurons is believed to cause neurodegeneration,Citation4,5 therefore, the inhibition of PrPSc formation is one of the therapeutic targets for prion diseases.
Screening of chemical libraries is one of the ways to identify therapeutic compounds for prion diseases. Cells persistently infected with prions provide a good platform for screening compound libraries to identify inhibitors of PrPSc formation and for analyzing the cell biology underlying prion propagation.Citation6-11 For example, Kocisko et al. Citation12 screened 2,000 FDA-approved drugs using prion-infected cells and obtained 17 potent inhibitors. Ghaemmaghami et al.Citation13 screened more than 10,000 compounds and identified 4 lead chemical scaffolds for PrPSc formation inhibitors on the basis of structure-activity relationships among 121 compounds that inhibit PrPSc formation in cells.
As the deletion of PrP gene prevented clinical onset of the diseaseCitation14 and the reduction of the PrPC level by small interference RNA prolonged the survival of prion-infected mice,Citation15 reduction of the PrPC level is also a target for the inhibition of PrPSc formation. Karapetyan et al.Citation16 screened 1,280 drugs approved for use in human from the US Drug Collection for their ability to decrease PrPC expression in cells, and they identified Astemizole as a candidate for the treatment and prophylaxis of prion diseases. Silber et al. also screened large chemical libraries to find compounds that lower PrPC expression in T98G human glioblastoma and IMR 32 human neuroblastoma cells using a cell-based ELISA.Citation17 Alternatively, stabilization of PrPC, which inhibits the conformational transition of PrPC into PrPSc, is also a target for the inhibition of PrPSc formation. In silico screening of compounds that are capable of binding to PrPC using docking simulation has identified a new class of compounds that inhibit PrPSc formation in prion-infected cells.Citation18-20
Although screening for compounds that bind PrPC or affect PrPC expression is two of the ways to identify potential therapeutic compounds, the advantage of the use of prion-infected cells is that compounds that inhibit PrPSc formation through interaction with PrPC, PrPSc, or other cellular factors will be screened. However, one technical limitation of using prion-infected cells is the requirement for proteinase K (PK) treatment to remove PrPC from the cell lysates. It is well known that PrPSc comprises PK-sensitive PrPSc (PrPSc-sen) and PK-resistant PrPSc (PrPSc-res) Citation21,22 and that PrPSc-sen is reported to possess higher infectivity and conversion activity than PrPSc-res.Citation2 PK treatment is expected to digest PrPSc-sen; so when PK-treatment is used, the effect of compounds on PrPSc formation may be underestimated.
Recently, we reported that the anti-PrP monoclonal antibody (mAb) 132, which recognizes amino acids 119–127 of PrP, is useful for PrPSc detection in cells and frozen tissue sections of animals infected with prions.Citation23,24 Although fixation and treatment of cells or tissue sections with guanidinium salts are prerequisite, this method does not require PK treatment for the discrimination of PrPSc from PrPC. To take advantage of this, we established a cell-based ELISA in which PrPSc is directly detected in cells without preparation of a cell lysate and PK treatment. The cell-based ELISA established in this study provides a simple and practical method for the primary screening of anti-prion compounds.
RESULTS
MAb 132 Discriminates Prion-Infected Cells from Uninfected Cells in a Cell-Based ELISA
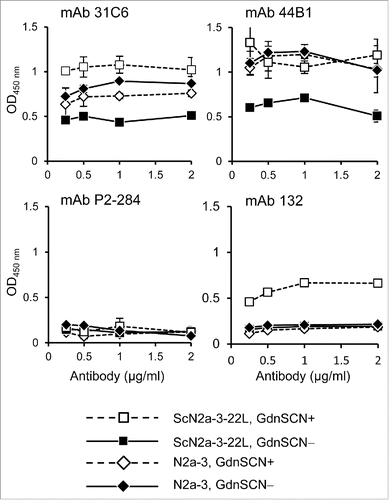
First we examined whether anti-PrP mAbs 31C6, 44B1, and 132 could discriminate prion-infected cells from prion-uninfected cells by direct staining of cells. Cells grown in 96-well plates were fixed with paraformaldehyde (PFA) and treated briefly with guanidine thiocyanate (GdnSCN) to denature PrPSc, and subsequently subjected to immunostaining. The mAbs 31C6 and 44B1 gave higher signals from ScN2a-3-22L cells, a subclone of Neuro2a mouse neuroblastoma (N2a-3) cells persistently infected with the 22L prion strain, than from prion-uninfected N2a-3 cells when these cells were pretreated with GdnSCN (). However, compared with the negative control mAb, these mAbs also showed positive signals from prion-uninfected N2a-3 cells, indicating that these mAbs are not suitable for the specific detection of PrPSc in prion-infected cells. In contrast, mAb 132 showed a positive reaction only when prion-infected cells were pretreated with GdnSCN. Absorbances obtained by mAb 132 staining from uninfected cells or GdnSCN-untreated prion-infected cells remained at levels comparable with the corresponding signals obtained using the negative control antibody. Thus, among the antibodies tested, only mAb 132 could distinguish prion-infected cells from uninfected cells in a cell-based ELISA.
Figure 1. MAb 132 discriminates prion-infected cells from uninfected cells in cell-based ELISA. Prion-infected cells (ScN2a-3-22L) and uninfected cells (N2a-3) were cultured in 96-well plates for 72 h. Cells were then fixed with PFA, permeabilized with Trion X-100, and treated with (GdnSCN+) or without (GdnSCN−) 5 M GdnSCN. After blocking, the cells were subjected to immunostaining with various anti-PrP antibodies at the indicated concentrations. MAb P2-284 was used as a negative control.
