ABSTRACT
Scrapie, the prion disease of sheep and goats, is a devastating malady of small ruminants. Due to its infectious nature, epidemic outbreaks may occur in flocks/herds consisting of highly susceptible animals. Field studies identified scrapie-protective caprine PrP variants, harboring specific single amino acid changes (Met-142, Arg-143, Asp-146, Ser-146, His-154, Gln-211 and Lys-222). Their effects are under further evaluation, and aim to determine the most protective allele. We assessed some of these variants (Asp-146, His-154, Gln-211 and Lys-222), after their exogenous expression as murine-caprine chimeras in a scrapie- infected murine cell line. We report that exogenously expressed PrPs undergo conformational conversion upon interaction with the endogenous pathological murine prion protein (PrPSC), which results in the detection of goat-specific and partially PK-resistant moieties. These moieties display a PK-resistance pattern distinct from the one detected in natural goat scrapie cases. Within this cellular model, distinct conformational conversion potentials were assigned to the tested variants. Molecules carrying the Asp-146, His-154 and Gln-211 alleles showed significantly lower conversion levels compared to wild type, confirming their protective effects against scrapie. Although we utilized a heterologous conversion system, this is to our knowledge, the first study of caprine PrP variants in a cellular context of scrapie, that confirms the protective effects of some of the studied alleles.
INTRODUCTION
Scrapie, the prion disease of sheep and goats, is a fatal neurodegenerative disorder caused, according to the predominate ‘protein-only hypothesis,Citation1 by a ‘proteinaceous infectious’ agent. Further substantiated by accumulating experimental evidence,Citation2-4 the ‘protein-only’ hypothesis postulates that the infectious agent corresponds to PrPSC, a β-sheet rich and partially Proteinase K resistant isoform of the normally occurring prion protein, PrPC. PrPSC interacts with PrPC and imposes a conformational rearrangement on the normal protein, resulting in the production of more aberrant, β-sheet rich molecules.Citation5
Disease manifestation is influenced by variations in the Prnp gene, which encodes PrPC.Citation6 Non-synonymous Single Nucleotide Polymorphisms (SNPs) present in the ovine Prnp have been strongly associated with resistance and were targeted by selective breeding strategies, resulting in significant reduction of disease prevalence.Citation7,8 Unfortunately, this has not yet been applied in the case of goats, as the most promising scrapie-protective allele(s) has not been determined yet.
One of the main goals in the field of small ruminant prion research during the past 5 y has been the study of caprine Prnp variability in correlation to scrapie. Similarly to sheep, the caprine Prnp gene is highly polymorphic.Citation6,9-16 Field studies, conducted in different countries on a variety of goat breeds, assigned some degree of protection against scrapie to the following alleles Met-142, Arg-143, Asp-146, Ser-146, His-154, Gln-211 and Lys-222.Citation11,13,14,17-23 Among these, Met-142 is linked to prolonged incubation period,Citation13,17,21,23 while Arg-143Citation18 and His-154 Citation14,18,19,21,23 are associated with moderate protection. Ser-146 or Asp-146,Citation14,19,20 Gln-211Citation11,21,23 and Lys-222 Citation11,21-23 are associated with higher degrees of protection, suggesting that they could be used as candidate alleles, targeted by a potential selective breeding strategy.
Both in vitro and in vivo experimental approaches have been implemented to further evaluate these alleles' protective effects prior to making an optimal selection of the best allele candidates. The in vitro approach utilized a cell-free system to estimate the conversion potential of several alleles.Citation24 In parallel, in vivo studies were initialized on either transgenic mice harboring caprine Prnp alleles,Citation25 or on experimentally challenged goats carrying the genotypes of interest.Citation26-28
In the present work we report the in vitro study of murine-caprine PrP chimeras consisting of the mature caprine Prnp sequence and carrying the alleles of interest, in a murine cell model of scrapie. Upon interaction of exogenously expressed proteins with endogenous murine PrPSC, conformational conversion of the studied variants occurs. This results in the detection of partially PK-resistant moieties. Interestingly, tested alleles display distinct conversion potentials, which in most of the cases, can be linked to some degree of protection against scrapie.
RESULTS
Cellular Distribution of the Expressed PrP Variants in the 22LN2a#58 Cell Line
Our study aimed at the assessment of selected caprine PrP variants in relation to scrapie, within a cellular model of the disease. We used a murine neuroblastoma cell line (22LN2a#58), which: a) resembles neurons, the main targets of scrapie and b) propagates the scrapie agent (PrPSC), due to its original infection with the mouse-adapted 22L scrapie strain. Thus, this cell line fosters/establishes a cellular environment that simulates disease conditions, as opposed to the corresponding non-infected cell line (N2a#58), where only the normal murine PrPC is detected (Figure S1).
PrP elements associated with processing and cellular trafficking of newly synthesized proteins, such as the N-terminal and the C-terminal GPI-anchor signal peptides, display differences between mice and goats (Figure S2). Consequently, murine-caprine chimeric PrPs, consisting of the mature caprine PrP sequence flanked by murine PrP elements, were expressed in the selected cell lines to ensure their correct synthesis, trafficking and anchorage onto the plasma membrane.
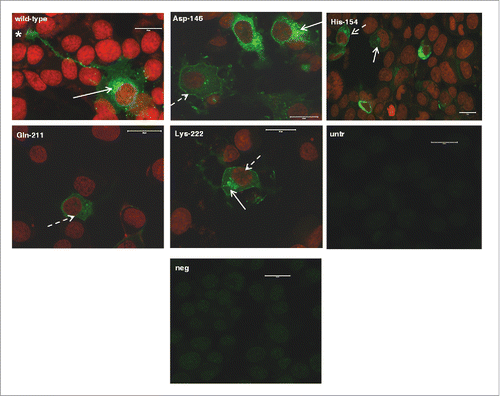
Normal trafficking and plasma membrane binding was confirmed by immunocytochemistry, using a goat PrP specific antibody (P4). All protein variants under study displayed endo-cellular, mainly peri-nuclear distribution. They were also detected on the plasma membrane and in neuronal axons (), suggesting that they followed the normal biosynthetic route.
FIGURE 1. Localization of murine-caprine chimeric PrPC molecules harboring either the wild type mature caprine PrP sequence or each one of the tested caprine alleles, within 22LN2a#58 cells. Chimeric murine –caprine PrP molecules consisting of the mature caprine PrP sequence and harboring either the wild type (wt), or the Asp-146, His-154, Gln-211 and Lys-222 variants were expressed in scrapie-infected 22LN2a#58 cells and detected by indirect immunofluoresence (green). Primary antibody: P4 (dilution 1/250), secondary antibody: GAM-FITC (dilution 1/1,000). Nuclei were counter-stained with propidium iodide (red). Primary antibody specificity was determined by processing untransfected cells (untr) as described before. Secondary antibody specificity was determined by processing transfected cells expressing the wild type chimeric PrP only with the secondary antibody (neg). Samples were observed under a Confocal Laser Scanning Microscope (Zeiss, Observer Z1), using the following filters: emission 488 nm - detection 515/530 nm (green), emission 543 nm - detection 570 nm (red). Magnification: x40. Scale bar: 20 μm. Peri-nuclear (white arrows), plasma membrane (white dashed arrows), neurite and neuronal axon localization (star) was detected.
Assessment of the 22LN2a#58 Cell Line Suitability for Heterologous PrP Conversion Studies
Impaired interspecies prion diseases transmission, known as the “species barrier” phenomenon, is detected due to altered interaction among heterologous PrPs or diminished PrPSC ability to impose conformational conversions onto the host PrPC, resulting in little or no PrPC conversion. Since we utilized a scrapie infected murine cell line for expression of caprine Prnp variants, interaction between murine PrPSC and exogenously expressed PrPC could result in the occurrence of an absolute ‘species barrier’ phenomenon.
