ABSTRACT
Extracellular vesicles (EVs) are actively secreted, membrane-bound communication vehicles that exchange biomolecules between cells. EVs also serve as dissemination vehicles for pathogens, including prions, proteinaceous infectious agents that cause transmissible spongiform encephalopathies (TSEs) in mammals. Increasing evidence accumulates that diverse protein aggregates associated with common neurodegenerative diseases are packaged into EVs as well. Vesicle-mediated intercellular transmission of protein aggregates can induce aggregation of homotypic proteins in acceptor cells and might thereby contribute to disease progression. Our knowledge of how protein aggregates are sorted into EVs and how these vesicles adhere to and fuse with target cells is limited. Here we review how TSE prions exploit EVs for intercellular transmission and compare this to the transmission behavior of self-templating cytosolic protein aggregates derived from the yeast prion domain Sup 35 NM. Artificial NM prions are non-toxic to mammalian cell cultures and do not cause loss-of-function phenotypes. Importantly, NM particles are also secreted in association with exosomes that horizontally transmit the prion phenotype to naive bystander cells, a process that can be monitored with high accuracy by automated high throughput confocal microscopy. The high abundance of mammalian proteins with amino acid stretches compositionally similar to yeast prion domains makes the NM cell model an attractive model to study self-templating and dissemination properties of proteins with prion-like domains in the mammalian context.
Many, if not all cells, release a repertoire of vesicles in the extracellular milieu. Secreted vesicles shed from the plasma membrane or produced by the endosomal system are collectively termed extracellular vesicles (EVs).Citation1 EVs are important mediators of intercellular communication and transfer proteins, RNAs and other cellular components between cells, thereby modulating diverse cellular processes in acceptor cells. As biomolecules incorporated into exosomes reflect the physiological state of their donor cells, they are also intensely surveyed as biomarker sources. Interestingly, pathogens such as viruses exploit exosomes for intercellular dissemination.Citation2 EVs have received further attention for their proposed role as transfer vehicles for pathologic proteins in neurodegenerative diseases, including prions, SOD1, TDP-43, Aβ peptides, α-synuclein or Tau.Citation3,4
Prions - Proteinaceous Infectious Particles
The first pathogenic protein aggregates identified in exosomes were prions, self-templating protein particles that cause devastating neurodegenerative diseases in mammals. TSEs in mammals occur mostly sporadic, but can also be of genetic or iatrogenic origin and can be infectious. Scrapie in sheep and goats and chronic wasting disease in deer, elk and moose constitute prion diseases that naturally transmit horizontally. The extreme resistance to inactivation procedures that destroy nucleic acid and the discovery that the host-encoded prion protein PrP was the main component of the infectious particle led to the proposal that TSE agents are solely protein-based and devoid of coding nucleic acid.Citation5 The cellular PrP (PrPC) is a highly glycosylated, glycosylphosphatidyl-inositol (GPI)-anchored protein enriched in lipid raft microdomains on neuronal and non-neuronal cell membranes. In a seeded polymerization reaction, PrPSc serves as a template that induces the structural rearrangement of PrPC monomers into β-sheet rich prion polymers.Citation6 Accumulation of PrPSc in the central nervous system is associated with astrogliosis and spongiform degeneration. Remarkably, PrPC cannot only take on one but a variety of self-templating conformations that are associated with different pathologies in their host. Substantial biophysical evidence supports the hypothesis that these prion strain properties are enciphered within the 3-dimensional fold of the prion polymer.Citation7
While initially coined for TSE agents,Citation5 the term “prion” was later adopted to describe proteinaceous particles that confer non-Mendelian traits in yeast.Citation8 Prions in lower eukaryotes are insoluble, self-perpetuating amyloid-like polymers that act as epigenetic elements of inheritance.Citation9 Unlike mammalian prions attached at the plasma membrane by a GPI-anchor, yeast prions are predominately cytoplasmic. Depending on the genetic makeup of the host and environmental factors, yeast prions can either be detrimental, benign or advantageous to their host.Citation10,11 De novo yeast prion induction and replication involve rare spontaneous nucleation events followed by growth and fragmentation of highly ordered protein fibrils, a process similar to the proposed propagation mechanism of mammalian prions.Citation12 The nucleation phase can be bypassed by exposure of yeast to in vitro formed prion aggregatesCitation13 or cytosolic “propagons” extracted from prion-containing strains.Citation14 Yeast prion proteins share little sequence homology with PrP. Instead, prion activity is governed by so-called prion domains, disordered regions often enriched in uncharged residues such as glutamine, asparagine and glycine.Citation15
In 1982, Prusiner defined prions as “small proteinaceous infectious particles which are resistant to inactivation by most procedures that modify nucleic acid.”Citation5 This original definition also holds true for protein aggregates in lower eukaryotes. We use the term “prion” to describe a biological process by which biologic information is enciphered, amplified and disseminated through protein conformation. To avoid any confusion in terminology, we will refer to prions causing TSEs as TSE prions, while we will term self-templating protein aggregates identified in yeast as “yeast prions.” Here, we specifically focus on the intercellular dissemination strategies of TSE prions and compare these to the surprising self-propagating and dissemination properties of a yeast prion domain in mammalian cells. Remarkably, prion-like domains (PrLDs) compositionally similar to annotated yeast prion domains are present in 1% of mammalian proteins, including proteins forming pathogenic aggregates in Amyotrophic Lateral Sclerosis (ALS) or Frontotemporal Dementia (FTD).Citation16 Prions derived from the yeast prion domain of Sup35 are not homologous to mammalian proteins and thus allow us to study protein aggregation and dissemination in the absence of a loss-of-function phenotype. As such, the yeast prion domain Sup35 constitutes an excellent tool to model general aggregation and dissemination propensities of proteins with related domains.