Performance of Cell-Based ELISA Using mAb 132
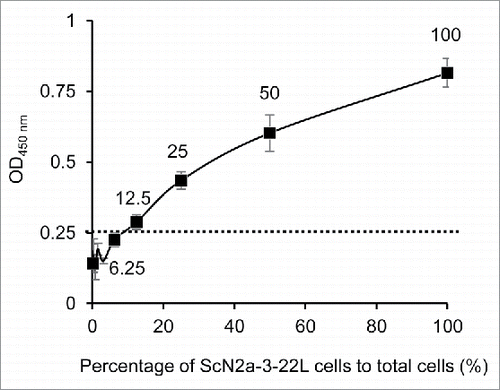
Next we examined the dynamic range of the cell-based ELISA. ScN2a-3-22L cells were 2-fold serially diluted with N2a-3 cells, and a total of 10,000 cells per well were seeded in a 96-well plate. The reaction became positive when 12.5% of cells in the well were prion-infected cells, and the signal was still in the linear range when all the cells were prion-infected cells (). These results indicate that the cell-based ELISA has approximately a 1 log dynamic range. Although it was expected that some uninfected cells became infected during the 72-h incubation, the spread of infection did not influence the interpretation because the linear increase of OD450 was observed in parallel with the ratio of infected and uninfected cells.
Figure 2. Dynamic range of PrPSc detection in cell-based ELISA. ScN2a-3-22L cell suspension (1.0 × 105 cells/ml) was 2-fold serially diluted with a N2a-3 cell suspension of the same concentration. Cell suspensions (10,000 cells/100μl/well) were added to wells and incubated for 72 h. After the incubation, the cells were subjected to PrPSc detection with mAb 132. The cutoff value (dotted line) was determined as the mean plus 3 × SD of the N2a-3 signal. Numbers with plots indicate percentages of ScN2a-3-22L cells to total cells in well.
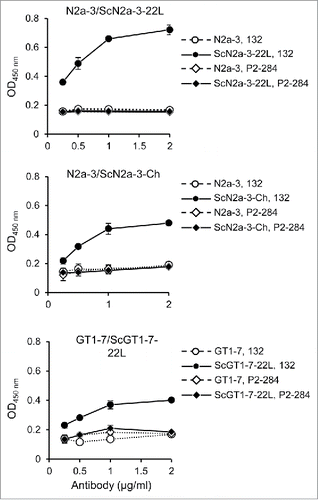
To examine whether mAb 132 is applicable to other prion strains or other cells, we used N2a-3 cells infected with the Chandler prion strain (ScN2a-3-Ch) and GT1-7 mouse immortalized hypothalamic neurons infected with the 22L prion strain (ScGT1-7-22L). The mAb 132 showed positive reaction to ScN2a-3-Ch and to ScGT1-7-22L cells (). These results suggested that the cell-based ELISA is applicable to different prion strains and cell types.
Figure 3. Detection of PrPSc of other prion strains and from other cell types. N2a-3 cells persistently infected with the 22L (ScN2a-3-22L), and the Chandler prion strain (ScN2a-3-Ch), and GT1-7 cells infected with the 22L prion strain (ScGT1-7-22L), were cultured in 96-well plates. After 72 h incubation, the cells were subjected to PrPSc detection with mAb 132 (open and closed circles). MAb P2-284 (open and closed diamonds) was used as a negative control mAb.
To evaluate the performance of the cell-based ELISA, basic indices of the PrPSc detection were analyzed (). The coefficient of variation (CV), which is the standard index for measuring variability, and is ideally < 10% when measuring intra-assay variability, was approximately 10%. The signal-to-background ratio (S/B), which is the standard index for acceptable breadth for determining negative and positive responses, and which is ideally > 2, was 2.6–3.6. The signal-to-noise ratio (S/N), which is index for signal intensity level to background fluctuations, and which should desirably be > 10, was 14.7–27.3. These indices demonstrate the reproducibility of the cell-based ELISA.
TABLE 1. Reliability of novel cell-based ELISA.
Utility of Cell-Based ELISA in Combination with Cytotoxicity Assay
If the cytotoxicity of test compounds could be assessed before PrPSc detection, all the procedures of the cell-based ELISA, from the cell culture, assessment of cytotoxicity, to the immunological detection of PrPSc, could be completed with in the same plate. Thus, we examined if the cytotoxicity assay (WST assay) affects the following PrPSc detection. Cells were treated with U18666A Citation25 or Pentosan polysulfate (PPS),Citation26 which are known as inhibitors for PrPSc formation, for 48 h and the WST assay was performed immediately before PrPSc detection. When ScN2a-3-22L cells were treated with U18666A, cell viability gradually decreased and obvious cytotoxicity was observed at U18666A concentrations > 5 μM. PrPSc detection using mAb 132 also decreased with increasing U18666A concentration and became lower than cutoff value at U18666A concentrations > 5 μM (). However, no remarkable differences in levels of PrPSc detection were observed with (ScN2a-3-22L, WST+) or without (ScN2a-3-22L, WST−) the WST assay (, see < 5 μM U18666A). This result indicates the WST assay has little effect on the subsequent PrPSc detection. In contrast to U18666A, PPS treatment showed no cytotoxicity even at the highest concentration tested, while PPS drastically decreased PrPSc levels to below the cutoff value at low concentrations (). These results demonstrate that the effect of test compounds on PrPSc formation and their cytotoxicity can be assessed in the same plate. In , low dose PPS treatment appeared to enhance cell viability; however those were among experimental variations and we confirmed that the low dose PPS treatment did not influence the viability of N2a-3 and ScN2a-3-22L cells (data not shown).
