ABSTRACT
Chronic wasting disease (CWD) is a prion disease found in deer, elk and moose in North America and since recently, wild reindeer in Norway. Caribou are at-risk to encounter CWD in areas such as Alberta, Canada, where the disease spreads toward caribou habitats. CWD susceptibility is modulated by species-specific polymorphisms in the prion protein gene (Prnp). We sequenced Prnp of woodland caribou from 9 Albertan populations. In one population (Chinchaga) a significantly higher frequency of the 138N allele linked to reduced CWD susceptibility was observed. These data are relevant for developing CWD management strategies including conservation of threatened caribou populations.
INTRODUCTION
Chronic wasting disease (CWD) is a prion disease which utterly occurs in wild and captive cervids, including mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus), Rocky Mountain elk (Cervus canadensis) and Shira's moose (Alces alces shirasi).Citation1,2 Just recently, the first CWD case in free-ranging reindeer (Rangifer tarandus tarandus) was diagnosed in southern Norway.Citation3 CWD was first recognized in the 1960s in Colorado and has been burgeoning across North America, in at least 23 states in the US and 2 provinces of Canada.Citation1 Prion diseases are a group of fatal neurodegenerative disorders that affect humans and animals and are caused by the conformational conversion of the cellular prion protein (PrPC) into the pathogenic isoform PrPSc. PrPSc is the sole component of prions and accumulates in the brains of infected individuals.Citation4,5 However, in CWD prions are not retained within the central nervous system but are shed into a wide range of peripheral tissues, body fluids and excreta.Citation6-10 In addition, environmental prions have been experimentally demonstrated to retain infectivity for years which contributes to horizontal transmission among cervids.Citation11 Like in prion diseases of other species, species-specific amino acid polymorphisms in the cervid PrP are associated with susceptibility, incubation time, and pathology of CWD. In most cases, the occurrence of at least one non-wild type allele of Prnp is correlated with a reduced susceptibility to CWD infection and prolonged incubation times.Citation12-14 The caribou prion protein primary structure is identical to the mule and white-tailed deer PrP, except for the positions of species-specific polymorphisms.
Caribou are threatened and declining in Alberta,Citation15 whereas mule and white-tailed deer populations are stable or expanding.Citation16 Considering the geographic spread of CWD and the proof of susceptibility of reindeer to CWD,Citation3,17 it is likely that eventually caribou encounter CWD, which will be detrimental for the already declining populations.
Four polymorphisms were discovered in 3 Alaskan caribou (Rangifer tarandus granti) herds at amino acid positions 2 (valine [V] or methionine [M]), 129 (glycine [G] or serine [S]), 138 (serine [S] or asparagine [N]) and 169 (valine [V] to methionine [M]), respectively [18]. Given the relevance of Prnp polymorphisms for CWD transmission and pathogenesis, it is important to determine the distribution of PrP polymorphisms among caribou populations that are likely to overlap with CWD positive herds in the future. Since CWD has been now well-established in Alberta, Canada, we analyzed the Prnp coding sequence of animals from 9 Albertan woodland caribou (Rangifer tarandus caribou) populations, including both the mountain and the boreal ecotypes. We did not identify novel polymorphic codons; however, we found a significantly increased frequency of the 138N allele in the Chinchaga population (boreal ecotype). These results allow predictions on the disease susceptibility of caribou populations on a genetic basis, and add important information regarding conservation management of this declining species.
RESULTS
Of the 5 polymorphic sites identified previously in Alaska caribou Citation18 the single base change at base 438 is a synonymous substitution at codon 146 (N to N) and codon 2 locates within the N-terminal signal peptide (amino acid 1 to 22) which is cleaved upon post-translational modification. Therefore, we focused on variations in the sequence encoding mature PrP (without N- and C-terminal signal peptides), specifically at codon 129, 138 and 169.