Extracellular Vesicles Are Involved in Intercellular Communication in Mammals
EVs are heterogeneous and differ in their biogenesis. Most vesicles that bud off the cell membrane (referred to as microvesicles) fall in the range of 200–500 nm, but smaller and larger membrane-bound particles have been described. Although EVs are discriminated by marker proteins, size and density, substantial overlap in all 3 parameters has been observed.Citation17,18 Exosomes are EVs in the range of 40–100 nm, which arise through inward budding into specialized late endosomal structures, referred to as multivesicular bodies (MVBs). Fusion of MVBs with the plasma membrane liberates the intraluminal bodies (ILVs) as exosomes into the extracellular space. MVB are not only intermediates of exosome release but also subject to autophagosomal degradation. Although the selection mechanisms that define the fate of cargo proteins remain elusive, accumulating evidence suggests that cells secrete subpopulations of exosomes that differ in cargo composition, size, subcellular distribution and biogenesis.Citation19 Recent research has highlighted some mechanisms that sort membrane associated proteins and cytosolic proteins into ILVs. These processes can act independently or collaboratively. Protein sorting into exosomes involves “endosomal sorting complex required for transport” (ESCRT)- dependent and -independent processes. The ESCRT complex and additional regulatory proteins support sorting of ubiquitinated cargo into MVBs.Citation20 Several other posttranslational cargo modifications have been reported, such as sumyolation, phosphorylation or specific carbohydrate signatures.Citation21 There is direct evidence showing that the number of N-linked glycans is a determinant for exosomal cargo sorting.Citation22 Membrane microdomains enriched in ceramides were also shown to be involved in cargo sorting.Citation23,24 Lipid components of raft-like domains, including cholesterol, ceramide, sphingomyelin, glycosphingolipids and phosphatidylcholine, are highly enriched in exosomes. The raft-like domain not only provides the platform for the ILVs budding, but is directly involved in cargo sorting. Specific lipids and integral membrane proteins such as tetraspanin interact with cargo.Citation19,25-27 Furthermore, aggregation of proteins or lipids might serve as a general sorting signal for exosomes, as antibody-mediated aggregation of cell surface receptors induces their sorting into exosomes.Citation28 Along these lines, higher-order oligomerization of plasma membrane associated retrovirus Gag protein is sufficient to target it to exosomes for hijacking exosome biogenesis for virus production.Citation29
Key to the function of EVs is attachment and membrane fusion to deliver biologically active cargo to the target cell. Importantly, exosomes selectively adhere to specific cells, a tropism defined by ligand-receptor interactions. While some receptor and ligand pairs mediating this interaction have been identified, most have not been explored so far. Specific integrins and cell adhesion molecules abundant on EV surfaces can facilitate attachment onto target cells and mediate host cell tropism.Citation30 Heparan sulfate proteoglycans,Citation31 phosphatidylserin receptorsCitation32 and lectinsCitation33 can serve as EV receptors. EVs can fuse directly with the plasma membrane and release the vesicle content into the cytoplasm.Citation34 Alternatively, EVs can be taken up by endocytosis or macropinocytosis.Citation35 Clathrin-, caveolin/lipid raft- dependent endocytosis or independent entry routes have been described for EV entry.Citation36,37 The size limit of cargo that can be internalized by certain pathways might influence the preferred uptake route for EVs.Citation38 It is possible that EVs use more than one entry route or use alternative pathways. One alternative route requires fusogenic proteins that mediate docking and direct fusion with host membranes, which has been shown for enveloped viruses and exosomes secreted by placenta.Citation2 Moreover, certain exosomal tetraspanin compositions can also mediate EV-host cell adhesion and membrane fusion.Citation39 How this fusion process is regulated for other EVs is so far unclear. Endocytosed EVs are either delivered to the lysosome or fuse with the limiting membrane of the late endosome to release their cargo into the cytosol.
Exosomes as Vehicles for Intercellular Dissemination of Transmissible Spongiform Encephalopathy Agents
TSE infection usually occurs through the intestinal route.Citation6 The spreading of prions from the gut through the lymphoreticular system and peripheral nerves to the brain involves intercellular dissemination of infectious entities.Citation40 How exactly TSE prions spread from cell to cell in vivo is only poorly understood. Routes for prion transmission have been mainly studied in cell culture. The formation of the infectious PrP isoform occurs after PrPC has reached the plasma membrane, either directly on the cell surface or within recycling endosomes, endolysosomal vesicles and / or MVBs.Citation6,41,42 Different dissemination strategies can be used by TSE prions, including direct cell contact,Citation43,44 for example via tunneling nanotubes,Citation45 or secretion of prions in EVs, such as microvesicles and exosomes.Citation46-50 Cell culture derived exosomes containing PrPSc are infectious to permissive cell lines and produce clinical disease in mice.Citation47,49 The observed differences in dissemination strategies might be related to prion strain differences or infected cell types.Citation43-45,48,49,51,52 As EVs extracted from body fluids also contain prion activity, they are likely to contribute to prion dissemination in vivo.Citation53
Exosomal sorting is not restricted to PrPSc, as PrPC is a normal constituent of intraluminal bodies in MVBs,Citation47,54 and is found in exosomal preparations of immortalized cell lines and primary cells of diverse origins.Citation42,48,55-60 Exosomes and microvesicles isolated from body fluids are also decorated with PrPC, suggesting that PrPC is a normal constituent of EVs.Citation61,62 PrPC expression has been shown to stimulate exosome secretion in primary astrocytes and fibroblasts.Citation63 As both PrPC and PrPSc are partitioned into intraluminal vesicles destined for secretion, protein polymerization is not a required trigger for secretion. Interestingly, the PrPSc glycosylation pattern often differs between cell extract and exosomes, arguing that specific subpopulations of PrPSc are selectively sorted into exosomes.Citation64 The contribution of different sorting pathways is less clear and might be cell type or strain dependent.
The presence of PrPC and PrPSc in lipid raft microdomains suggests that PrP isoforms are sorted to exosomes in association with lipid rafts.Citation26,65 Both ceramide dependent and Tsg101-ESCRT mediated pathways contribute to exosomal prion secretion in 2 cellular TSE models.Citation42,52 While ESCRT Tsg101 subunit silencing directly affected exosome and PrPSc secretion, a compound inhibiting the ceramide-dependent exosome pathway only marginally affected exosome secretion but led to selective exclusion of PrPSc and infectivity from exosomes derived from a neuroglial cell line.Citation42 This is in contrast to a study using a murine hypothalamic cell line where chemical impairment of the ceramide-dependent pathway reduced exosomes and exosome-associated PrPC and PrPSc 52.