Figure 4. Utility of cell-based ELISA in combination with cytotoxicity assay. (A and B) Influence of the cytotoxicity assay on subsequent PrPSc detection. ScN2a-3-22L cells were cultured in 96-well plates for 24 h and then incubated with U18666A (A) or PPS (B) for 48 h at the indicated concentrations. Immediately before fixation, cell viability was measured with the WST assay using CCK-8. After removal of the CCK-8 reagent, cells were subjected to PrPSc detection in the same plate. Regarding PrPSc detection, PrPSc signals detected after the WST assay (WST+, open diamonds and circles) or without WST assay (WST−, closed diamonds and circles) are shown. The dashed lines in WST assay indicate survival rate at 80%. The PrPSc signal detected from N2a-3 cells was used to calculate cutoff values (mean plus 3 × SD) for PrPSc detection (dashed lines). (C) IFA for PrPSc detection. ScN2a-3-22L cells were cultured in a chambered coverglass for 24 h and then treated or with U18666A or PPS for 48 h at the indicated concentrations. ScN2a-3-22L and N2a-3 cells untreated with these compounds were used as positive and negative controls for PrPSc, respectively. The cells were subjected to PrPSc-specific straining with mAb 132 (green), and cell nuclei were counterstained with 4', 6-diamidino-2-phenylindole (DAPI) (blue). (D) Expression of PrPC in N2a-3 and ScN2a-3-22L cells. Cells were stained with mAb 31C6 (green) without pretreatment of GdnSCN for the detection of PrPC. Nuclei were counterstained with DAPI (blue). P2-284: negative control mAb. Scale bar: 20 μm. (E) Immunoblot analysis. ScN2a-3-22L cells were cultured in 24-well plates for 24 h and subsequently treated with U18666A or PPS for 48 h at the indicated concentrations. Cells were then subjected to immunoblot analysis for the detection of PrPSc-res. N2a-3 cells untreated with these compounds were used as negative controls for PrPSc-res. PrPSc-res levels relative to untreated ScN2a-3-22L cells are shown at the bottom. Scale bar: 20 μm.
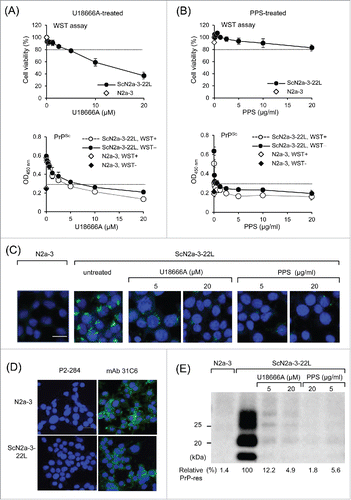
To confirm the accuracy of PrPSc detection after U18666A or PPS treatment in the cell-based ELISA, we analyzed PrPSc in PPS- or U18666A-treated cells by immunofluorescence assay (IFA) () and PrPSc-res by immunoblot analysis (). In IFA, PrPSc signals were decreased but were still detected after treatment with 5 μM U18666A and further weakened at 20 μM U18666A (). The expressions of PrPC in N2a-3 and ScN2a-3-22L cells were confirmed by mAb 31C6 without pretreatment of cells with GdnSCN (). The result of the immunoblot analysis of U18666A-treated cells was consistent with that of the IFA (). In contrast, treatment of cells with 5 μg/ml PPS decreased PrPSc signals to levels comparable with those seen in cells treated with 20 μg/ml PPS. The gradual decrease in PrPSc levels with the increase of U18666A but drastic decrease in PrPSc levels at PPS concentrations < 5 μg/ml of PPS, is consistent with those obtained with the cell-based ELISA, demonstrating the utility of the cell-based ELISA for screening anti-prion compounds.
Detection of PrPSc-sen and PrPSc-res by mAb 132
Since the cell-based ELISA established here does not require PK treatment for PrPSc detection, it is possible that both PrPSc-res and PrPSc-sen could be detected using mAb 132, even if not all the PrP species are detected. To assess this possibility, lysates from N2a-3 and ScN2a-3-22L cells were blotted onto polyvinylidene difluoride (PVDF) membranes, and after PK treatment and subsequent GdnSCN exposure (or in the reverse order), the membranes were probed with mAb 132.
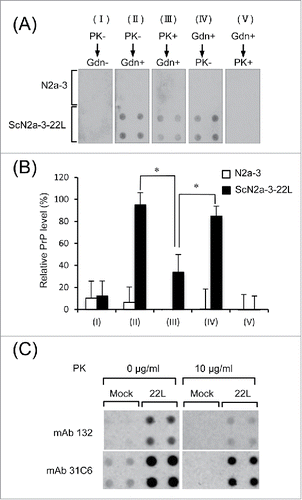
No PrP was detected from the lysates of N2a-3 or ScN2a-3-22L cells lacking both PK and GdnSCN treatment (, I). PrP was detected in the lysate of ScN2a-3-22L cells when the membrane was not treated with PK but treated with GdnSCN (, II). These signals represent PrPSc because no PrP was detected from the lysate of N2a-3 cells under the same condition. However, the intensity of PrPSc signals decreased to approximately 40% of those seen in II when the PVDF membrane was first treated with PK and then with GdnSCN (, III, and ). The PrPSc signals detected under these conditions were considered to be those from PrPSc-res and the decrease in the PrPSc signal accounted for the digestion of PrPSc-sen by PK. When the PVDF membrane was treated with GdnSCN but not PK (, IV), PrPSc levels were as high as those obtained in II, suggesting that the signal accounted for the sum of PrPSc-sen and PrPSc-res. When the membrane was first treated with GdnSCN and then treated with PK, no PrP signals were obtained, since PrPSc denatured by GdnSCN was digested with PK (, V). The presence of PrPC in the lysate of N2a-3 was confirmed with mAb 31C6 ().