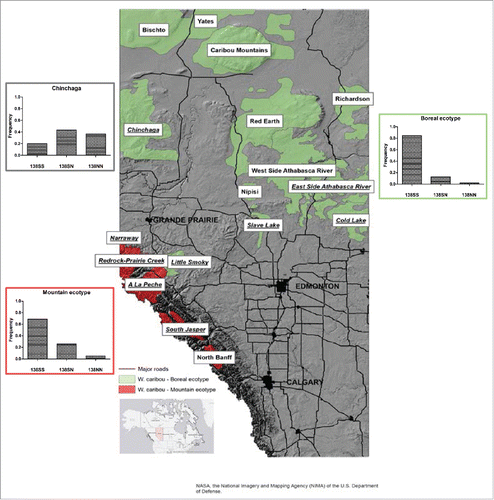
There are 16 populations of woodland caribou (Rangifer tarandus caribou) currently existing in Alberta, Canada, which can be further classified into 2 ecotypes based upon geography and migration patterns: the boreal ecotype (with individuals typically sedentary) and the mountain ecotype (with some individuals sedentary and other migratory).Citation15 We analyzed a total of 118 samples collected from 9 woodland caribou populations. No polymorphism was detected at codon 169 in any of the samples; thus, the PrP genotypes at codon 129 and codon 138 were categorized by individual populations (). A significant difference in allele frequencies at codon 138 between populations was detected by Fisher's Exact test (p = 1.056e-08). Interestingly, a stastically significant difference was observed when frequencies of the 138N and 138S alleles, respectively, were compared using Fisher's Exact test between the Chinchaga population and all other populations, grouped into the boreal or mountain ecotype (). If the caribou populations excluding Chinchaga population were classified into either mountain or boreal ecotypes, in populations of the mountain ecotype (Jasper, Narraway, Redrock Prairie Creek and A La Peche), the allele frequencies of 129G and 138S are 1.0 and 0.821, respectively; in caribou populations of the boreal ecotype (Cold Lake, East-side Athabasca River, Slave Lake and Little Smoky), the allele frequencies of 129G and 138S are 0.989 and 0.913, respectively. The Chinchaga population is also classified as boreal ecotype. Whereas the 129G frequency is with 0.967 similar to the other populations, the frequency of the 138S allele is 0.417, which is significantly lower than in the other populations of either ecotype. Furthermore, we observed that as many as 36.7% of the animals of the Chinchaga population are homozygous for the 138N allele (). In summary, we have identified a woodland caribou population (Chinchaga) with an uncommonly high frequency of the 138N Prnp allele.
TABLE 1. Allele frequencies in woodland caribou populations.
FIGURE 1. Geographic distribution and Prnp allele frequencies at codon 138 of woodland caribou populations. The map indicates the distribution of all woodland caribou populations in Alberta, those classified as boreal ecotype are depicted in green, the populations of the mountain ecotype in red. Populations included in our study are underlined. Bar graphs indicate the frequency of 138SS, 138SN and 138NN genotypes of all analyzed population of the mountain and boreal ecotype excluding Chinchaga, which is shown in a separate bar graph.
DISCUSSION
To date, 16 polymorphic sites in the Prnp gene have been discovered among the cervid family.Citation21 Amino acid 138 (S/N) has been found to be polymorphic in the Prnp gene of caribou (Rangifer tarandus spp.),Citation17 18 and fallow deer (Dama dama) Citation22 but not in white-tailed or mule deer which only carry the allele encoding serine. In mule and white-tailed deer a Prnp pseudogene (Prnpψ) was identified which exclusively encoded N at position 138.Citation12 However, Prnpψ was not found in old world deer such as reindeer, elk, fallow deer Citation12 or caribou,Citation18 arguing against the possibility that the higher frequency of the 138N allele found in caribou of the Chinchaga population is due to Prnpψ.
Asparagine at codon 138 has been associated with at least partial protection against CWD infection as in reindeer experimentally inoculated with CWD prions animals that harbored at least one 138N allele did not develop prion disease until termination of the experiment, suggesting genetic protection.Citation17 Similarly, fallow deer (Dama dama) with the 138N PrP genotype did not develop CWD when they were exposed to infected mule deer and environmental CWD prions.Citation22 Along this line, in vitro conversion of cervid PrP-138N into a protease-resistant form reminiscent of PrPSc was less efficient than the conversion of PrP-138S.Citation23
We demonstrate that caribou of the Chinchaga population exhibit significantly higher frequencies of the 138SN and 138NN Prnp genotypes in comparison to the other 4 boreal populations, the mountain populations of woodland caribou (Rangifer tarandus caribou), and 3 herds of Alaskan caribou (Rangifer tarandus grantii;Citation18). The Chinchaga is a large natural foothills region in north-western Alberta, with a large portion of this area disturbed by industrial development. Most likely due to human activities, the Chinchaga caribou population is not self-sustaining; the estimated population size is about 250 animals.Citation15 It is not clear why this high frequency of the 138N-encoding Prnp allele is only confined to the Chinchaga population but not to other boreal woodland caribou populations. Gene flow from other caribou populations can be one possible explanation, and an extensive microsatellite assignment test of 808 individuals revealed that the genetic characteristics of 3 diverse caribou subspecies, including Grant's caribou (Rangifer tarandus granti), barren-ground caribou (Rangifer tarandus groenlandicus) and woodland caribou (Rangifer tarandus caribou) are equally mixed in the Chinchaga population.Citation24 This suggests genetic admixture Citation25 in this herd. This population is provincially classified as woodland caribou (Rangifer tarandus caribou); however, from a genetic perspective it cannot be simply fitted in any subspecies.