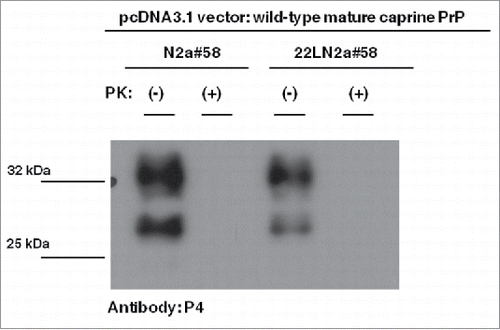
Thus, we first tested whether exogenously expressed PrPs carrying the wild-type mature caprine Prnp sequence, the most susceptible caprine variant, are efficiently processed by endogenous murine PrPSC to produce partially PK-resistant moieties of caprine origin. Western blot analysis using a goat PrP specific antibody (L42), revealed PK-resistant moieties when PK-treated cell lysates from 22LN2a#58 cells expressing the wild-type caprine PrP chimera were analyzed, in contrast to similarly processed N2a#58 cell lysates (). This indicates that within the 22LN2a#58 cellular environment, exogenously expressed murine-caprine chimeric proteins interact with murine PrPSC and convert to aberrant PrP. Interestingly, these PK-resistant moieties were not recognized by another goat specific PrP antibody, P4, which binds within the last octa-repeat at the N-terminal region of the protein and very close to the PK cleavage site (). Since this antibody detects PrPSC from natural goat scrapie cases,Citation29 its inability to detect caprine converted alleles within our experimental system suggests that molecules in the latter case are characterized form a different PK-resistant core compared to natural goat scrapie cases. All the above indicate that even though the 22LN2a#58 cell line does not absolutely mimick all aspects of natural disease, it can be utilized for assessment of different caprine PrP variant convertibility.
FIGURE 2. Murine-caprine chimeric PrP molecules harboring the wild type mature caprine PrP sequence are converted to partially resistant to Proteinase K treatment moieties in the murine scrapie-infected 22LN2a#58 cells. Western blot analysis on cell lysates from scrapie-infected (22LN2a#58) or non-scrapie infected (N2a#58) cells expressing murine-caprine chimeric PrP molecules harboring the wild type mature caprine PrP sequence after transient transfection with the corresponding vector. Two days after transfection cells were lysed; lysates were either treated with Proteinase K (PK+) or not (PK-), and proteins were methanol precipitated. Protein pellets were resuspended in sample buffer, analyzed by SDS-PAGE, electrotransferred on a PVDF membrane and probed with the sheep/goat PrP-specific L42 antibody (dilution 1/10). The secondary antibody (GAM-HRP) was used at a 1/10,000 dilution. The blot was developed on autoradiography film, with enhanced chemiluminesence (ECL). For PK treatment 0.75 μgs PK/mg total protein were used for 1 h at 37°C, with agitation; reaction was stopped with addition of PMSF. Analyzed protein loads correspond to 10 μgs for PK(-) samples and to the equivalent of 100 μgs before PK-treatment for PK(+) samples. Pertinent Molecular Weight Markers are indicated. An L42 immunopositive band at ∼25 kDa is detected in PK-treated lysates from scrapie-infected cells, while it is absent from scrapie-free cells, indicating that the exogenously expressed PrP undergoes conformational rearrangement in the scrapie-infected cells, which results in partial resistance to PK.
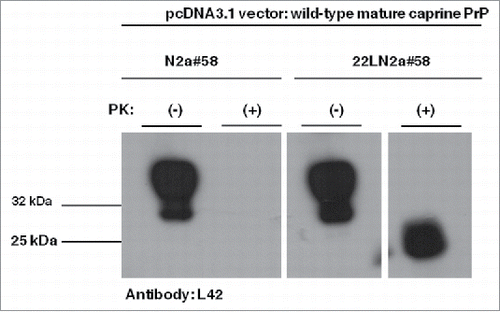
FIGURE 3. Goat specific Proteinase K-resistant moieties produced in the 22LN2a#58 cells are different from PrPSC molecules of naturally occurring goat scrapie cases. Western blot analysis on cell lysates from scrapie-infected (22LN2a#58) or non-scrapie infected (N2a#58) cells expressing murine-caprine chimeric PrP molecules harboring the wild type mature caprine PrP sequence. The same samples as in were subjected to SDS-PAGE, electrotransferred onto a PVDF membrane and probed with the P4 antibody (1/250 dilution), which recognizes a PrP region close to the PK cleavage site. Pertinent Molecular Weight Markers are indicated on the left. Absence of a P4 immunopositive band at ∼25 kDa in PK-treated lysates from both scrapie-infected and scrapie-free cells, indicates that the goat specific Proteinase K-resistant moieties produced in the scrapie-infected cells () are different from the PrPSC molecules detected in natural scrapie cases.Citation29
To further characterize the glycosylation pattern of the goat-specific PK-resistant moiety detected in the utilized cell line, we analyzed scrapie-infected cells, either untransfected or transfected to express the caprine wild-type chimera, with a panel of antibodies that either recognize goat-only (L42) or both murine and caprine PrPs (6H4). Acquired results (Figure S3A and S3B) indicate that the major goat-specific band, detected at approximately 25 kDa, corresponds to the double-glycosylated caprine-PrP species. Additional analysis of PK-treated lysates from scrapie-infected cells expressing the caprine wild-type chimera and PK-treated brain tissue material originating from ovine scrapie using the L42 antibody also suggested that the goat-specific PK-resistant moiety mainly corresponds to the double-glycosylated form (Figure S3C).
Assessment of Caprine PrP Variants' Conversion Potential in the 22LN2a#58 Cell Line
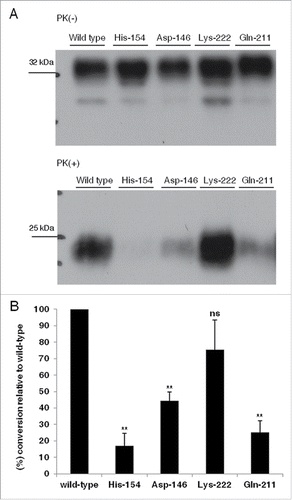
We reasoned that allelic variants displaying reduced conversion efficiencies compared to wild-type within the utilized cellular context, could be assigned as scrapie-protective. Conversion efficiencies for each allelic variant were estimated after Western blot analysis (L42 antibody) on both PK-treated and non PK-treated fractions of lysates from cells expressing the variants under study (). Conversion efficiencies were estimated as the ratio of band intensities detected in the PK-treated fraction (PrPSC) to band intensities of the corresponding non-PK treated sample (total PrP: PrPC and PrPSC). To acquire comparable results, all samples were processed simultaneously using lysate volumes including similar levels of exogenously expressed proteins and a standard PK to total protein ratio. Given that the wild-type allele is the most susceptible, all conversion efficiencies were expressed as a percent relative to this allele. Statistical analysis (unpaired student t-test) performed on the mean values of percent conversion efficiencies determined for each allele relative to wild type (3 independent experiments), revealed that as opposed to Lys-222, variants carrying the Asp-146, His-154 and Gln-211 alleles display significantly lower convertibility compared to wild-type ().
FIGURE 4 (see next page). Murine-caprine chimeric PrP molecules harboring the Asp-146, His-154 and Gln-211 caprine PrP variants display reduced conversion efficiency to partially Proteinase K- resistant moieties in murine scrapie-infected 22LN2a#58 cells. (A) Representative Western Blot on cell lysates from 22LN2a#58 cells, expressing murine-caprine PrP chimeras carrying either the wild type mature caprine PrP sequence or the Asp-146, His-154, Gln-211 and Lys-222 variants, after transient transfections with the corresponding vectors. Cells were lysed 2 d after transfection and lysates were either subjected to PK-treatment (PK+) or not (PK-). Protein samples were analyzed by SDS-PAGE, electrotransferred onto a PVDF membrane and probed with the sheep/goat PrP-specific L42 antibody (1/10 dilution). The secondary antibody (GAM-HRP) was used at a 1/10,000 dilution. For development, enhanced chemiluminescence (ECL) was used. Protein loads correspond to lysate volumes containing similar amounts of the exogenously expressed proteins. PK treatment was preformed with 0.75 μgs PK/mg total protein, at 37°C, for 1 h, with agitation, and was stopped with the addition of PMSF. Protein loads for PK(+) samples are 10-fold higher than the corresponding loads of PK(-) samples. PK treated samples were subjected to methanol precipitation before analysis. Pertinent Molecular Weight Markers are indicated. Immunopositive L42 bands of ∼25 kDa in PK-treated samples correspond to partially PK-resistant moieties. (B) Histograms depicting the mean and standard errors of the tested alleles' conversion efficiencies relative to wild type, determined by 3 independent experiments. Conversion efficiencies were estimated as the ratio of L42 immunopositive band intensities at ∼25 kDa in PK-treated samples (PrPSC) to immunopositive band intensities at ∼32 kDa in non-PK treated samples (total PrP, PrPC and PrPSC), and were expressed as a percent of the conversion efficiency of the most susceptible allele (wild type). Band intensities were determined by densitometric analysis performed on corresponding Western blots (A), using the ImageJ software. Unpaired student t-test was used for statistical analysis. **: p <0.01, ns: not statistically significant
Prediction of Structural Changes Imposed on the Caprine PrP by the Allelic Variants Under Study
Possible alterations in hydrogen bond formation within the caprine PrP variants under study and their effects on the molecules' regional stabilities were predicted based on the wild type ovine PrP globular domain crystallographic data.Citation30 A structural model of the molecule's globular domain depicting tested alleles' localization, generated using the pdb.viewer software, is shown in . Predictions of amino acid alteration effects on hydrogen bond formation relative to wild type, identified 2 groups of variants: a) variants with no impact on hydrogen bond formation relative to wild type (Asp-146, His-154) and b) variants diminishing hydrogen bonds present in the wild type allele (Gln-211 and Lys-222). depicts snapshots of the predicted molecules' structure carrying either the wild type or any of the tested alleles.