Figure 5. Detection of PrPSc-sen and PrPSc-res by mAb 132 in a dot-blot analysis. (A) Dot-blot analysis. Lysates from N2a-3 or ScN2a-3-22L cells were transferred onto PDVF membranes using a dot-blotter (quadruplicates). The membranes were treated with PK (10 μg/ml; PK+) (III) or not (PK−) (I and II) for 1 h at RT and subsequently with (Gdn+) (II and III) or without (Gdn−) 3 M GdnSCN (II) for 30 min at RT, or in the reverse order (IV and V). Then, the membranes were stained with mAb 132, and chemiluminescence signals were detected and quantified with a LAS 3000 chemiluminescence image analyzer. The PrP levels (%) relative to the PrP level in II, detected using mAb 132, are shown in (B). *, p < 0.05, Student's t-test). (C) Confirmation of PrPC in dot-blot analysis. Lysates from N2a-3 (Mock) or ScN2a-3-22L (22L) cells were blotted onto PDVF membranes (quadruplicates). The membranes were treated with PK (10 μg/ml) or not (0 μg/ml) for 1 h at RT. After the termination of PK digestion, the membranes were treated with DNase I and then with 3 M GdnSCN for 30 min at RT as described in Materials and Methods. Finally membranes were stained with mAbs 132 and 31C6 for chemiluminescence detection.
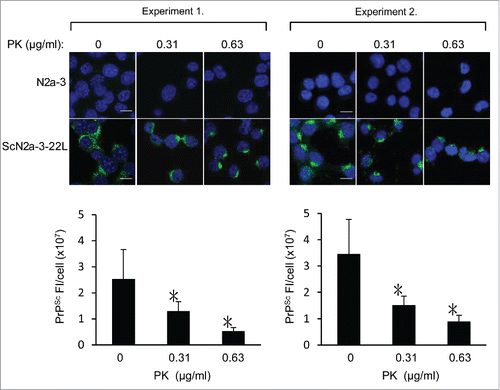
To further examine whether mAb 132 detects PrPSc-sen, ScN2a-3-22L cells were treated with low concentrations of PK and then subjected to PrPSc-specific immunostaining. The fluorescent intensities of PrPSc signals in PK-treated ScN2a-3-22L cells decreased to less than a half of those in PK-untreated ScN2a-3-22L cells, even at the lowest PK concentration tested (, 0.31 μg/ml). The decrease in fluorescence intensities of PrPSc signals by PK treatment supported the idea that mAb 132 detected both PrPSc-sen and PrPSc-res.
Figure 6. Possible detection of PrPSc-sen and PrPSc-res using mAb 132 in IFA. N2a-3 and ScN2a-3-22L cells and were cultured in a chambered coverglass for 24 h. Cells were fixed with 4% paraformaldehyde in PBS for 10 min and then treated with 0.1% Triton X-100 and 0.1 M glycine in PBS. Subsequently, cells were treated with PK at the indicated concentrations at 4°C for 30 min. Cells were then treated with 5 M GdnSCN for 10 min at RT and subjected to PrPSc-specific immunofluorescence staining. Representative fluorescence images of two independent experiments are shown at the top. Graphs show the corresponding quantification result of PrPSc fluorescence intensities (FI) per ScN2a-3-22L cell. Fluorescence intensities from perinuclear regions of N2a-3 were subtracted as background. Experiment 1: mean fluorescence intensities and standard deviations of 16 cells from 5 microscopic fields (3–4 cells/field). Experiment 2: mean fluorescence intensities and standard deviations of 16–24 cells from 4 to 5 microscopic fields (3–5 cells/field). *, p < 0.001, Welch's t-test). Scale bars: 10 μm.
DISCUSSION
We have reported that mAb 132, which recognizes an epitope consisting of mouse PrP aa 119–127, can specifically detect PrPSc from prion-infected cells or tissues without the removal of PrPC by PK treatment.Citation23,24 This feature of mAb 132 facilitated the establishment of a novel cell-based ELISA in which PrPSc levels in prion-infected cells are assessed without the removal of PrPC. As anticipated, mAb 132 was the only anti-PrP mAbs tested that could distinguish prion-infected cells from uninfected cells (). Signals obtained from uninfected cells and GdnSCN-untreated prion-infected cells probed with mAb 132 were comparable with signals obtained using a negative control mAb, providing a suitable S/B ratio (). MAb 132 reacted poorly with PrPC on the cell surface,Citation27 but reacted with PrPSc, PrPC and recombinant PrP in immunoblot analysis.Citation28 Thus, mAb 132 appears to recognize a linear epitope that becomes antibody-accessible after denaturation of the PrP molecule. However, mAb 132 did not show a positive reaction to uninfected cells, even after GdnSCN treatment. We do not have any clear explanation for this phenomenon, one possibility is that once the region containing the mAb 132 epitope on PrPC was exposed by GdnSCN treatment, the region may refold into antibody-inaccessible form after the removal of GdnSCN. Surface plasmon resonance analysis revealed that the binding of monovalent mAb132 (e.g., recombinant Fab) was significantly weaker than bivalent mAb 132 (e.g., recombinant IgG), indicating that the bivalent binding is required for the efficient binding to the epitope (A.S. & M.H., manuscript in preparation). Reaction of mAb 132 to PrPC expressed in the cells will be a monovalent binding, whereas that to PrPSc will occur as bivalent binding because PrPSc exists as oligomer/aggregate of PrP molecules. Thus the binding kinetics of mAb 132 may partly explain the inefficient binding of mAb 132 to PrPC: monovalent binding is not enough to stain PrPC efficiently in IFA. However, further studies are still required for the elucidation of the mechanism of PrPSc-specific staining by mAb 132.