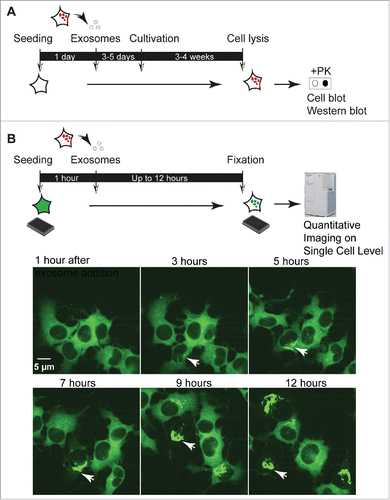
Little is known if TSE prion-containing exosomes derived from different cell types are equally infectious to different recipient cells. Generally, very few cell lines are permissive to TSE prions, and prion strains exhibit selective infectivity for specific cell lines and even subclones thereof.Citation6,66 However, when tested in permissive cell cultures, exosomes isolated from different persistently infected cell lines proved infectious to recipient cells of different origin.Citation42,49 TSE prion-containing exosomes might thus be taken up by recipient cells unspecifically or via ligand-receptor pairs functional in the tested donor-recipient cell combinations. A problem in defining cellular pathways that mediate prion internalization and infection is that TSE prion infection takes days to weeks to be detectable in cell culture. The currently used assays rely on detection of newly formed PrPSc weeks post infection by cell colony blot or western blot.Citation42,47-49,51 These assays do not measure single cell events and cannot discriminate between early events following internalization and subsequent secondary amplification and spreading events (). Cellular uptake of PrPSc can also be visualized by confocal microscopy, but also non-permissive cells internalize PrPSc.Citation67 Thus, these studies do not allow drawing conclusions on the internalization pathways that lead to productive infections.
FIGURE 1. Cell culture assays to study prion infection by exosomes. (A) Cell culture assays to study exosome-mediated TSE prion infection. Published exosome-mediated TSE infection assays are time consuming und rely on the detection of newly formed, proteinase K (PK) resistant PrPSc. Naive cells permissive to infection with the respective TSE prion strain are exposed to exosomal preparations isolated from prion-infected cells for 4–5 days, followed by several weeks of culture. Read-out is PK-resistant PrPSc detected by cell blot or western blot.Citation42,47-49,51 (B) Quantitative imaging of exosome-mediated NM aggregate induction. Recipient NM-GFPsol cells are seeded on a 384 well plate for 1 hour. Exosomes isolated from conditioned medium of donor cells are added to the wells. Life or fixed cells are subjected to automated high throughput confocal microscopy. Read-out is induction of NM-GFP aggregates in recipient cells. Life imaging analysis demonstrates the appearance of NM-GFP aggregates as soon as 3 hours post exosome addition. The arrowhead marks cells with exosome-induced NM-GFP aggregates. The assay can also reveal bidirectional inheritance of NM aggregates by daughter cells, a characteristic of TSE prions replicating in cellular models.Citation6
A Yeast Prion Domain as a Model Protein to Study Dissemination Pathways of Cytosolic, Self-Templating Protein Aggregates in Mammalian Cells
Yeast prions have been studied extensively in the past to unravel basic principles of conformational templating. The translation termination factor Sup35 of S. cerevisiae is the best-studied yeast prion. Under rare circumstances, Sup35 adopts an inactive amyloid fold that induces heritable nonsense suppression in progeny and mating partners. Its prion propensity is governed by the prion domain N. The N domain together with a highly charged M domain are modular but otherwise dispensable for the termination function of the carboxyterminal C domain. Like most yeast prion domains, the N domain is enriched in uncharged amino acids, such as glutamine, asparagine, tyrosine, serine and glycine.Citation15
Interestingly, the prionogenic properties of the Sup35 prion domain are conserved when it is expressed in bacteriaCitation68 and mammalian cell models.Citation69 Investigating prion-like propagation and dissemination mechanism by using S. cerevisiae Sup35 can thus help to understand basic principles of cytosolic prion-like behavior in heterologous systems. Consistent with the finding that the Sup35 prion state can be induced in prion-free yeast cells by in vitro formed prion aggregates,Citation13,70 we recently demonstrated that cytosolically expressed NM stays soluble in neuroblastoma cells but can be induced to aggregate upon addition of recombinant NM amyloid fibrils.Citation69,71 Once induced, NM aggregates are faithfully propagated to daughter cells over multiple cell divisions. Furthermore, Sup35 NM protein aggregates in mammalian cells not only transmit vertically to progeny but also horizontally to naive cells in coculture. In analogy to the transmission pathways of TSE prions in mammalian cells, we found evidence for NM aggregate transmission to adjacent cells, potentially via actin-containing cytonemes,Citation9,71 and via EVs.Citation72 Although S. cerevisiae also secretes infectious prions in extracellular vesicles, so far it is unclear if these vesicles naturally transmit the prion state to bystander cells.Citation14,73 Different N2a clones all produced NM-containing EVs that were taken up by recipient cells and induced aggregation of GFP-tagged NM in the cytosol. Induction efficiency was, however, low, compared with aggregate induction efficiency when cells were in close proximity, suggesting that direct cell contact is the most efficient way of NM aggregate dissemination in our model.Citation71
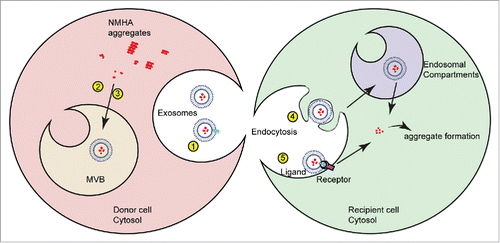
As limiting dilution cloning had been successfully used in the past to isolate cell clones with increased susceptibility to TSE prions,Citation74 we used the same strategy to isolate cell clones that secrete EVs capable of efficiently shuttling prion infectivity to recipient cells (). Through sequential centrifugation and Optiprep gradients, prion activity could be traced to vesicle fractions that fall in the size and density range of exosomes. NM released via exosomes was protected from proteolysis, arguing that at least a fraction of NM was present in the exosomal lumen72.