FIGURE 5. Hydrogen bond formation prediction within caprine PrP variants harboring the tested alleles compared to wild type. (A) Structural model of the caprine PrP globular domain, based on ovine PrP crystallographic data [PDB entry 1uw3]Citation30 and visualized using the pdb.viewer software. Amino acid residues corresponding to allelic variants of interest are shown in red. Helixes are shown in orange and β-sheet in green. (B-E) Snapshots of the caprine PrP molecule, depicting hydrogen bonds formed in wild type and in variants harboring amino acid substitutions. Residues of interest are shown in red; hydrogen bonds are depicted as dashed light green lines. (B) Prediction of the Asp-146 allele effect on hydrogen bond formation relative to wild type (wt). No changes are predicted by the substitution of Asparagine by Aspartic acid at position 146, (C) Prediction of the His-154 allele effect on hydrogen bond formation compared to wild type (wt). No changes in hydrogen bonds formation are predicted by the substitution of Arginine by Histidine at position 154, (D) Prediction of the Gln-211 allele effect on hydrogen bond formation compared to wild type (wt). Substitution of Arginine by Glutamine at residue 211, diminishes the hydrogen bond formed between Arg-211 and His-143 and ablates the connection between helix3 and the S1H1 loop. Note that for easier depiction, parts of helix3 (orange surfaces) are not displayed, (E) Prediction of the Lys-222 allele effect on hydrogen bond formation relative to wild type (wt). Substitution of Gln-222 by Lysine, abolishes the hydrogen bond formed between Gln-222 and Gln-175. For easier depiction, parts of helix3 are not displayed.
![FIGURE 5. Hydrogen bond formation prediction within caprine PrP variants harboring the tested alleles compared to wild type. (A) Structural model of the caprine PrP globular domain, based on ovine PrP crystallographic data [PDB entry 1uw3]Citation30 and visualized using the pdb.viewer software. Amino acid residues corresponding to allelic variants of interest are shown in red. Helixes are shown in orange and β-sheet in green. (B-E) Snapshots of the caprine PrP molecule, depicting hydrogen bonds formed in wild type and in variants harboring amino acid substitutions. Residues of interest are shown in red; hydrogen bonds are depicted as dashed light green lines. (B) Prediction of the Asp-146 allele effect on hydrogen bond formation relative to wild type (wt). No changes are predicted by the substitution of Asparagine by Aspartic acid at position 146, (C) Prediction of the His-154 allele effect on hydrogen bond formation compared to wild type (wt). No changes in hydrogen bonds formation are predicted by the substitution of Arginine by Histidine at position 154, (D) Prediction of the Gln-211 allele effect on hydrogen bond formation compared to wild type (wt). Substitution of Arginine by Glutamine at residue 211, diminishes the hydrogen bond formed between Arg-211 and His-143 and ablates the connection between helix3 and the S1H1 loop. Note that for easier depiction, parts of helix3 (orange surfaces) are not displayed, (E) Prediction of the Lys-222 allele effect on hydrogen bond formation relative to wild type (wt). Substitution of Gln-222 by Lysine, abolishes the hydrogen bond formed between Gln-222 and Gln-175. For easier depiction, parts of helix3 are not displayed.](/cms/asset/e3cf6e52-8445-4670-ba6b-1ad846c5801d/kprn_a_1199312_f0005_oc.gif)
DISCUSSION
Scrapie presents a nearly worldwide distribution of harmful effects to animal welfare, by extent the agricultural sector, and subsequently economy. Affected animals, apart from suffering from this fatal disorder, pose the risk of transmitting the disease within a flock/herd, either though milk (transmission from scrapie-affected mothers to offspringsCitation31,32 or via infectious agent dissemination in the environment through affected animals' placenta shedsCitation33 and/or excretions).Citation34 Moreover, binding of the scrapie agent to the soil and preservation of its infectivity for a long time,Citation35 constitutes a further source of disease dissemination to animals (sheep and goats) sharing the same pasture.
It is therefore evident that there is an urgent need to control scrapie spread and work toward its elimination. In sheep, recruitment of genetically resistant animals in selective breeding programs, targeting the almost absolute protective Ala-136,Arg-154,Arg-171 allele,Citation6 yielded very promising results and significantly reduced disease prevalence.Citation7,8 Even though there is increasing interest, similar strategies have not yet been applied in goats. This is mainly due to the lack of evidence that points to a credible equivalent to the ovine Ala-136,Arg-154,Arg-171, caprine Prnp allele, and to the until recently limited information on the genetics of caprine Prnp variants in association to scrapie.
Field studies enabled identification of caprine Prnp variants associated with some degree of protection against scrapie, albeit not absolute. These variants include Met-142,Citation13,17,21,23 His-154,Citation14,18,19,21,23 Arg-143,Citation18 and alleles with stronger protective effects, such as Ser-146, Asp-146,Citation14,19,20 Gln-211Citation11,21,23 and Lys-222.Citation11,21-23
These alleles' relative low occurrence frequency among herds and/or goat breeds examined in field studies, suggests that additional investigations within the context of a well defined experimental scheme, including sufficient test samples and controls, are required prior to gaining further conclusive information on their effects. Different experimental approaches, utilizing either in vitroCitation24 or in vivoCitation25-28 methodologies, have been used toward this direction.
In vitro studies significantly contribute to an initial evaluation and present the advantage of delivering fast results. Up to date, only a single in vitro study has been reportedCitation24 in the field, which utilized a cell-free conversion system to determine conversion efficiencies of recombinant caprine PrPs carrying scrapie-protective substitutions in the presence of the mouse-adapted Me7 strain.
In a further effort to study selected caprine Prnp alleles in an in vitro system that would simulate the conversion process, we expressed murine-caprine chimeras in a murine cellular model of scrapie propagating the 22L mouse-adapted scrapie strain and studied their potential to be converted into aberrant PrP molecules upon interaction with endogenous PrPSC.
We utilized the cellular biosynthetic machinery to establish post-translationally modified and properly folded proteins as substrates for conversion. Since the conversion process requires interaction between PrPC and PrPSC,Citation5 proper substrate folding is highly desired. Glycosylation of the expressed proteins within our experimental system enabled both their proper folding and cellular trafficking.Citation36 Normal PrPC cellular trafficking is also critical for proper foldingCitation37 and conversion occurrence. Indeed, physiological PrPC trafficking results in its localization on the plasma membrane, in sub-cellular compartments and vehicles,Citation38 and enables its co-localization with PrPSC on the plasma membrane as well as in endo-cellular compartments with low pH,Citation39 where conversion takes place.Citation37,40 We ensured physiological cell trafficking of the studied protein variants and their subsequent binding to the plasma membrane, by expressing murine-caprine chimeras consisting of the mature caprine PrP sequence, flanked by murine PrP regulatory elements, to enable Endoplasmatic Reticulum lumen targeting and GPI-anchor addition.
Efficient conversion depends on the compatibility of the interacting PrP isoforms and is associated with both their primary sequence and structural similarity.Citation41 Initial physical binding of the interacting PrP isoforms and/or PrPC rearrangement, both critical steps in the conversion process,Citation42 are expected to be affected by the degree of PrPC- PrPSC homology.