Conformation-dependent immunoassay (CDI) has demonstrated the existence of PrPSc-sen and PrPSc-res in the brains of prion-affected humans and animals.Citation29 The proportion of PrPSc-sen is believed to be high; for example, CDI revealed that PrPSc-sen constituted approximately 50–90% and 90% of PrPSc in the brains of hamsters infected with hamster-adapted prion strains and CJD patients, respectively.Citation29,30 Also immuno-electron microscopic analysis of mice infected with the RML let to an estimate that > 85% of the PrPSc in the brain was PK sensitive.Citation31 The PK-sensitive fraction of PrPSc is reported to possess higher infectivity and higher conversion activity per PrP molecule than the PK-resistant fraction.Citation2 Taken together, these results suggest that PrPSc-sen may be the more substantial entity of prions. Thus, evaluation of the effect of compounds on PrPSc-sen may be important for screening anti-prion compounds. Screening methods of anti-prion compounds using prion-infected cells reported to date included PK treatment for the removal of PrPC.Citation12,13,32,33 However, effect of compounds on PrPSc-sen cannot be assessed or may be underestimated if PK treatment is included during the analysis.
MAb 132 discriminated PrPSc from PrPC without PK treatment, suggesting that mAb could detect both PrPSc-sen and PrPSc-res; Citation23,24,34 however, this had not yet been directly demonstrated. In a dot-blot analysis performed using cell lysates prepared with non-ionic detergent, the PrPSc level detected after PK digestion and subsequent GdnSCN treatment was much lower than that detected after GdnSCN treatment alone (). Consistent with the result of dot-blot analysis, the fluorescence intensities of PrPSc decreased after treatment of the cells with even low concentrations of PK (). Quantification of the result of dot-blot analysis and the IFA implies that > 50% of PrPSc in N2a-3 cells infected with the 22L strain are sensitive to PK, which is similar to the proportion of PK-sensitive PrPSc in the brains of prion-affected humans and animals.Citation29,30,35 Taken together, these results demonstrate that mAb 132 detects not only PrPSc-res, but at least a certain fraction of PrPSc-sen if not all.
The absolute sensitivity for the detection of PrPSc in the cell-based ELISA is not expected to be high because of the lack of procedures for PrPSc concentration such as phosphotungstic acid precipitation.Citation13,36 However, a 1 log dynamic range for direct PrPSc detection in prion-infected cells is enough to evaluate the anti-prion effect exerted by a 50% effective dose as a primary screening method. The remarkable advantage of the cell-based ELISA is its simplicity: all the procedures, from cell culture to PrPSc detection, can be completed in the same plate without making cell lysates or PK treatment. Probing both PrPSc-res and PrPSc-sen is another technical improvement, which is difficult to achieve using procedures that include PK treatment. It is believed that human cells or human PrPC-expressing cells infected with human prions are a better platform for screening for potential therapeutic reagents than prion-infected cells of other species. The epitope for mAb 132 is well conserved from mammals to chickens,Citation37,38 so mAb 132 will be applicable to the detection of PrPSc from human prion-infected cells using a protocol similar to the one developed in this study.
MATERIALS AND METHODS
Antibodies and Regents
The anti-PrP mAbs 132, 31C6, and 44B1, which recognize amino acids 119–127, 143–149, and discontinuous epitopes of mouse PrP, respectively, were used.Citation28 Anti-feline parvovirus subgroup mAb P2-284 was used as a negative control antibody.Citation39 ECL™ anti-mouse IgG, horseradish peroxidase (HRP)-linked F(ab')2 fragment from sheep was purchased from GE Healthcare. The Alexa Fluor 488 F(ab')2 fragment of goat anti-mouse IgG (H+L) was purchased from Life Technologies. PPS was purchased from Dainippon Sumitomo Pharma and U18666A was purchased from Sigma-Aldrich.
Cell Culture
N2a-3 cells, a subclone of the N2a mouse neuroblastoma cell lineCitation40 and N2a-3 cells persistently infected with the 22L prion strain (ScN2a-3-22L)Citation41 or the Chandler prion strain (ScN2a-3-Ch)Citation40 were used. GT1-7 mouse immortalized hypothalamic neuronsCitation42 that are persistently infected with the 22L prion strain (ScGT1-7-22L) Citation23 were also used.
Cell-Based ELISA
N2a-3 and ScN2a-3-22L cells were seeded into 96-well plates (Thermo Scientific) at a density of 1 × 104 cells/100 μl/well and cultured for 24 h. Cells were then freshly fed with the medium with or without anti-prion compounds and incubated for 48 h. After the removal of medium, cells were fixed with 50 μl/well of 4% PFA in phosphate-buffered saline (PBS) for 10 min at room temperature (RT). After the removal of PFA, 100 μl of 0.1% Triton X-100 and 0.1 M glycine in PBS was added to each well to permeabilize the cell membrane and quench residual PFA. The plates were then incubated for 10 min at RT. After the removal of glycine and Triton X-100, the cells were treated with 50 μl of 5 M GdnSCN for 10 min at RT. Cells were then washed once with PBS and blocked with 5% skim milk in PBS for 30 min at RT. Immunostaining was carried out using anti-PrP mAb 132 (1 μg/ml) as the primary antibody for at least 6 h at 4°C and diluted (1:5,000) HRP-conjugated anti-mouse IgG as the secondary antibody for 1 h at RT. Finally, antigen-antibody complexes were detected with the colorimetric HRP substrate 3,3′,5,5′ tetramethylbenzidine (Sigma). After incubation for 15 min at RT, the reaction was stopped by adding sulfuric acid to 0.25 M and optical density at 450 nm was measured using a microplate reader (Infinite M200 Pro, Tecan). In some cases, the cytotoxicity assay described below was carried out immediately before fixation of the cells.