How is NM prion activity packaged into exosomes? We found the neutral sphingomyelinase inhibitor Spiroepoxide significantly reduced exosome and NM release, suggesting that ceramide-mediated exosome biogenesis is involved in NM secretion. Both soluble and insoluble protein was packaged into exosomes, and no correlation existed between NM aggregation state and exosome numbers.Citation72 Donor cells expressing soluble NM secreted even more vesicle-associated NM than donor clones containing NM prions, arguing that aggregation per se was not a required trigger for incorporation into exosomes. NM shares no sequence homology with mammalian proteins, so it is unlikely that specific recognition signals mediated selective recruitment.
The finding that different cell clones secreting exosomes with distinct infectivity can be isolated correlates with findings for TSE infected cells.Citation51 NM prion producing cell clones had been originally derived from a bulk population of N2a cells transduced with lentivirus coding for NM that were subsequently exposed to recombinant NM fibrils.Citation69 Cell clones differ in NM expression levels and show phenotypic variation of NM aggregates. The morphological phenotype of NM prions is remarkably persistent and does not change even over prolonged culture.Citation69 We compared 2 cell clones for their secretion of NM aggregates via EVs. Interestingly, cell clone 1C expressed more total NMCitation69 and also secreted more total NM in association with exosomes than cell clone s2E.Citation72 Cell clone s2E selected for its production of highly infectious conditioned medium secreted approximately 6 x more exosomes than 1C clone, but exhibited a seeding activity which was approximately 280 x higher than that of exosomes derived from clone 1C. Secreted EVs from clones 1C and s2E did not differ in size. Filter trap assay and SDD AGE demonstrated that a considerable amount of aggregated NM was present in exosomal preparations of both cell clones. However, comparison of the aggregation states of exosome-packaged NM revealed that lower-order NM oligomers were preferentially sorted into exosomes by the clone s2E.Citation72 The finding that the aggregation state of NM within exosomes was distinct from that seen in whole cell extracts suggests that NM aggregate sorting into exosomes is a selective process. Our data are in line with the hypothesis that lower-order oligomers constitute highly active templates for seeded polymerization.Citation75 Notably, rupture of exosomal membranes by sonication left NM oligomers relatively unaffected but drastically reduced the infectivity of the preparation, strongly arguing that only intact exosomes efficiently deliver NM aggregates to target cells.Citation72
Further evidence that distinct exosomes are released from different donor cell populations comes from new experiments with human HEK cells engineered to express NM. Similar to our N2a model, exposure of engineered HEK cells to recombinant NM fibrils turned soluble cytoplasmic NM into morphologically heterogeneous, self-templating protein aggregates that were stably propagated by individual cell clones (). Also HEK cells released soluble and aggregated NM in association with exosomes (). Consistent with our previous results, we did not observe increased exosome release in cells with aggregated NM-HA (). HEK donor cells secreted significantly less exosomes compared with N2a donor clones 1C and s2E (). While we expected to achieve lower induction rates due to lower exosome numbers, exosomes derived from HEK NM-HAagg cells were basically non-infectious to HEK NM-GFPsol recipient cells (data not shown). Comparison of the NM aggregation states in donor cell populations revealed that the oligomerization state of NM in the cell lysates of all donor cell populations was remarkably similar (). Exosome-associated NM from HEK NM-HAagg cells was also enriched for lower-order oligomers comparable to the exosomes produced by highly efficient donor clone s2E (). The lack of aggregate induction by NM oligomer-bearing exosomes in the HEK system argues that there is considerable difference in composition and activity of EVs isolated from different cell lines and even cell clones. Possible differences in the seeding activities of exosome populations are likely related to the relative number of secreted exosomes, the relative expression level of amyloidogenic protein, and the relative amount and oligomerization state of incorporated aggregated protein (). Another intriguing possibility is that subsequent exosome-target cell interactions could influence the biologic activity of the NM cargo. This possibility, however, needs further elucidation.
FIGURE 2. Human HEK cells secrete both soluble and aggregated Sup35 NM in association with exosomes. HEK cells expressing HA-epitope tagged NM (NM-HA) before (A) and after (B) NM aggregate induction by recombinant NM fibrils. NM-HAsol: soluble NM-HA. NM-HAagg: aggregated NM-HA. NM-HA was stained with anti-HA antibody (red) and nuclei were counterstained with Hoechst (blue). Maximum intensity projections were generated from Z-stacks. (C) Western blot analysis of exosomes from HEK NM-HAsol, NM-HAagg and N2a NM-HAagg s2E cell clones for exosomal marker Alix and NM-HA. Exosomes were isolated according to a previously described method.Citation72 (D) Exosome numbers released from HEK NM-HAsol and NM-HAagg were determined using ZetaView PMX 110-SZ-488 Nano Particle Tracking Analyzer with the same measurement setting. Results shown are means ± SD (n = 3; *** p < 0.001; unpaired student t test). (E) Exosome numbers released from HEK NM-HAagg, N2a NM-HAagg clone s2E (selected for high aggregate inducing activity in recipient cells) and N2a NM-HAagg clone 1C. Results shown are means ± SD (n = 3; *** p<0.001; one-way ANOVA). (F) Glutaraldehyde cross-linking of proteins in cell lysates or exosomes from HEK NM-HAsol, HEK NM-HAagg and N2a NM-HAagg clones s2E or 1C to determine the oligomerization state of NM-HA. Cross-linking was done as described previously.Citation72
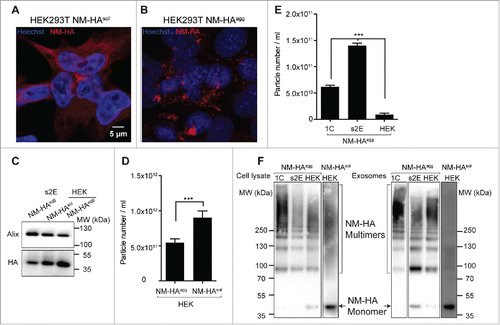
FIGURE 3. Factors influencing seeding activity of protein aggregates incorporated into exosomes. Exosome-mediated secretion of NM-HA by donor cells and subsequent uptake and seeding of NM-GFP prions in recipient cells. Infectivity of NM-HA bearing exosomes is likely determined by the following parameters: (1) Enhanced secretion of exosomes, (2, 3) Selective sorting of low-order oligomers, (4, 5) Specific exosome-target cell interaction (ligand-receptor recognition). This could include cell specific ligand-receptor interactions or differences in the intracellular fate of endocytosed exosomes. After internalization, the NM-HA aggregates contained in exosomes are released and induce new aggregate formation in N2a NM-GFP cells. The mechanism of NM-HA release into the cytosol is so far unknown.