The Chinchaga region has been included in the Alberta Woodland Caribou Recovery Plan for planning and implementing recovery actions. Our data suggest a reduced genetic susceptibility of Chinchaga caribou to CWD, therefore indicating lower risk of disease when compared with other populations that are also declining. Overall, our results also highlight the importance of investigating Prnp allele frequencies to inform authorities on potential CWD-resistant caribou herds. Currently there is no efficient approach available to eliminate environmental contamination and spread of CWD, however, the possibility of preserving CWD-protective gene lines needs to be considered.
MATERIALS AND METHODS
Blood Samples
We used a total number of 118 anticoagulated whole blood samples of 9 Albertan caribou populations which were collected at different time points during caribou capture conducted between 2002–2008 and were stored at -80°C. The protocol for sample collection was approved by the University of Calgary Life & Environmental Sciences Animal Care Committee (LESACC; Study Id: AC16–0195). Of these 9 populations, Chinchaga (n = 30), Cold Lake (n = 10), East-side Athabasca River (n = 15), Slave Lake (n = 10) and Little Smokey (n = 11) belong to the boreal ecotype; the other 4 populations: Japer (n = 10), Narraway (n = 13), Redrock Prairie Creek (n = 10) and A La Peche (n = 9) are classified as the mountain ecotype. Numbers of samples were chosen to represent about 10% of individuals of each population.Citation19,20
Genomic DNA Extraction, PCR and Sequencing
Genomic DNA extraction from blood was done by using a commercial kit (DNeasy Blood and Tissue Kit, Qiagen) according to the manufacturer's recommendation. One hundred ng of genomic DNA were used as a template in PCR to amplify the sequence encoding the mature PrP in the third exon of the Prnp gene. The PCR primers were designed to complement sequences of the N- and C-terminal signal peptides (amino acid 1 to 24 and 25 to 256) of the caribou (Rangifer tarandus ssp.) prion protein gene. PCR cocktail components were as follows: 5 μl of 10X Pfu reaction buffer (Agilent), 2 μl of 5mM dNTP, 2 μl of 10 μM forward primer (5′-CCT AGT TCT CTT TGT GGC CAT GTG-3′), 2 μl of 10 μM reverse primer (5′-TGA GGA AAG AGA TGA GGA GGA TCA C-3′), and 0.5 μl of Pfu polymerase. The PCR reactions were performed by following an initial denaturation at 94°C for 4 minutes, and 39 cycles of denaturation (94°C, 30 seconds), annealing (62°C, 30 seconds), extension (72°C, 1 minute) and a final extension at 72°C for 10 minutes. PCR products were analyzed on agarose gel, purified and sequenced using the forward and reverse primers used for PCR amplification (University of Calgary CoreDNA service or Eton Bioscience, San Diego). In addition, several PCR products which showed no polymorphisms at codon 129, 138, 146 and 169 were chosen for cloning (Zero Blunt™ TOPO™ PCR Cloning Kit; Invitrogen). Inserts of 10 single clones per PCR product were sequenced to confirm the absence of polymorphic residues.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
FUNDING
This work was supported by a research grant from the Alberta Conservation Association and by a Genome Canada Large Scale Applied Research Program grant to SG and a NSERC Discovery Grant to MM. SG is supported by the Canada Research Chair program. We also thank Alberta Environment and Parks, Government of Alberta and Parks Canada for continued collaboration. We are grateful to Grace Kwong for help with biostatistics.
REFERENCES
- Gilch S, Chitoor N, Taguchi Y, Stuart M, Jewell JE, Schatzl HM. Chronic wasting disease. Top Curr Chem 2011; 305:51-77; PMID:21598099
- Williams ES, Young S. Neuropathology of chronic wasting disease of mule deer (Odocoileus hemionus) and elk (Cervus elaphus nelsoni). Vet Pathol 1993; 30:36-45; PMID:8442326; http://dx.doi.org/10.1177/030098589303000105
- Benestad SL, Mitchell G, Simmons M, Ytrehus B, Vikoren T. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res 2016; 47:88; PMID:27641251; http://dx.doi.org/10.1186/s13567-016-0375-4
- Prusiner SB. Prions. Proc Natl Acad Sci U S A 1998; 95:13363-83
- Prusiner SB. The prion diseases. Brain Pathol 1998; 8:499-513; PMID:9669700; http://dx.doi.org/10.1111/j.1750-3639.1998.tb00171.x