Primary sequence homology, especially in critical amino acid positions, dictates the efficiency of interspecies prion transmission. Such a critical amino acid has been linked with the mouse-hamster species barrier, since the mutated murine PrP, carrying the hamster-specific Methionine at position 138 instead of the mouse-specific Isoleucine, did not undergo conversion when expressed in scrapie infected cells.Citation43 Amino acid residues within the PrP β-sheet2-helix2 (S2H2) loop, especially at positions 170 and 174, are also linked with interspecies prion transmission. The S2H2 loop is involved in the initial PrPC unfolding, which precedes the actual conformational conversion event and results in the separation of the β-sheet1-helix1-β-sheet2 (S1H1S2) from the helix2-helix3 PrPC region.Citation44 Residues at positions 170 and 174 affect the S2H2 loop rigidness and allow efficient conversion only in cases of regional structural homology between the interacting PrP isoforms.Citation45 The significance of the H2S2 loop region in interspecies prion transmission has been highlighted by another study, which points out the importance of primary sequence rather than structural similarities.Citation46 Furthermore, correlation of multiple amino acid residues with the murine-rabbit ‘species barrier’, suggests that primary sequence differences between interacting PrP isoforms may be associated with conversion-incompatible structures.Citation47
Studies on heterologous PrP interactions revealed altered molecular associationsCitation42,48 and lower conversion efficiencies. Other researchers determined PrPC regions involved in the initial bindingCitation49-51 and in the subsequent conversion process.Citation51-54 According to these studies, amino acid residues 101-110 (murine PrP numbering) are associated with the initial binding of interacting PrP isoforms,Citation49 while a partial subset of this region, involving residues 99-104, plays an active role in the actual conversion process.Citation52 Similarly, amino acid residues 136-140 (Arg-Pro-Met-Ile-His) have been recognized as another interaction interface, also critical for the conversion process.Citation49 Finally, the charge distribution of amino acids consisting helix1 (residues 143-151) is critical for efficient conversion.Citation54
Our experimental approach utilized a heterologous conversion system to study different caprine PrPC variants after their interaction with PrPSC molecules produced by scrapie-infected murine neuroblastoma cells. Even though primary mature PrP sequences of mice and goats differ in 23 positions (Figure S4), no differences are detected in critical positions associated with the initial binding of the interacting molecules and subsequent PrPC conversion. This observation would predict that initial binding and further conformational conversion could occur in the chosen cellular context, suggesting the occurrence of an in-complete “species barrier”.
Moreover, the strain effect is expected to influence the conversion process, as different strains are associated with different PrPSC conformations.Citation55 According to the ‘conformational selection model’,Citation56 PrP molecules from a given species accommodate a range of different PrPSC conformations, and enable efficient conversion upon interaction with heterologous PrP molecules only in cases of substantial overlap among the permissible conformations between interacting species. Also, during the strain adaptation process in a new host, a different PrPSC conformation, present in the original pool, may be preferentially propagated, resulting in strain shifts.Citation57 In this study we utilized the 22L mouse-adapted scrapie strain, which originated from sheep scrapie brain pool (SSBP/1) inoculated in mice without any other intermediate host.Citation58 Absence of an intermediate host could suggest that the majority of the naturally occurring strain characteristics were preserved after passaging in mice, which could enable us to predict that the mouse-adapted strain would be able to impose conformational conversions on caprine PrPs. These predictions were confirmed by our experimental findings, which enabled the detection of partially PK-resistant moieties of caprine origin.
Nevertheless, the caprine PK-resistant moieties produced within the utilized cellular context were different from PrPSC molecules detected in natural scrapie cases, as proven from the inability to detect them using the P4 antibody. Since the epitope of this antibody is close to the PK cleavage site and it is present in PrPSC molecules from natural goat-scrapie cases while absent from the in vitro produced caprine PK-resistant moieties, it follows that these caprine PK-resistant moieties are characterized by a different PK cleavage site compared to natural occurring caprine PrPSC. This deviation could reflect conformational differences between the naturally occurring and the in vitro produced PrPSC molecules, possibly associated with the adaptation process that the original strain underwent in the murine cell line and the subsequent selection of PrPSC conformations compatible with the conformations of the expressed murine-caprine chimeras.
The cellular conversion system described before was used to study variants of murine-caprine chimeras carrying single nucleotide polymorphisms in the mature caprine Prnp sequence. Some of the previously reported scrapie-protective caprine Prnp variants (Asp-146, His-154, Gln-211 and Lys-222) were tested. Since conversion of PrPC to PrPSC is the key event in prion diseases, we reasoned that decreased conversion potentials relative to the most susceptible wild type allele would be associated with scrapie-protective variants. Our experimental results determined significantly lower conversion potentials relative to wild type for the Asp-146, His-154 and Gln-211 alleles, in contrast to Lys-222, which could not be distinguished from wild type by statistical analysis.
Similar to another cell-free conversion study,Citation24 our data indicate that the Asp-146, His-154 and Gln-211 variants are linked with protection against scrapie, albeit they are assigned with different conversion efficiencies in each experimental approach. These differences may reflect strain-specific (22L and Me7) effects on the tested alleles' conversion. Structural studies on PrPSC molecules representing either the 22L or the Me7 strain, indicated conformational differences mainly associated with beta sheets,Citation59 suggesting potentially different interactions with the variants under study, that could possibly influence the effectiveness of subsequent conversion. Moreover, the profound discrepancy regarding the Lys-222 variant performance, which did not show significantly lower conversion relative to wild-type in our study as opposed to almost complete absence of conversion in the study of Eiden and colleagues,Citation24 could also be explained by previously reported differences in prion strains' behavior upon interaction with PrPs of the same sequence.Citation60 This further points out the importance of assessing a specific variant relative to several strains, in order to safely assess its protective effect against scrapie.
Nevertheless as noted before, conclusions acquired form in vitro studies, using mouse-adapted scrapie strains, need to be validated by in vivo observations. Recently published data on transgenic mice harboring the caprine Lys-222 allele after challenge with goat-scrapie and other TSE isolates, assigned strong protection to this allele.Citation25 Similarly, studies on experimental transmission (oralCitation28 or intracerebralCitation27) of natural goat scrapie isolates in heterozygous Lys-222 goats, concluded that Lys-222 significantly extends survival time after inoculation,Citation28 with no signs of PrPSC deposition up to 4,5 y post-inoculation.Citation27 Further, protective effects were assigned to the His-154, Gln-211 and Lys-222 alleles after oral or intracerebral inoculation of goats (wild type, heterozygous or homozygous for each of these alleles) with a single naturally occurring goat scrapie isolate. Based on the outcomes of intracerebral inoculations, Lys-222 was associated with the highest, albeit not absolute, degree of protection.Citation26
The molecular basis of scrapie-protective PrP variants has not been elucidated yet. However, substantial evidence attributes reduced conversion efficiency to resistant PrP variants, as the highly protective Ala-136,Arg-154,Arg-171 variant was found to constitute only a minor fragment of the PrPSC molecules detected in the brains of scrapie-affected sheep carrying the Ala-136,Arg-154,Arg-171/Val-136,Arg-154,Gln-171 genotype.Citation61,62 Further, susceptibility to prion diseases is linked with the propensity of PrP variants to transit from the a-helical to a β-sheet structure state, where the latter is affected by interactions between critical amino acid residues.Citation63 Moreover, single amino acid substitutions may introduce local structural alterations and affect PrPC stability.Citation64 Also, scrapie-protective variants display lower thermal stability relatively to the susceptible ones.Citation65,66
In light of these data, evaluation of our experimental results after assessment of the tested alleles' effects on the corresponding molecules' regional stabilities, as judged by hydrogen bond formation and/or ionic interactions alterations relative to wild type (according to structural prediction models, ), further provides mechanistic explanations in most of the cases.
Indeed, all tested variants were predicted to result in reduced regional stabilities of the corresponding molecules, by either diminishing hydrogen bond formation or altering ionic interactions with neighboring residues.
More specifically, even though no hydrogen bond alterations could be predicted in the case of the His-154 and Asp-146 substitutions, altered ionic interactions with neighboring amino acid residues, due to introduced charge changes, were predicted. In particular, substitution of the wild type and positively charged Arg-154 by Histidine, would be expected to reduce the strength of the ionic interaction between residue at position 154 and the negatively charged Asp-150.Citation67 Since codon 154 resides within helix1,Citation30 introduction of the His-154 polymorphism is expected to reduce its regional stability. Moreover, this polymorphism results in altered charge distribution at the C-terminal part of helix1, which has been shown to affect conversion efficiency.Citation54
Moreover, introduction of the negatively charged Aspartic acid in place of Asparagine at position 146 could result in a repulsive ionic interaction with the neighboring Asp-147. Taking into account that residue 146 is located just before helix1,Citation30 its predicted repulsive interaction with the first residue of this helix is expected to reduce helix1 stability.