Cytotoxicity Assay
After removal of the medium, the cells were washed with Opti-MEM without phenol red (Gibco) and subjected to the WST cytotoxicity assay using Cell Counting Kit 8 (CCK-8; DojinDo). CCK-8 reagent was diluted 1:100 with Opti-MEM (Thermo) and added to each well (100 μl/well). After incubation at 37°C for 1 h, the absorbance at 450 nm was measured using a microplate reader.
Immunoblot Analysis
SDS-PAGE and immunoblotting for PrPSc detection were carried out as described elsewhere.Citation40,41
Dot-Blotting
N2a-3 and ScN2a-3-22L cells were cultured for 72 h in 6-well plates. Cells were lysed with 200 μl/well of lysis buffer (0.5% Triton X-100, 0.5% sodium deoxycholate, 150 mM NaCl, 5 mM EDTA, and 10 mM Tris-HCl [pH 7.5]) and the protein concentration of the lysate was measured using a DC protein assay kit (Bio-Rad). Cell lysates equivalent to 40 μg of total protein were transferred onto a PVDF membrane using a dot-blotter (Bio-Rad). The PVDF membrane was treated with PK (10 μg/ml), or not, for 1 h at RT, and then PK digestion was terminated by the addition of Pefabloc (Roche) to 1 mM for 15 min at 4°C. The membrane was then treated with 50 μg/ml DNase I for 15 min and subsequently with 3 M GdnSCN for 30 min at RT. For the detection of PrP, the membrane was incubated with mAb 132 (1 μg/ml) in 1% skim milk-PBS containing 0.1% Tween 20 (PBST) at 4°C overnight. After washing the membrane with PBST, HRP-conjugated anti-mouse IgG was used as the secondary antibody for 1 h at RT. ECL Western Blotting Detection Reagents (GE Healthcare) and a LAS-3000 chemiluminescence image analyzer (Fujifilm) were used to visualize the immune-reactive proteins.
IFA
PrPSc-specific immunofluorescence staining using mAb 132 and quantitative analysis of the signals were performed as described previously.Citation23,34
Calculation of CV, S/B, and S/N
Forty-eight wells were seeded with N2a-3 cells and another 48 wells were seeded with ScN2a-3-22L at the density of 1 × 104 cells/100 μl/well. After 72 h incubation, the PrP signals were detected as described in the section entitled “Cell-based ELISA.” Twenty-well sections were assigned for staining with mAb 132 or mAb P2-284 (negative control mAb), and the remaining 8 wells were assigned as blank wells. Four independent experiments were carried out to calculate CV, S/B, and S/N.
ABBREVIATIONS
| aa | = | amino acid(s) |
| CDI | = | Conformation-dependent immunoassay |
| CJD | = | Creutzfeld-Jakob disease |
| CNS | = | central nervous system |
| CV | = | coefficient of variation |
| DAPI | = | 4',6-diamidino-2-phenylindole |
| ELISA | = | enzyme-linked immunosorbent assay |
| FDA | = | Food and Drug Administration |
| FI | = | fluorescence intensities |
| GdnSCN | = | guanidine thiocyanate |
| IFA | = | immunofluorescence assay |
| HRP | = | horseradish peroxidase |
| mAb | = | monoclonal antibody |
| N2a | = | Neuro2a |
| PBS | = | phosphate buffered saline |
| PBST | = | PBS containing 0.1% Tween 20 |
| PFA | = | paraformaldehyde |
| PK | = | proteinase K |
| PPS | = | Pentosan polysulfate |
| PrPC | = | cellular isoform of prion protein |
| PrPSc | = | abnormal isoform of prion protein |
| PrPSc-res | = | PK-resistant PrPSc |
| PrPSc-sen | = | PK-sensitive PrPSc |
| PVDF | = | polyvinylidene difluoride |
| RML | = | Rocky mountain laboratories |
| RT | = | room temperature |
| S/B | = | signal-to-background ratio |
| ScGT1-7-22L | = | GT1-7 mouse immortalized hypothalamic neurons infected with the 22L strain |
| ScN2a-3-22L | = | N2a-3 cells persistently infected with the 22L prion strain |
| ScN2a-3-Ch | = | N2a-3 cells persistently infected with the Chandler prion strain |
| S/N | = | signal-to-noise ratio |
| WST | = | 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt |
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors have no conflict of interest to declare. The authors also declare no competing financial interests.
Funding
This work was supported by a Grant-in-Aid for Science Research (A) (grant no. 15H02475), a grant from the Program for Leading Graduate Schools (F01), from the Ministry of Education, Culture, Sports, Science, and Technology, Japan. This work was also supported by grants for TSE research (H26-Shokuhin-Ippan-004) and Research on Measures for Intractable Diseases from the Ministry of Health, Labour and Welfare of Japan. This work was also supported by the Global Institution for Collaborative Research and Education (GI-CoRE). We thank Zensho Co., Ltd, for the BSL3 facility.