Evidence for Secretion of Human Proteins with PrLDs in Association with Exosomes
Algorithms devised to identify novel prion proteins predict that approximately 1% of mammalian proteins contain PrLDs.Citation15,16,76 The majority of mammalian proteins with PrLDs are nucleic acid binding proteins. The PrLDs play critical roles in protein function by mediating protein-protein interactions or phase transition required for the formation of physiologically relevant membrane-less organelles, such as stress granules.Citation16 A prominent protein known to contain a PrLD is the RNA-binding protein TIA-1, an essential component of stress granules. The finding that replacement of TIA-1 PrLD with the Sup35 prion domain restores its normal function argues that yeast prion domains and predicted PrLDs are indeed functionally related.Citation77
Importantly, aberrant aggregation of proteins with PrLDs might be the underlying cause of degeneration in several neurodegenerative diseases and myopathies.Citation78-81 ALS is a fatal motor neuron disease that is mostly sporadic. Ten percent of cases are genetic and have been linked to mutations in a variety of proteins, such as SOD1, VCP, OPTN, TDP-43, hnRNPA1, hnRNPA2 and FUS, many of which form insoluble pathological inclusions. FUS, TDP-43, hnRNPA1 and hnRNPA2 contain putative PrLDs similar to annotated yeast prion domains. Systematic screens in yeast recently identified TAF15 and EWSR1 as further aggregation-prone PrLD-bearing proteins linked to neurodegenerative diseases.Citation82,83 Several other proteins listed as PrLD-like proteins await further characterization. Deregulated PrLD-mediated protein assembly has been proposed to promote the formation of protein aggregates with self-templating and dissemination properties. Indeed, a recent study showed that replacement of the Sup35 prion domain with the human hnRNPA2B1 PrLD generates a protein with definite prion activity in yeast, arguing that PrLDs of human proteins can drive prion assembly at least in lower eukaryotes.Citation81
TDP-43 is a nuclear RNA-binding protein involved in transcription and splicing and is associated with cytoplasmic inclusions in ALS and FTD. The predicted PrLD of TDP-43 mediates its aggregation in vitro and in vivo.Citation84 Recombinant TDP-43 fibrils and TDP-43 aggregates extracted from ALS and / or FTD patients have seeding activity and cause mislocalization and aggregation of TDP-43 in cell culture.Citation85,86 TDP-43 oligomers or aggregates also transmit from donor to recipient cells in culture, either through tunneling nanotubes or exosomes.Citation86-89 Cell culture experiments suggest that the ceramide-dependent exosomal pathway is involved in exosomal TDP-43 release.Citation88 As TDP-43 is also present in exosomal fractions from brains and CSF of healthy controls, its assembly into disease-associated aggregates might not cause the sorting into EVs.Citation87,89 Still, exosome-associated TDP-43 was reported to be increased in ALS brains compared with controls.Citation88 Interestingly, mammalian proteins that harbor intrinsically disordered domains with amino acid compositions similar to yeast prion domainsCitation15,76 appear to be frequent constituents of exosomes. Of the human RNA-binding proteins with PrLDs,Citation80 71% have been previously reported in exosomal fractions (http://www.exocarta.org/). PrLD-containing proteins can even be actively involved in the selective sorting of specific microRNAs into EVs for secretion. A sumyolated form of hnRNPA2B1 controls the sorting of a subpopulation of microRNAs into exosomes.Citation90 The presence of PrLD containing proteins in exosomes could thus reflect the physiological function of the respective protein. Whether aberrantly folded proteins with PrLDs are generally sorted into exosomes and how this might contribute to intercellular aggregate spreading remains to be established.
CONCLUSION
Research over the last years has demonstrated that not only TSE prions are sorted into exosomes, but also pathogenic protein aggregates associated with more common neurodegenerative diseases. Among them, proteins with domains compositionally similar to yeast prion domains have been found associated with EVs, suggesting that EVs might contribute to their intercellular dissemination. Our knowledge on mechanisms that drive cargo sorting into EVs and uptake by recipient cells is limited. There is an urgent need for assays that monitor cargo delivery to target cells that are amenable to high throughput screening. Here we showed that the non-mammalian prion domain of Sup35 can serve as a versatile tool to study exosome-mediated induction of self-templating protein aggregates. The NM prion cell assay has been successfully adapted to automated high throughput confocal microscopy. The fast and accurate detection of aggregate induction in recipient cells will help to characterize general cellular pathways involved in aggregation and dissemination of protein aggregates.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
ACKNOWLEDGMENTS
We thank Sybille Krauss and Philip Denner for critical comments on this manuscript. We are grateful to Philip Denner and Birgit Kurkowsky for advice on automated imaging.
FUNDING
This work was supported by the Helmholtz Portfolio Wirkstoffforschung.