Reduced PrP regional stabilities were also predicted in the case of Gln-211 and Lys-222 polymorphisms residing in helix3,Citation30 due to charge changes and concurrent ablation of hydrogen bonds. In more detail, substitution of Arg-211 by Glutamine, diminishes the original positive charge of the wild type allele and in the same time is predicted to result in the ablation of the hydrogen bond formed between wild type Arg-211 and His-143. Since amino acid residue 143 is located within the PrP S1H1 loop,Citation30 ablation of the hydrogen bond under investigation due to the Gln-211 polymorphism, would abolish the connection between S1H1 and helix3, thus conferring reduced stability to the molecule. Finally, substitution of the wild type Glutamine at position 222 by the positively charged Lysine, is further predicted to abolish the hydrogen bond formed between Gln-222 and Gln-175 and result in lower regional stability of the corresponding PrP variant.
Collectively, our predictions assigned reduced regional stabilities to the Asp-146, His-154, Gln-211 and Lys-222 alleles which, with the exception of Lys-222, also displayed reduced conversion efficiencies in the utilized murine scrapie-infected cell line. The discrepancy referring to the predicted and experimental effects of the Lys-222 variant in our study could be explained by the observation that several folding pathways are associated with the conformational changes that helixes 2 and 3 undergo during PrP conversion.Citation68 This suggests that the prevalent pathway may modulate the result of the PrPC - PrPSC interaction, thus providing a structural approach of strain diversity. Moreover, studies on the production of intermediate structures during PrPC unfolding indicate that some polymorphic variants may acquire different intermediate conformations depending on their environment.Citation69 All the above could explain the failed attempt to detect reduced conversion of the Lys-222 variant within our experimental model.
In summary, we report the study of goat-scrapie protective variants within a cellular model of prion diseases, which confirms the protective effects of the Asp-146, His-154 and Gln-211 alleles shown in other in vitro and in vivo studies; we further provide mechanistic insights on the basis of these alleles' protective effects, using structural prediction models. The fact that within our experimental system the Lys-222 allele was not linked with resistance is probably related with its increased efficiency to be converted by the 22L mouse-adapted scrapie strain propagating in the chosen cell line. Even though not absolutely representative of naturally occurring scrapie strains, this observation highlights the importance of strain effects on the performance of protective alleles, and necessitates the evaluation of candidate alleles in relation to the circulating TSE strains at any given geographical area.
MATERIALS AND METHODS
Expression Vectors Construction
Eukaryotic expression vectors (pcDNA3.1/Zeo(+)) carrying the mature caprine Prnp sequence (amino acid residues 25-233), flanked by the N-terminal murine PrP signal peptide (moPrP aa 1-22) and the C-terminal murine PrP GPI-anchor signal peptide (moPrP amino acid residues 228-255) followed by the murine PrP 3′UTR, were produced by standard cloning methods. Different alleles (wild-type, Asp-146, His-154, Gln-211 and Lys-222) of the mature caprine Prnp were PCR amplified from corresponding pDrive vectors,Citation70 using the magoF_ΕcoRI: 5′ cag gga att ccg aag aag cga cca aaa c 3′ and magoR_XhoI: 5′ aac tcg agt gcc ccc ctt tgg taa taa g 3′ primers, to incorporate EcoRI and XhoI restriction sites at their 5′ and 3′ and were cloned into pZeTa (pZero vector modified at the EcoRV restriction site, to allow TA cloning). Inserts were excised from pZeTa vectors by EcoRI/XhoI digestion and ligated in the pcDNA3.1/Zeo(+) vector backbone, generated after EcoRI/XhoI digestion of previously constructed vectors.Citation71 Vector and insert integrity were confirmed with Sanger sequencing in both directions (Figure S5).
Cell Lines and Transfections
The murine neuroblastoma N2a#58 cell line, over-expressing murine PrP, and its corresponding scrapie-infected 22LN2a#58 cell line (propagating the 22L mouse-adapted scrapie strain), were used.Citation72 Cells were cultured in OPTIMEM (Gibco) supplemented with 10% (v/v) FBS and 1% (v/v) penicillin/streptomycin, inside a 5% CO2 atmosphere incubator maintained at 37°C. Media was exchanged every 3 d. Transient transfections were performed on 60% confluent cultures, using the XtermeGene9 reagent (Roche, Cat No 06 365 787 001), according to the manufacturer instructions and a 3:1 ratio of transfection reagent to plasmid DNA. All experiments were conducted on Mycoplasma-free cells.
Indirect Immunofluoresence
Cells grown on coverslips were transfected as described under the “cell lines and transfections” section. At 48 h post-transfection, cells were rinsed with PBS, fixed with 4% (w/v) paraformaldeyde and permeabilized with 0.3% (v/v) TritonX-100. After further PBS washes, samples were blocked with 10% normal goat serum in TBS for 30 min at room temperature, and then incubated with the primary antibody (P4,Citation29 at a 1/50 dilution). Following washes with TBS, samples were incubated with the secondary antibody, in the dark (Goat-anti-Mouse FITC-conjugated, Sigma-Aldrich, at a 1/1,000 dilution). All antibody incubations were done for 1 h, at room temperature. After washes with TBS, nuclei counter-staining was performed via a 15 min incubation with propidium iodide (3 μg/ml in PBS) in the presence of RNase (10 μg/ml). Coverslips were rinsed with TBS and mounted on microscope slides. Samples were observed under a Confocal Laser Scanning Microscope (Zeiss, Observer Z1), using the following filters: emission 488 nm - detection 515/530 nm (green), emission 543 nm - detection 570 nm (red). Control samples to determine primary (untransfected samples processed according to the above description) and secondary antibody (transfected cells incubated only with the secondary antibody) specificities were included in the analysis.
Cell Lysis and Lysate Processing
Regularly cultured N2a#58 and 22LN2a#58 cells were lysed on the fourth and fifth day after passaging respectively. Transfected cells were lysed 2 d after transfection. For lysis, cells cultured in 100 mm culture dishes (Corning) were rinsed once with PBS and incubated with 1 ml ice-cold cell lysis buffer (Tris 10 mM, pH 7.5, NaCl 100 mM, EDTA 10 mM, Triton-100 0.5% w/v, Na-Deoxycholate 0.5% w/v) for 10 min, at room temperature. Cell lysates were either stored at -80°C, or processed immediately. After cell debris sedimentation (centrifugation at 14,000 g for 1 min, at room temperature), total protein concentration for each lysate was determined. Equal amounts of total protein were either treated with PK (Merck, Cat No 1245680100) or not and subjected to methanol precipitation, unless otherwise noted. Transfection efficiencies were taken into account before processing lysates from transfected cells, to ensure analysis of samples with similar amounts of exogenously expressed murine-caprine chimeras. For PK treatment, 0.75 μgs PK/mg total protein were used. Digestion was performed at 37°C, for 1 h, with agitation (225 cycles/min), and was stopped with PMSF (5 mM final concentration, also used in non-PK treated samples). PK treated samples further contained 1% (w/v) Lauryl-sarcosyl. Proteins were precipitated by adding methanol to the samples and incubating at -80°C for 16 h. Methanol precipitated samples were centrifuged at 14,000 g for 15 min, at room temperature, to sediment the proteins. Protein pellets were resuspended in O'Farrell (50 mM Tris pH 6.8, 2% SDS (w/v), 3% β-mercaptoethanol (v/v), 10% Glycerol (v/v)), boiled for 5 min and subjected to SDS-PAGE and Western Blot analysis.