- John TR, Schatzl HM, Gilch S. Early detection of chronic wasting disease prions in urine of pre-symptomatic deer by real-time quaking-induced conversion assay. Prion 2013; 7:253-8; PMID:23764839; http://dx.doi.org/10.4161/pri.24430
- Angers RC, Browning SR, Seward TS, Sigurdson CJ, Miller MW, Hoover EA et al. Prions in skeletal muscles of deer with chronic wasting disease. Science 2006; 311:1117; PMID:16439622; http://dx.doi.org/10.1126/science.1122864
- Mathiason CK, Powers JG, Dahmes SJ, Osborn DA, Miller KV, Warren RJ et al. Infectious prions in the saliva and blood of deer with chronic wasting disease. Science 2006; 314:133-6; PMID:17023660; http://dx.doi.org/10.1126/science.1132661
- Cheng YC, Hannaoui S, John TR, Dudas S, Czub S, Gilch S. Early and non-invasive detection of chronic wasting disease prions in elk feces by real-time quaking induced conversion. PLoS One 2016; 11:e0166187
- Tamguney G, Miller MW, Wolfe LL, Sirochman TM, Glidden DV, Palmer C et al. Asymptomatic deer excrete infectious prions in faeces. Nature 2009; 461:529-32; PMID:19741608; http://dx.doi.org/10.1038/nature08289
- Saunders SE, Bartelt-Hunt SL, Bartz JC. Occurrence, transmission, and zoonotic potential of chronic wasting disease. Emerg Infect Dis 2012; 18:369-76; PMID:22377159; http://dx.doi.org/10.3201/eid1803.110685
- O'Rourke KI, Spraker TR, Hamburg LK, Besser TE, Brayton KA, Knowles DP. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol 2004; 85:1339-46; PMID:15105552; http://dx.doi.org/10.1099/vir.0.79785-0
- Jewell JE, Conner MM, Wolfe LL, Miller MW, Williams ES. Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J Gen Virol 2005; 86:2127-34; PMID:16033959; http://dx.doi.org/10.1099/vir.0.81077-0
- Schatzl HM, Wopfner F, Gilch S, von Brunn A, Jager G. Is codon 129 of prion protein polymorphic in human beings but not in animals? Lancet 1997; 349:1603-4; PMID:9174569; http://dx.doi.org/10.1016/S0140-6736(05)61632-7
- Hervieux D, Hebblewhite M, DeCesare NJ, Russell M, Smith K, Robertson S, Boutin S. Widespread declines in woodland caribou (Rangifer tarandus caribou) continue in Alberta. Canadian J Zool 2013; 91:872-82; http://dx.doi.org/10.1139/cjz-2013-0123
- Dawe KL, Bayne EM, Boutin S. Influence of climate and human land use on the distribution of white-tailed deer (Odocoileus virginianus) in the western boreal forest. Canadian J Zool 2014; 92:353-63; http://dx.doi.org/10.1139/cjz-2013-0262
- Mitchell GB, Sigurdson CJ, O'Rourke KI, Algire J, Harrington NP, Walther I et al. Experimental oral transmission of chronic wasting disease to reindeer (Rangifer tarandus tarandus). PLoS One 2012; 7:e39055; PMID:22723928; http://dx.doi.org/10.1371/journal.pone.0039055
- Happ GM, Huson HJ, Beckmen KB, Kennedy LJ. Prion protein genes in caribou from Alaska. J Wildl Dis 2007; 43:224-8; PMID:17495306; http://dx.doi.org/10.7589/0090-3558-43.2.224
- Environment Canada. Recovery Strategy for the Woodland Caribou, Boreal population (Rangifer tarandus caribou) in Canada Species at Risk Act Recovery Strategy Series. Environment Canada 2011; Ottawa. vi + 55 pp.
- Environment Canada. Recovery Strategy for the Woodland Caribou, Southern Mountain population (Rangifer tarandus caribou) in Canada. Species at Risk Act Recovery Strategy Series. Environment Canada 2014; Ottawa. viii + 103 pp.
- Robinson SJ, Samuel MD, O'Rourke KI, Johnson CJ. The role of genetics in chronic wasting disease of North American cervids. Prion 2012; 6:153-62; PMID:22460693; http://dx.doi.org/10.4161/pri.19640
- Rhyan JC, Miller MW, Spraker TR, McCollum M, Nol P, Wolfe LL et al. Failure of fallow deer (Dama dama) to develop chronic wasting disease when exposed to a contaminated environment and infected mule deer (Odocoileus hemionus). J Wildl Dis 2011; 47:739-44; PMID:21719844; http://dx.doi.org/10.7589/0090-3558-47.3.739
- Raymond GJ, Bossers A, Raymond LD, O'Rourke KI, McHolland LE, Bryant PK, III et al. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J 2000; 19:4425-30; PMID:10970836; http://dx.doi.org/10.1093/emboj/19.17.4425
- Weckworth BV, Musiani M, McDevitt AD, Hebblewhite M, Mariani S. Reconstruction of caribou evolutionary history in Western North America and its implications for conservation. Mol Ecol 2012; 21:3610-24; PMID:22612518; http://dx.doi.org/10.1111/j.1365-294X.2012.05621.x
- Pritchard JK, Falush D, Stephens M. Inference of population structure in recently admixed populations. Am J Hum Genetet 2002; 71:177