REFERENCES
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013; 200:373-83; PMID:23420871; http://dx.doi.org/10.1083/jcb.201211138
- van Dongen HM, Masoumi N, Witwer KW, Pegtel DM. Extracellular vesicles exploit viral entry routes for cargo delivery. Microbiol Mol Biol Rev 2016; 80:369-86; PMID:26935137; http://dx.doi.org/10.1128/MMBR.00063-15
- Howitt J, Hill AF. Exosomes in the pathology of neurodegenerative diseases. J Biol Chem 2016; 291:26589-97; PMID:27852825; http://dx.doi.org/10.1074/jbc.R116.757955
- Grad LI, Fernando SM, Cashman NR. From molecule to molecule and cell to cell: prion-like mechanisms in amyotrophic lateral sclerosis. Neurobiol Dis 2015; 77:257-65; PMID:25701498; http://dx.doi.org/10.1016/j.nbd.2015.02.009
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136-44; PMID:6801762; http://dx.doi.org/10.1126/science.6801762
- Grassmann A, Wolf H, Hofmann J, Graham J, Vorberg I. Cellular aspects of prion replication in vitro. Viruses 2013; 5:374-405; PMID:23340381; http://dx.doi.org/10.3390/v5010374
- Bessen RA, Kocisko DA, Raymond GJ, Nandan S, Lansbury PT, Caughey B. Non-genetic propagation of strain-specific properties of scrapie prion protein. Nature 1995; 375:698-700; PMID:7791905; http://dx.doi.org/10.1038/375698a0
- Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science 1994; 264:566-9; PMID:7909170; http://dx.doi.org/10.1126/science.7909170
- Hofmann J, Vorberg I. Life cycle of cytosolic prions. Prion 2013; 7:369-77; PMID:24021964; http://dx.doi.org/10.4161/pri.26414
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A 2011; 108:5337-41; PMID:21402947; http://dx.doi.org/10.1073/pnas.1102762108
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 2012; 482:363-8; PMID:22337056; http://dx.doi.org/10.1038/nature10875
- Knowles TP, Waudby CA, Devlin GL, Cohen SI, Aguzzi A, Vendruscolo M, Terentjev EM, Welland ME, Dobson CM. An analytical solution to the kinetics of breakable filament assembly. Science 2009; 326:1533-7; PMID:20007899; http://dx.doi.org/10.1126/science.1178250
- King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature 2004; 428:319-23; PMID:15029195; http://dx.doi.org/10.1038/nature02391
- Kabani M, Melki R. Sup35p in its soluble and prion states is packaged inside extracellular vesicles. MBio 2015; 6(4):pii: e01017-15; PMID:26286691; http://dx.doi.org/10.1128/mBio.01017-15
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146-58; PMID:19345193; http://dx.doi.org/10.1016/j.cell.2009.02.044
- March ZM, King OD, Shorter J. Prion-like domains as epigenetic regulators, scaffolds for subcellular organization, and drivers of neurodegenerative disease. Brain Res 2016; 1647:9-18; PMID:26996412; http://dx.doi.org/10.1016/j.brainres.2016.02.037
- Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 2016; 36:301-12; PMID:27053351; http://dx.doi.org/10.1007/s10571-016-0366-z
- Maas SL, Breakefield XO, Weaver AM. Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol 2016; 27(3):172-88; PMID:27979573
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell 2011; 21:708-21; PMID:21962903; http://dx.doi.org/10.1016/j.devcel.2011.08.019
- Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 2009; 458:445-52; PMID:19325624; http://dx.doi.org/10.1038/nature07961
- Moreno-Gonzalo O, Villarroya-Beltri C, Sanchez-Madrid F. Post-translational modifications of exosomal proteins. Front Immunol 2014; 5:383; PMID:25157254; http://dx.doi.org/10.3389/fimmu.2014.00383
- Liang Y, Eng WS, Colquhoun DR, Dinglasan RR, Graham DR, Mahal LK. Complex N-linked glycans serve as a determinant for exosome/microvesicle cargo recruitment. J Biol Chem 2014; 289:32526-37; PMID:25261472; http://dx.doi.org/10.1074/jbc.M114.606269
- Wubbolts R, Leckie RS, Veenhuizen PT, Schwarzmann G, Mobius W, Hoernschemeyer J, Slot JW, Geuze HJ, Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J Biol Chem 2003; 278:10963-72
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008; 319:1244-7; PMID:18309083; http://dx.doi.org/10.1126/science.1153124
- Verweij FJ, van Eijndhoven MA, Hopmans ES, Vendrig T, Wurdinger T, Cahir-McFarland E, Kieff E, Geerts D, van der Kant R, Neefjes J, et al. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-kappaB activation. The EMBO journal 2011; 30:2115-29; PMID:21527913; http://dx.doi.org/10.1038/emboj.2011.123
- de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood 2003; 102:4336-44; PMID:12881314; http://dx.doi.org/10.1182/blood-2003-03-0871
- Janas T, Janas MM, Sapon K, Janas T. Mechanisms of RNA loading into exosomes. FEBS Lett 2015; 589:1391-8; PMID:25937124; http://dx.doi.org/10.1016/j.febslet.2015.04.036
- Vidal M, Mangeat P, Hoekstra D. Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation. J Cell Sci 1997; 110(Pt 16):1867-77; PMID:9296387
- Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol 2007; 5:e158; PMID:17550307; http://dx.doi.org/10.1371/journal.pbio.0050158
- Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015; 527:329-35; PMID:26524530; http://dx.doi.org/10.1038/nature15756
- Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A 2013; 110:17380-5; PMID:24101524; http://dx.doi.org/10.1073/pnas.1304266110
- Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 2011; 124:447-58; PMID:21242314; http://dx.doi.org/10.1242/jcs.074088
- Saunderson SC, Dunn AC, Crocker PR, McLellan AD. CD169 mediates the capture of exosomes in spleen and lymph node. Blood 2014; 123:208-16; PMID:24255917; http://dx.doi.org/10.1182/blood-2013-03-489732
- Montecalvo A, Larregina AT, Shufesky WJ, Stolz DB, Sullivan ML, Karlsson JM, Baty CJ, Gibson GA, Erdos G, Wang Z, et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012; 119:756-66; PMID:22031862; http://dx.doi.org/10.1182/blood-2011-02-338004
- Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem 2014; 289:22258-67; PMID:24951588; http://dx.doi.org/10.1074/jbc.M114.588046
- Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010; 11:675-87; PMID:20136776; http://dx.doi.org/10.1111/j.1600-0854.2010.01041.x
- Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Mörgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 2013; 288:17713-24; PMID:23653359; http://dx.doi.org/10.1074/jbc.M112.445403
- Amano A, Takeuchi H, Furuta N. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect 2010; 12:791-8; PMID:20685339; http://dx.doi.org/10.1016/j.micinf.2010.05.008
- Rana S, Yue S, Stadel D, Zoller M. Toward tailored exosomes: the exosomal tetraspanin web contributes to target cell selection. Int J Biochem Cell Biol 2012; 44:1574-84; PMID:22728313; http://dx.doi.org/10.1016/j.biocel.2012.06.018
- Prinz M, Heikenwalder M, Junt T, Schwarz P, Glatzel M, Heppner FL, Fu YX, Lipp M, Aguzzi A. Positioning of follicular dendritic cells within the spleen controls prion neuroinvasion. Nature 2003; 425:957-62; PMID:14562059; http://dx.doi.org/10.1038/nature02072
- Yim YI, Park BC, Yadavalli R, Zhao X, Eisenberg E, Greene LE. The multivesicular body is the major internal site of prion conversion. J Cell Sci 2015; 128:1434-43; PMID:25663703; http://dx.doi.org/10.1242/jcs.165472
- Vilette D, Laulagnier K, Huor A, Alais S, Simoes S, Maryse R, Provansal M, Lehmann S, Andreoletti O, Schaeffer L, et al. Efficient inhibition of infectious prions multiplication and release by targeting the exosomal pathway. Cell Mol Life Sci 2015; 72:4409-27; PMID:26047659; http://dx.doi.org/10.1007/s00018-015-1945-8
- Paquet S, Langevin C, Chapuis J, Jackson GS, Laude H, Vilette D. Efficient dissemination of prions through preferential transmission to nearby cells. J Gen Virol 2007; 88:706-13; PMID:17251590; http://dx.doi.org/10.1099/vir.0.82336-0
- Kanu N, Imokawa Y, Drechsel DN, Williamson RA, Birkett CR, Bostock CJ, Brockes JP. Transfer of scrapie prion infectivity by cell contact in culture. Curr Biol 2002; 12:523-30; PMID:11937020; http://dx.doi.org/10.1016/S0960-9822(02)00722-4
- Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J, et al. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol 2009; 11:328-36; PMID:19198598; http://dx.doi.org/10.1038/ncb1841
- Leblanc P, Alais S, Porto-Carreiro I, Lehmann S, Grassi J, Raposo G, Darlix JL. Retrovirus infection strongly enhances scrapie infectivity release in cell culture. EMBO J 2006; 25:2674-85; PMID:16724107; http://dx.doi.org/10.1038/sj.emboj.7601162
- Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci U S A 2004; 101:9683-8; PMID:15210972; http://dx.doi.org/10.1073/pnas.0308413101
- Alais S, Simoes S, Baas D, Lehmann S, Raposo G, Darlix JL, Leblanc P. Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles. Biol Cell 2008; 100:603-15; http://dx.doi.org/10.1042/BC20080025
- Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol 2007; 211:582-90; PMID:17334982; http://dx.doi.org/10.1002/path.2145
- Mattei V, Barenco MG, Tasciotti V, Garofalo T, Longo A, Boller K, Löwer J, Misasi R, Montrasio F, Sorice M. Paracrine diffusion of PrP(C) and propagation of prion infectivity by plasma membrane-derived microvesicles. PLoS One 2009; 4:e5057; PMID:19337375; http://dx.doi.org/10.1371/journal.pone.0005057
- Arellano-Anaya ZE, Huor A, Leblanc P, Lehmann S, Provansal M, Raposo G, Andréoletti O, Vilette D. Prion strains are differentially released through the exosomal pathway. Cell Mol Life Sci 2015; 72:1185-96; PMID:25227242; http://dx.doi.org/10.1007/s00018-014-1735-8
- Guo BB, Bellingham SA, Hill AF. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J Biol Chem 2015; 290:3455-67; PMID:25505180; http://dx.doi.org/10.1074/jbc.M114.605253
- Saa P, Yakovleva O, de Castro J, Vasilyeva I, De Paoli SH, Simak J, Cervenakova L. First demonstration of transmissible spongiform encephalopathy-associated prion protein (PrPTSE) in extracellular vesicles from plasma of mice infected with mouse-adapted variant Creutzfeldt-Jakob disease by in vitro amplification. J Biol Chem 2014; 289:29247-60; PMID:25157106; http://dx.doi.org/10.1074/jbc.M114.589564
- Veith NM, Plattner H, Stuermer CA, Schulz-Schaeffer WJ, Burkle A. Immunolocalisation of PrPSc in scrapie-infected N2a mouse neuroblastoma cells by light and electron microscopy. Eur J Cell Biol 2009; 88:45-63; PMID:18834644; http://dx.doi.org/10.1016/j.ejcb.2008.08.001
- Wang G, Zhou X, Bai Y, Zhang Z, Zhao D. Cellular prion protein released on exosomes from macrophages binds to Hsp70. Acta Biochim Biophys Sin (Shanghai) 2010; 42:345-50; PMID:20458448; http://dx.doi.org/10.1093/abbs/gmq028
- Fevrier B, Vilette D, Laude H, Raposo G. Exosomes: a bubble ride for prions? Traffic 2005; 6:10-7; PMID:15569241; http://dx.doi.org/10.1111/j.1600-0854.2004.00247.x
- Wang GH, Zhou XM, Bai Y, Yin XM, Yang LF, Zhao D. Hsp70 binds to PrPC in the process of PrPC release via exosomes from THP-1 monocytes. Cell Biol Int 2011; 35:553-8; PMID:20964628; http://dx.doi.org/10.1042/CBI20090391
- Wik L, Klingeborn M, Willander H, Linne T. Separate mechanisms act concurrently to shed and release the prion protein from the cell. Prion 2012; 6:498-509; PMID:23093798; http://dx.doi.org/10.4161/pri.22588
- Lazar I, Clement E, Ducoux-Petit M, Denat L, Soldan V, Dauvillier S, Balor S, Burlet-Schiltz O, Larue L, Muller C, et al. Proteome characterization of melanoma exosomes reveals a specific signature for metastatic cell lines. Pigment Cell Melanoma Res 2015; 28:464-75; PMID:25950383; http://dx.doi.org/10.1111/pcmr.12380