SDS-PAGE and Western Blot Analysis
Protein samples were analyzed by SDS-PAGE on 12% polyacrylamide gels. For samples originating from regularly cultured, un-transfected N2a#58 or 22LN2a#58 cells, 100 μgs of non-PK treated and 400 μgs collected prior to PK treatment for PK-treated samples, were loaded. For non PK-treated samples originating from transfected cells, variable total protein amounts characterized by similar levels of exogenously expressed murine-caprine PrP chimeras, and 10 times higher amounts of each of the corresponding PK-treated samples were analyzed. For Western blot analysis, proteins were electro-transferred onto PVDF membranes. Membranes were blocked in 5% powder milk in PBST, before being probed with specific antibodies. PrP of either murine or caprine origin was detected after probing membranes with the 6H4 monoclonal antibody (Prionics, Cat No 01-011), at a 1/5,000 dilution. For the detection of exogenously expressed PrPs of caprine origin, the goat/sheep specific monoclonal antibody L42Citation73 was used at a 1/10 dilution. Another goat/sheep specific antibody, P4, was also used at a 1/250 dilution in some cases. Following primary antibody incubation and PBST washes, membranes were incubated with a Horse Radish Peroxidase-conjugated secondary antibody (Goat Anti Mouse-HRP, Sigma-Aldrich, Cat No Α0168), at a 1/10,000 dilution. All incubations were performed for 1 h at room temperature. After final washes with PBST, membranes were developed on autoradiography films using Enhanced Chemiluminescence (Chemiluminescent HRP substrate kit, GenScript Inc., Cat No L00221V500).
Densitometric Analysis and Conversion Efficiency Estimation
Relative intensities of immunopositive bands corresponding to the proteins under study were estimated using the ImageJ software (v1.45s, available on http://imagej.nih.gov/ij). Conversion efficiencies of exogenously expressed PrPs were calculated as the percent ratio of the PK-resistant fragment detected for these proteins (determined after densitometric analysis of the PK-treated immunoreactive bands) to the total PrP (PrPC and PrPSC) present in the corresponding non-PK treated samples. For conversion efficiencies estimation, loading differences between untreated and PK-treated samples were taken into account. For each experiment, conversion efficiency of the wild-type murine-caprine PrP chimera was set to 100 and the conversion efficiencies for all the tested variants were estimated relative to wild-type.
Statistical Analysis
Conversion efficiencies determined for each allelic variant from 3 independent experiments were analyzed relative to wild type using the unpaired student t-test (http://graphpad.com/quickcalcs/ttest1/?Format=SEM).
Caprine PrP Variants Structure Predictions
The DeepView/Swiss-Pdb Viewer (v4.1.0, available at http://www.expasy.org/spdbv/) was used to predict structural changes imposed by the selected allelic variants on the caprine PrP relative to wild type. Since both sheep and goat PrPs are almost identical (Figure S6), ovine wild-type PrP crystallographic data, retrieved from the PDB entry 1uw3,Citation30 were used as reference for the predictions.
ABBREVIATIONS
| PrP | = | Prion protein |
| PrPC | = | Normal |
| cellular | = | PrP |
| PrPSC | = | Pathogenic |
| Scrapie | = | PrP |
| N-terminal | = | Amino-terminal region of a protein |
| C-terminal | = | C-terminal region of a protein |
| moPrP | = | murine PrP |
| PK | = | Proteinase K |
| UTR | = | untranslated region |
| PBST | = | Phosphate Buffered Saline containing 0.05% Tween 20 |
| HRP | = | Horseradish peroxidase |
| GPI-anchor | = | Glycosylphosphatidylinositol-anchor |
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed
ACKNOWLEDGMENTS
The authors would like to thank Dr. Sylvain Lehmann (Institute for Research in Biotherapy, INSERM, Montpellier, France) for providing the murine cell lines N2a58 and 22LN2a58; Dr. Christos Panagiotidis (School of Pharmacy, Aristotle University of Thessaloniki, Greece) for providing the pZeTa vector; Dr. Martin Groschup (Institute for Novel and Emerging Infectious Diseases, Friedrich-Loeffler Institut, Germany) for providing the L42 and P4 antibodies and Dr. Emmanuel Panteris (School of Biology, Aristotle University of Thessaloniki, Greece) for his valuable help on the acquisition of Confocal Laser Scanning Microscope images. We are grateful to Dr. Cynthia Humphreys Panagiotidis (School of Pharmacy, Aristotle University of Thessaloniki, Greece), for her valuable contribution to the experimental set-up and the final revision of the manuscript. This work is dedicated in memory of Captain Niko Lillios towards his endless trip to the lost Atlantis.
Funding
This research has been co-financed by the European Union (European Social Fund – ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) - Research Funding Program: Heracleitus II. Investing in knowledge society through the European Social Fund. The study was supported by the EU project Goat-BSE (FOOD-CT-2006-36353).
REFERENCES
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136-44; http://dx.doi.org/10.1126/science.6801762
- Weber P, Giese A, Piening N, Mitteregger G, Thomzig A, Beekes M, Kretzschmar HA. Generation of genuine prion infectivity by serial PMCA. Vet Microbiol 2007; 123:346-57; PMID:17493773; http://dx.doi.org/10.1016/j.vetmic.2007.04.004
- Makarava N, Kovacs GG, Bocharova O, Savtchenko R, Alexeeva I, Budka H, Rohwer RG, Baskakov IV. Recombinant prion protein induces a new transmissible prion disease in wild-type animals. Acta Neuropathol 2010; 119:177-87; PMID:20052481; http://dx.doi.org/10.1007/s00401-009-0633-x
- Kim J-I, Cali I, Surewicz K, Kong Q, Raymond GJ, Atarashi R, Race B, Qing L, Gambetti P, Caughey B, et al. Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem 2010; 285:14083-7; PMID:20304915; http://dx.doi.org/10.1074/jbc.C110.113464
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363-83; http://dx.doi.org/10.1073/pnas.95.23.13363
- Goldmann W. PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res 2008; 39:30; PMID:18284908; http://dx.doi.org/10.1051/vetres:2008010
- Hagenaars TJ, Melchior MB, Bossers A, Davidse A, Engel B, van Zijderveld FG. Scrapie prevalence in sheep of susceptible genotype is declining in a population subject to breeding for resistance. BMC Vet Res 2010; 6:25; PMID:20470415; http://dx.doi.org/10.1186/1746-6148-6-25
- Nodelijk G, van Roermund HJW, van Keulen LJM, Engel B, Vellema P, Hagenaars TJ. Breeding with resistant rams leads to rapid control of classical scrapie in affected sheep flocks. Vet Res 2011; 42:5; PMID:21314971; http://dx.doi.org/10.1186/1297-9716-42-5
- Babar ME, Abdullah M, Nadeem A, Haq A. U. Prion protein gene polymorphisms in four goat breeds of Pakistan. Mol Biol Rep 2009; 36:141-4; PMID:17934795; http://dx.doi.org/10.1007/s11033-007-9162-7
- Vaccari G, Panagiotidis CH, Acin C, Peletto S, Barillet F, Acutis P, Bossers A, Langeveld J, van Keulen L, Sklaviadis T, et al. State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res 2009; 40:48; PMID:19505422; http://dx.doi.org/10.1051/vetres/2009031
- Bouzalas IG, Dovas CI, Banos G, Papanastasopoulou M, Kritas S, Oevermann A, Papakostaki D, Evangelia C, Papadopoulos O, Seuberlich T, et al. Caprine PRNP polymorphisms at codons 171, 211, 222 and 240 in a Greek herd and their association with classical scrapie. J Gen Virol 2010; 91:1629-34; PMID:20107013; http://dx.doi.org/10.1099/vir.0.017350-0