REFERENCES
- Aguzzi A, Baumann F, Bremer J. The prion's elusive reason for being. Annu Rev Neurosci 2008; 31:439-77; PMID:18558863; http://dx.doi.org/10.1146/annurev.neuro.31.060407.125620
- Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature 2005; 437:257-61; PMID:16148934; http://dx.doi.org/10.1038/nature03989
- Wang F, Wang X, Yuan CG, Ma J. Generating a prion with bacterially expressed recombinant prion protein. Science 2010; 327:1132-5; PMID:20110469; http://dx.doi.org/10.1126/science.1183748
- Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science 2003; 302:871-4; PMID:14593181; http://dx.doi.org/10.1126/science.1090187
- Chesebro B, Trifilo M, Race R, Meade-White K, Teng C, LaCasse R, Raymond L, Favara C, Baron G, Priola S, et al. Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 2005; 308:1435-9; PMID:15933194; http://dx.doi.org/10.1126/science.1110837
- Marijanovic Z, Caputo A, Campana V, Zurzolo C. Identification of an intracellular site of prion conversion. PLoS Pathog 2009; 5:e1000426; PMID:19424437; http://dx.doi.org/10.1371/journal.ppat.1000426
- Veith NM, Plattner H, Stuermer CA, Schulz-Schaeffer WJ, Burkle A. Immunolocalisation of PrPSc in scrapie-infected N2a mouse neuroblastoma cells by light and electron microscopy. Eur J Cell Biol 2009; 88:45-63; PMID:18834644; http://dx.doi.org/10.1016/j.ejcb.2008.08.001
- Caughey B, Raymond GJ. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem 1991; 266:18217-23; PMID:1680859
- Taraboulos A, Raeber AJ, Borchelt DR, Serban D, Prusiner SB. Synthesis and trafficking of prion proteins in cultured cells. Mol Biol Cell 1992; 3:851-63; PMID:1356522; http://dx.doi.org/10.1091/mbc.3.8.851
- Goold R, Rabbanian S, Sutton L, Andre R, Arora P, Moonga J, Clarke AR, Schiavo G, Jat P, Collinge J, et al. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat Commun 2011; 2:281; PMID:21505437; http://dx.doi.org/10.1038/ncomms1282
- Yamasaki T, Baron GS, Suzuki A, Hasebe R, Horiuchi M. Characterization of intracellular dynamics of inoculated PrP-res and newly generated PrP(Sc) during early stage prion infection in Neuro2a cells. Virology 2014; 450-451:324-35; PMID:24503096; http://dx.doi.org/10.1016/j.virol.2013.11.007
- Kocisko DA, Baron GS, Rubenstein R, Chen J, Kuizon S, Caughey B. New inhibitors of scrapie-associated prion protein formation in a library of 2000 drugs and natural products. J Virol 2003; 77:10288-94; PMID:12970413; http://dx.doi.org/10.1128/JVI.77.19.10288-10294.2003
- Ghaemmaghami S, May BC, Renslo AR, Prusiner SB. Discovery of 2-aminothiazoles as potent antiprion compounds. J Virol 2010; 84:3408-12; PMID:20032192; http://dx.doi.org/10.1128/JVI.02145-09
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell 1993; 73:1339-47; PMID:8100741; http://dx.doi.org/10.1016/0092-8674(93)90360-3
- White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci U S A 2008; 105:10238-43; PMID:18632556; http://dx.doi.org/10.1073/pnas.0802759105
- Karapetyan YE, Sferrazza GF, Zhou M, Ottenberg G, Spicer T, Chase P, Fallahi M, Hodder P, Weissmann C, Lasmezas CI. Unique drug screening approach for prion diseases identifies tacrolimus and astemizole as antiprion agents. Proc Natl Acad Sci U S A 2013; 110:7044-9; PMID:23576755; http://dx.doi.org/10.1073/pnas.1303510110
- Silber BM, Gever JR, Rao S, Li Z, Renslo AR, Widjaja K, Wong C, Giles K, Freyman Y, Elepano M, et al. Novel compounds lowering the cellular isoform of the human prion protein in cultured human cells. Bioorg Med Chem 2014; 22:1960-72; PMID:24530226; http://dx.doi.org/10.1016/j.bmc.2014.01.001
- Kuwata K, Nishida N, Matsumoto T, Kamatari YO, Hosokawa-Muto J, Kodama K, Nakamura HK, Kimura K, Kawasaki M, Takakura Y, et al. Hot spots in prion protein for pathogenic conversion. Proc Natl Acad Sci U S A 2007; 104:11921-6; PMID:17616582; http://dx.doi.org/10.1073/pnas.0702671104
- Hosokawa-Muto J, Kamatari YO, Nakamura HK, Kuwata K. Variety of antiprion compounds discovered through an in silico screen based on cellular-form prion protein structure: Correlation between antiprion activity and binding affinity. Antimicrob Agents Chemother 2009; 53:765-71; PMID:19015328; http://dx.doi.org/10.1128/AAC.01112-08
- Ferreira NC, Marques IA, Conceicao WA, Macedo B, Machado CS, Mascarello A, Chiaradia-Delatorre LD, Yunes RA, Nunes RJ, Hughson AG, et al. Anti-prion activity of a panel of aromatic chemical compounds: in vitro and in silico approaches. PLoS One 2014; 9:e84531; PMID:24400098; http://dx.doi.org/10.1371/journal.pone.0084531
- Tzaban S, Friedlander G, Schonberger O, Horonchik L, Yedidia Y, Shaked G, Gabizon R, Taraboulos A. Protease-sensitive scrapie prion protein in aggregates of heterogeneous sizes. Biochemistry 2002; 41:12868-75; PMID:12379130; http://dx.doi.org/10.1021/bi025958g
- Pastrana MA, Sajnani G, Onisko B, Castilla J, Morales R, Soto C, Requena JR. Isolation and characterization of a proteinase K-sensitive PrPSc fraction. Biochemistry 2006; 45:15710-7; PMID:17176093; http://dx.doi.org/10.1021/bi0615442