- Hussain A, Babar ME, Imran M, Haq IU, Javed MM. Detection of four novel polymorphisms in PrP gene of Pakistani sheep (Damani and Hashtnagri) and goats (Kamori and Local Hairy) breeds. Virol J 2011; 8:246; PMID:21595993; http://dx.doi.org/10.1186/1743-422X-8-246
- Goldmann W, Ryan K, Stewart P, Parnham D, Xicohtencatl R, Fernandez N, Saunders G, Windl O, González L, Bossers A, et al. Caprine prion gene polymorphisms are associated with decreased incidence of classical scrapie in goat herds in the United Kingdom. Vet Res 2011; 42:110; PMID:22040234; http://dx.doi.org/10.1186/1297-9716-42-110
- Papasavva-Stylianou P, Windl O, Saunders G, Mavrikiou P, Toumazos P, Kakoyiannis C. PrP gene polymorphisms in Cyprus goats and their association with resistance or susceptibility to natural scrapie. Vet J 2011; 187:245-50; PMID:20093056; http://dx.doi.org/10.1016/j.tvjl.2009.10.015
- Fragkiadaki EG, Vaccari G, Ekateriniadou LV, Agrimi U, Giadinis ND, Chiappini B, Esposito E, Conte M, Nonno R. PRNP genetic variability and molecular typing of natural goat scrapie isolates in a high number of infected flocks. Vet Res 2011; 42:104; PMID:21961834; http://dx.doi.org/10.1186/1297-9716-42-104
- Acín C, Martín-Burriel I, Monleón E, Lyahyai J, Pitarch JL, Serrano C, Monzón M, Zaragoza P, Badiola JJ. Prion protein gene variability in Spanish goats. Inference through susceptibility to classical scrapie strains and pathogenic distribution of peripheral PrP(sc.). PLoS One 2013; 8:e61118; PMID:23580248; http://dx.doi.org/10.1371/journal.pone.0061118
- Goldmann W, Martin T. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. J Gen Virol 1996; 77:2885-91; PMID:8922485; http://dx.doi.org/10.1099/0022-1317-77-11-2885
- Billinis C, Panagiotidis CH, Psychas V, Argyroudis S, Nicolaou A, Leontides S, Papadopoulos O, Sklaviadis T. Prion protein gene polymorphisms in natural goat scrapie. J Gen Virol 2002; 83:713-21; PMID:11842266; http://dx.doi.org/10.1099/0022-1317-83-3-713
- Papasavva-Stylianou P, Kleanthous M, Toumazos P, Mavrikiou P, Loucaides P. Novel polymorphisms at codons 146 and 151 in the prion protein gene of Cyprus goats, and their association with natural scrapie. Vet J 2007; 173:459-62; PMID:16314132; http://dx.doi.org/10.1016/j.tvjl.2005.09.013
- Ortiz-Pelaez A, Georgiadou S, Simmons MM, Windl O, Dawson M, Arnold ME, Neocleous P, Papasavva-Stylianou P. Allelic variants at codon 146 in the PRNP gene show significant differences in the risk for natural scrapie in Cypriot goats. Epidemiol Infect 2015; 143:1304-10; PMID:25140573; http://dx.doi.org/10.1017/S0950268814002064
- Barillet F, Mariat D, Amigues Y, Faugeras R, Caillat H, Moazami-Goudarzi K, Rupp R, Babilliot JM, Lacroux C, Lugan S, et al. Identification of seven haplotypes of the caprine PrP gene at codons 127, 142, 154, 211, 222 and 240 in French Alpine and Saanen breeds and their association with classical scrapie. J Gen Virol 2009; 90:769-76; PMID:19218225; http://dx.doi.org/10.1099/vir.0.006114-0
- Acutis PL, Bossers A, Priem J, Riina MV, Peletto S, Mazza M, Casalone C, Forloni G, Ru G, Caramelli M. Identification of prion protein gene polymorphisms in goats from Italian scrapie outbreaks. J Gen Virol 2006; 87:1029-33; PMID:16528054; http://dx.doi.org/10.1099/vir.0.81440-0
- Corbière F, Perrin-Chauvineau C, Lacroux C, Costes P, Thomas M, Bremaud I, Martin S, Lugan S, Chartier C, Schelcher F, et al. PrP-associated resistance to scrapie in five highly infected goat herds. J Gen Virol 2013; 94:241-5; PMID:23100359; http://dx.doi.org/10.1099/vir.0.047225-0
- Eiden M, Soto EO, Mettenleiter TC, Groschup MH. Effects of polymorphisms in ovine and caprine prion protein alleles on cell-free conversion. Vet Res 2011; 42:30; PMID:21324112; http://dx.doi.org/10.1186/1297-9716-42-30
- Aguilar-Calvo P, Espinosa JC, Pintado B, Gutiérrez-Adán A, Alamillo E, Miranda A, Prieto I, Bossers A, Andreoletti O, Torres JM. Role of the Goat K222-PrPC Polymorphic Variant in Prion Infection Resistance. J Virol 2014; 88:2670-6; PMID:24352451; http://dx.doi.org/10.1128/JVI.02074-13
- Lacroux C, Perrin-Chauvineau C. Genetic resistance to scrapie infection in experimentally challenged goats. J Virol 2014; 88:2406-13. Available from: PMID:24284317; http://dx.doi.org/10.1128/JVI.02872-13
- Acutis P, Martucci F, D'Angelo A, Peletto S. Resistance to classical scrapie in experimentally challenged goats carrying mutation K222 of the prion protein gene. Vet Res 2012; 43:8; PMID:22296670; http://dx.doi.org/10.1186/1297-9716-43-8
- White S, Reynolds J, Waldron D. Extended scrapie incubation time in goats singly heterozygous for PRNP S146 or K222. Gene 2012; 501:49-51; PMID:22516690; http://dx.doi.org/10.1016/j.gene.2012.03.068
- Harmeyer S, Pfaff E, Groschup MH. Synthetic peptide vaccines yield monoclonal antibodies to cellular and pathological prion proteins of ruminants. J Gen Virol 1998; 79:937-45; PMID:9568991; http://dx.doi.org/10.1099/0022-1317-79-4-937
- Haire LF, Whyte SM, Vasisht N, Gill AC, Verma C, Dodson EJ, Dodson GG, Bayley PM. The crystal structure of the globular domain of sheep prion protein. J Mol Biol 2004; 336:1175-83; PMID:15037077; http://dx.doi.org/10.1016/j.jmb.2003.12.059
- Konold T, Moore SJ, Bellworthy SJ, Terry LA, Thorne L, Ramsay A, Salguero FJ, Simmons MM, Simmons HA. Evidence of effective scrapie transmission via colostrum and milk in sheep. BMC Vet Res 2013; 9:99; PMID:23651710; http://dx.doi.org/10.1186/1746-6148-9-99
- Konold T, Simmons HA, Webb PR, Bellerby PJ, Hawkins SAC, González L. Transmission of classical scrapie via goat milk. Vet Rec 2013; 172:455; PMID:23625249; http://dx.doi.org/10.1136/vr.f2613
- Schneider DA, Madsen-Bouterse SA, Zhuang D, Truscott TC, Dassanayake RP, O'Rourke KI. The placenta shed from goats with classical scrapie is infectious to goat kids and lambs. J Gen Virol 2015; 96:2464-9; PMID:25888622; http://dx.doi.org/10.1099/vir.0.000151
- Gough KC, Maddison BC. Prion transmission: Prion excretion and occurrence in the environment. Prion 2010; 4:275-82; PMID:20948292; http://dx.doi.org/10.4161/pri.4.4.13678
- Johnson CJ, Phillips KE, Schramm PT, McKenzie D, Aiken JM, Pedersen JA. Prions adhere to soil minerals and remain infectious. PLoS Pathog 2006; 2:e32; PMID:16617377; http://dx.doi.org/10.1371/journal.ppat.0020032
- Ermonval M, Mouillet-Richard S, Codogno P, Kellermann O, Botti J. Evolving views in prion glycosylation: functional and pathological implications. Biochimie 2003; 85:33-45; PMID:12765773; http://dx.doi.org/10.1016/S0300-9084(03)00040-3
- Grassmann A, Wolf H, Hofmann J, Graham J, Vorberg I. Cellular aspects of prion replication in vitro. Viruses 2013; 5:374-405; PMID:23340381; http://dx.doi.org/10.3390/v5010374
- Linden R, Martins V. Physiology of the prion protein. Physiol Rev 2008; 88:673-728; PMID:18391177; http://dx.doi.org/10.1152/physrev.00007.2007
- Veith NM, Plattner H, Stuermer CAO, Schulz-Schaeffer WJ, Bürkle A. Immunolocalisation of PrPSc in scrapie-infected N2a mouse neuroblastoma cells by light and electron microscopy; PMID:18834644
- Noinville S, Chich JF, Rezaei H. Misfolding of the prion protein: Linking biophysical and biological approaches. Vet Res 2008; 39; PMID:18533092; http://dx.doi.org/10.1051/vetres:2008025