- Yamasaki T, Suzuki A, Shimizu T, Watarai M, Hasebe R, Horiuchi M. Characterization of intracellular localization of PrP(Sc) in prion-infected cells using a mAb that recognizes the region consisting of aa 119-127 of mouse PrP. J Gen Virol 2012; 93:668-80; PMID:22090211; http://dx.doi.org/10.1099/vir.0.037101-0
- Sakai K, Hasebe R, Takahashi Y, Song CH, Suzuki A, Yamasaki T, Horiuchi M. Absence of CD14 delays progression of prion diseases accompanied by increased microglial activation. J Virol 2013; 87:13433-45; PMID:24089559; http://dx.doi.org/10.1128/JVI.02072-13
- Klingenstein R, Lober S, Kujala P, Godsave S, Leliveld SR, Gmeiner P, Peters PJ, Korth C. Tricyclic antidepressants, quinacrine and a novel, synthetic chimera thereof clear prions by destabilizing detergent-resistant membrane compartments. J Neurochem 2006; 98:748-59; PMID:16749906; http://dx.doi.org/10.1111/j.1471-4159.2006.03889.x
- Caughey B, Raymond GJ. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J Virol 1993; 67:643-50; PMID:7678300
- Kim CL, Karino A, Ishiguro N, Shinagawa M, Sato M, Horiuchi M. Cell-surface retention of PrPC by anti-PrP antibody prevents protease-resistant PrP formation. J Gen Virol 2004; 85:3473-82; PMID:15483265; http://dx.doi.org/10.1099/vir.0.80113-0
- Kim CL, Umetani A, Matsui T, Ishiguro N, Shinagawa M, Horiuchi M. Antigenic characterization of an abnormal isoform of prion protein using a new diverse panel of monoclonal antibodies. Virology 2004; 320:40-51; PMID:15003861; http://dx.doi.org/10.1016/j.virol.2003.10.026
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157-65; PMID:9771749; http://dx.doi.org/10.1038/2654
- Safar JG, Geschwind MD, Deering C, Didorenko S, Sattavat M, Sanchez H, Serban A, Vey M, Baron H, Giles K, et al. Diagnosis of human prion disease. Proc Natl Acad Sci U S A 2005; 102:3501-6; PMID:15741275; http://dx.doi.org/10.1073/pnas.0409651102
- Godsave SF, Wille H, Kujala P, Latawiec D, DeArmond SJ, Serban A, Prusiner SB, Peters PJ. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J Neurosci 2008; 28:12489-99; PMID:19020041; http://dx.doi.org/10.1523/JNEUROSCI.4474-08.2008
- Heal W, Thompson MJ, Mutter R, Cope H, Louth JC, Chen B. Library synthesis and screening: 2,4-diphenylthiazoles and 2,4-diphenyloxazoles as potential novel prion disease therapeutics. J Med Chem 2007; 50:1347-53; PMID:17305326; http://dx.doi.org/10.1021/jm0612719
- Leidel F, Eiden M, Geissen M, Kretzschmar HA, Giese A, Hirschberger T, Tavan P, Schatzl HM, Groschup MH. Diphenylpyrazole-derived compounds increase survival time of mice after prion infection. Antimicrob Agents Chemother 2011; 55:4774-81; PMID:21746938; http://dx.doi.org/10.1128/AAC.00151-11
- Yamasaki T, Suzuki A, Hasebe R, Horiuchi M. Comparison of the anti-prion mechanism of four different anti-prion compounds, anti-PrP monoclonal antibody 44B1, pentosan polysulfate, chlorpromazine, and U18666A, in prion-infected mouse neuroblastoma cells. PLoS One 2014; 9:e106516; PMID:25181483; http://dx.doi.org/10.1371/journal.pone.0106516
- Cronier S, Gros N, Tattum MH, Jackson GS, Clarke AR, Collinge J, Wadsworth JD. Detection and characterization of proteinase K-sensitive disease-related prion protein with thermolysin. Biochem J 2008; 416:297-305; PMID:18684106; http://dx.doi.org/10.1042/BJ20081235
- Wadsworth JD, Joiner S, Hill AF, Campbell TA, Desbruslais M, Luthert PJ, Collinge J. Tissue distribution of protease resistant prion protein in variant Creutzfeldt-Jakob disease using a highly sensitive immunoblotting assay. Lancet 2001; 358:171-80; PMID:11476832; http://dx.doi.org/10.1016/S0140-6736(01)05403-4
- Ishiguro N, Inoshima Y, Sassa Y, Takahashi T. Molecular characterization of chicken prion proteins by C-terminal-specific monoclonal antibodies. Vet Immunol Immunopathol 2009; 128:402-6; PMID:19118905; http://dx.doi.org/10.1016/j.vetimm.2008.11.025
- Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, Schwarz TF, Werner T, Schatzl HM. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol 1999; 289:1163-78; PMID:10373359; http://dx.doi.org/10.1006/jmbi.1999.2831
- Horiuchi M, Mochizuki M, Ishiguro N, Nagasawa H, Shinagawa M. Epitope mapping of a monoclonal antibody specific to feline panleukopenia virus and mink enteritis virus. J Vet Med Sci 1997; 59:133-6; PMID:9070987; http://dx.doi.org/10.1292/jvms.59.133
- Uryu M, Karino A, Kamihara Y, Horiuchi M. Characterization of prion susceptibility in Neuro2a mouse neuroblastoma cell subclones. Microbiol Immunol 2007; 51:661-9; PMID:17641468; http://dx.doi.org/10.1111/j.1348-0421.2007.tb03954.x
- Nakamitsu S, Kurokawa A, Yamasaki T, Uryu M, Hasebe R, Horiuchi M. Cell density-dependent increase in the level of protease-resistant prion protein in prion-infected Neuro2a mouse neuroblastoma cells. J Gen Virol 2010; 91:563-9; PMID:19812263; http://dx.doi.org/10.1099/vir.0.016287-0
- Schatzl HM, Laszlo L, Holtzman DM, Tatzelt J, DeArmond SJ, Weiner RI, Mobley WC, Prusiner SB. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J Virol 1997; 71:8821-31; PMID:9343242