- Hagiwara K, Hara H, Hanada K. Species-barrier phenomenon in prion transmissibility from a viewpoint of protein science. J Biochem 2013; 153:139-45; PMID:23284000; http://dx.doi.org/10.1093/jb/mvs148
- Horiuchi M, Priola SA, Chabry J, Caughey B. Interactions between heterologous forms of prion protein: binding, inhibition of conversion, and species barriers. Proc Natl Acad Sci U S A 2000; 97:5836-41; PMID:10811921; http://dx.doi.org/10.1073/pnas.110523897
- Priola SA, Chesebro B. A single hamster PrP amino acid blocks conversion to protease-resistant PrP in scrapie-infected mouse neuroblastoma cells. J Virol 1995; 69:7754-8; PMID:7494285
- Prigent S, Rezaei H. PrP assemblies: Spotting the responsible regions in Prion propagation. Prion 2011; 5:69-75; PMID:21788728; http://dx.doi.org/10.4161/pri.5.2.16383
- Sigurdson C. A molecular switch controls interspecies prion disease transmission in mice. J Clin Invest 2010; 120:2590-9; PMID:20551516; http://dx.doi.org/10.1172/JCI42051
- Bett C, Fernández-Borges N, Kurt TD, Lucero M, Nilsson KPR, Castilla J, Sigurdson CJ. Structure of the β2-α2 loop and interspecies prion transmission. FASEB J 2012; 26:2868-76; PMID:22490928; http://dx.doi.org/10.1096/fj.11-200923
- Vorberg I, Groschup M, Pfaff E, Priola S. Multiple amino acid residues within the rabbit prion protein inhibit formation of its abnormal isoform. J Virol 2003; 77:2003-9; PMID:12525634; http://dx.doi.org/10.1128/JVI.77.3.2003-2009.2003
- Mallik S, Yang W, Norstrom EM, Mastrianni JA. Live cell fluorescence resonance energy transfer predicts an altered molecular association of heterologous PrPSc with PrPC. J Biol Chem 2010; 285:8967-75; PMID:20086009; http://dx.doi.org/10.1074/jbc.M109.058107
- Abalos GC, Cruite JT, Bellon A, Hemmers S, Akagi J, Mastrianni JA, Williamson RA, Solforosi L. Identifying key components of the PrPC-PrPSc replicative interface. J Biol Chem 2008; 283:34021-8; PMID:18826953; http://dx.doi.org/10.1074/jbc.M804475200
- Norstrom EM, Mastrianni JA. The AGAAAAGA palindrome in PrP is required to generate a productive PrPSc-PrPC complex that leads to prion propagation. J Biol Chem 2005; 280:27236-43; PMID:15917252; http://dx.doi.org/10.1074/jbc.M413441200
- Taguchi Y, Schätzl HM. Identifying critical sites of PrPc -PrPSc interaction in prion-infected cells by dominant-negative inhibition. Prion 2013; 7:452-456; PMID:24401595; http://dx.doi.org/10.4161/pri.27500
- Hara H, Okemoto-Nakamura Y, Shinkai-Ouchi F, Hanada K, Yamakawa Y, Hagiwara K. Mouse prion protein (PrP) segment 100 to 104 regulates conversion of PrP(C) to PrP(Sc) in prion-infected neuroblastoma cells. J Virol 2012; 86:5626-36; PMID:22398286; http://dx.doi.org/10.1128/JVI.06606-11
- Chen J, Thirumalai D. Helices 2 and 3 are the initiation sites in the PrP C→ PrP SC transition. Biochemistry 2012; 52:310-319; PMID:23256626; http://dx.doi.org/10.1021/bi3005472
- Norstrom EM, Mastrianni JA. The charge structure of helix 1 in the prion protein regulates conversion to pathogenic PrPSc. J Virol 2006; 80:8521-9; PMID:16912302; http://dx.doi.org/10.1128/JVI.00366-06
- Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med 1998; 4:1157-65; PMID:9771749; http://dx.doi.org/10.1038/2654
- Collinge J, Clarke A. A general model of prion strains and their pathogenicity. Science 2007; 318:930-6; PMID:17991853; http://dx.doi.org/10.1126/science.1138718
- Solforosi L, Milani M, Mancini N, Clementi M, Burioni R. A closer look at prion strains: Characterization and important implications. Prion 2013; 7:99-108; PMID:23357828; http://dx.doi.org/10.4161/pri.23490
- Béringue V, Vilotte J, Laude H. Prion agent diversity and species barrier. Vet Res 2008; 39:47; PMID:18519020; http://dx.doi.org/10.1051/vetres:2008024
- Baron GS, Hughson AG, Raymond GJ, Offerdahl DK, Barton KA, Raymond LD, Dorward DW, Caughey B. Effect of Glycans and GPI Anchor on Strain Dependent Conformations of Scrapie Prion Protein: Improved Purifications and IR Spectra. Biochemistry 2011; 50:4479-90; PMID:21539311; http://dx.doi.org/10.1021/bi2003907
- Atarashi R, Sim VL, Nishida N, Caughey B, Katamine S. Prion strain-dependent differences in conversion of mutant prion proteins in cell culture. J Virol 2006; 80:7854-62; PMID:16873242; http://dx.doi.org/10.1128/JVI.00424-06
- Morel N, Andréoletti O. Absolute and relative quantification of sheep brain prion protein (PrP) allelic variants by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 2007; 21:4093-100; PMID:18008391; http://dx.doi.org/10.1002/rcm.3317
- Jacobs JG, Bossers A , Rezaei H, van Keulen LJM, McCutcheon S, Sklaviadis T, Lantier I, Berthon P, Lantier F, van Zijderveld FG, et al. Proteinase K-Resistant Material in ARR/VRQ Sheep Brain Affected with Classical Scrapie Is Composed Mainly of VRQ Prion Protein. J Virol 2011; 85:12537-46; PMID:21917981; http://dx.doi.org/10.1128/JVI.00448-11
- Khan M, Sweeting B. Prion disease susceptibility is affected by β-structure folding propensity and local side-chain interactions in PrP. Proc Natl Acad Sci U S A 2010; 107:19808-13; PMID:21041683; http://dx.doi.org/10.1073/pnas.1005267107
- Kirby L, Agarwal S, Graham JF, Goldmann W, Gill AC. Inverse correlation of thermal lability and conversion efficiency for five prion protein polymorphic variants. Biochemistry 2010; 49:1448-59; PMID:20085368; http://dx.doi.org/10.1021/bi901855z
- Bujdoso R, Burke DF, Thackray AM. Structural differences between allelic variants of the ovine prion protein revealed by molecular dynamics simulations. Proteins 2005; 61:840-9; PMID:16252284; http://dx.doi.org/10.1002/prot.20755
- Gill AC, Agarwal S, Pinheiro TJT, Graham JF. Structural requirements for efficient prion protein conversion: cofactors may promote a conversion-competent structure for PrP(C). Prion 2010; 4:235-42; PMID:20864807; http://dx.doi.org/10.4161/pri.4.4.13394
- Pastore A, Zagari A. A structural overview of the vertebrate prion proteins. Prion 2007; 1:185-97; PMID:19164911; http://dx.doi.org/10.4161/pri.1.3.5281
- Chakroun N, Prigent S, Dreiss CA, Noinville S, Chapuis C, Fraternali F, Rezaei H. The oligomerization properties of prion protein are restricted to the H2H3 domain. FASEB J 2010; 24:3222-31; PMID:20410442; http://dx.doi.org/10.1096/fj.09-153924
- Rezaei H, Choiset Y, Eghiaian F, Treguer E, Mentre P, Debey P, Grosclaude J, Haertle T. Amyloidogenic Unfolding Intermediates Differentiate Sheep Prion Protein Variants. J Mol Biol 2002; 322:799-814; PMID:12270715; http://dx.doi.org/10.1016/S0022-2836(02)00856-2
- Kanata E, Humphreys-Panagiotidis C, Giadinis ND, Papaioannou N, Arsenakis M, Sklaviadis T. Perspectives of a scrapie resistance breeding scheme targeting Q211, S146 and K222 caprine PRNP alleles in Greek goats. Vet Res 2014; 45:43; PMID:24717012; http://dx.doi.org/10.1186/1297-9716-45-43
- Salta E, Kanata E, Ouzounis C, Gilch S, Schätzl H, Sklaviadis T. Assessing Proteinase K Resistance of Fish Prion Proteins in a Scrapie-Infected Mouse Neuroblastoma Cell Line. Viruses 2014; 6:4398-421; PMID:25402173; http://dx.doi.org/10.3390/v6114398
- Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J Virol 2000; 74:320-5; PMID:10590120; http://dx.doi.org/10.1128/JVI.74.1.320-325.2000
- Vorberg I, Buschmann A, Harmeyer S, Saalmüller A, Pfaff E, Groschup MH. A novel epitope for the specific detection of exogenous prion proteins in transgenic mice and transfected murine cell lines. Virology 1999; 255:26-31; PMID:10049818; http://dx.doi.org/10.1006/viro.1998.9561