- Carayon K, Chaoui K, Ronzier E, Lazar I, Bertrand-Michel J, Roques V, Balor S, Terce F, Lopez A, Salomé L, et al. Proteolipidic composition of exosomes changes during reticulocyte maturation. J Biol Chem 2011; 286:34426-39; PMID:21828046; http://dx.doi.org/10.1074/jbc.M111.257444
- Robertson C, Booth SA, Beniac DR, Coulthart MB, Booth TF, McNicol A. Cellular prion protein is released on exosomes from activated platelets. Blood 2006; 107:3907-11; PMID:16434486; http://dx.doi.org/10.1182/blood-2005-02-0802
- Vella LJ, Greenwood DL, Cappai R, Scheerlinck JP, Hill AF. Enrichment of prion protein in exosomes derived from ovine cerebral spinal fluid. Vet Immunol Immunopathol 2008; 124:385-93; PMID:18501435; http://dx.doi.org/10.1016/j.vetimm.2008.04.002
- Dias MV, Teixeira BL, Rodrigues BR, Sinigaglia-Coimbra R, Porto-Carreiro I, Roffe M, Hajj GN, Martins VR. PRNP/prion protein regulates the secretion of exosomes modulating CAV1/caveolin-1-suppressed autophagy. Autophagy 2016; 12:2113-28; PMID:27629560; http://dx.doi.org/10.1080/15548627.2016.1226735
- Vella LJ, Sharples RA, Nisbet RM, Cappai R, Hill AF. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur Biophys J 2008; 37:323-32; PMID:18064447; http://dx.doi.org/10.1007/s00249-007-0246-z
- Shen B, Wu N, Yang JM, Gould SJ. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem 2011; 286:14383-95; PMID:21300796; http://dx.doi.org/10.1074/jbc.M110.208660
- Klohn PC, Stoltze L, Flechsig E, Enari M, Weissmann C. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc Natl Acad Sci U S A 2003; 100:11666-71; PMID:14504404; http://dx.doi.org/10.1073/pnas.1834432100
- Greil CS, Vorberg IM, Ward AE, Meade-White KD, Harris DA, Priola SA. Acute cellular uptake of abnormal prion protein is cell type and scrapie-strain independent. Virology 2008; 379:284-93; PMID:18692214; http://dx.doi.org/10.1016/j.virol.2008.07.006
- Garrity SJ, Sivanathan V, Dong J, Lindquist S, Hochschild A. Conversion of a yeast prion protein to an infectious form in bacteria. Proc Natl Acad Sci U S A 2010; 107:10596-601; PMID:20484678; http://dx.doi.org/10.1073/pnas.0913280107
- Krammer C, Kryndushkin D, Suhre MH, Kremmer E, Hofmann A, Pfeifer A, Scheibel T, Wickner RB, Schätzl HM, Vorberg I. The yeast Sup35NM domain propagates as a prion in mammalian cells. Proc Natl Acad Sci U S A 2009; 106:462-7; PMID:19114662; http://dx.doi.org/10.1073/pnas.0811571106
- Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature 2004; 428:323-8; PMID:15029196; http://dx.doi.org/10.1038/nature02392
- Hofmann JP, Denner P, Nussbaum-Krammer C, Kuhn PH, Suhre MH, Scheibel T, Lichtenthaler SF, Schätzl HM, Bano D, Vorberg IM. Cell-to-cell propagation of infectious cytosolic protein aggregates. Proc Natl Acad Sci U S A 2013; 110:5951-6; PMID:23509289; http://dx.doi.org/10.1073/pnas.1217321110
- Liu S, Hossinger A, Hofmann JP, Denner P, Vorberg IM. Horizontal transmission of cytosolic Sup35 prions by extracellular vesicles. MBio 2016; 7(4):pii: e00915-16
- Kabani M, Melki R. More than just trash bins? Potential roles for extracellular vesicles in the vertical and horizontal transmission of yeast prions. Curr Genet 2015; 62(2):265-70; PMID:26553335
- Klohn PC, Castro-Seoane R, Collinge J. Exosome release from infected dendritic cells: a clue for a fast spread of prions in the periphery? J Infect 2013; 67:359-68; PMID:23911964; http://dx.doi.org/10.1016/j.jinf.2013.07.024
- Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J 2001; 20:6236-45; PMID:11707395; http://dx.doi.org/10.1093/emboj/20.22.6236
- Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol 2010; 30:319-32; PMID:19884345; http://dx.doi.org/10.1128/MCB.01140-09
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 2004; 15:5383-98; PMID:15371533; http://dx.doi.org/10.1091/mbc.E04-08-0715
- Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell 2011; 147:498-508; PMID:22036560; http://dx.doi.org/10.1016/j.cell.2011.10.011
- Brettschneider J, Del Tredici K, Toledo JB, Robinson JL, Irwin DJ, Grossman M, Suh E, Van Deerlin VM, Wood EM, Baek Y, et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann Neurol 2013; 74:20-38; PMID:23686809; http://dx.doi.org/10.1002/ana.23937
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 2013; 201:361-72; PMID:23629963; http://dx.doi.org/10.1083/jcb.201302044
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013; 495:467-73; PMID:23455423; http://dx.doi.org/10.1038/nature11922
- Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, Oristano R, Liu AX, Ramos D, Jethava N, Hosangadi D, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A 2011; 108:20881-90; PMID:22065782; http://dx.doi.org/10.1073/pnas.1109434108
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet 2012; 21:2899-911; PMID:22454397; http://dx.doi.org/10.1093/hmg/dds116
- Budini M, Buratti E, Stuani C, Guarnaccia C, Romano V, De Conti L, Baralle FE. Cellular model of TAR DNA-binding protein 43 (TDP-43) aggregation based on its C-terminal Gln/Asn-rich region. J Biol Chem 2012; 287:7512-25; PMID:22235134; http://dx.doi.org/10.1074/jbc.M111.288720
- Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem 2011; 286:18664-72; PMID:21454603; http://dx.doi.org/10.1074/jbc.M111.231209
- Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, Yoshida M, Murayama S, Mann DM, Akiyama H, et al. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep 2013; 4:124-34; PMID:23831027; http://dx.doi.org/10.1016/j.celrep.2013.06.007
- Ding X, Ma M, Teng J, Teng RK, Zhou S, Yin J, Fonkem E, Huang JH, Wu E, Wang X. Exposure to ALS-FTD-CSF generates TDP-43 aggregates in glioblastoma cells through exosomes and TNTs-like structure. Oncotarget 2015; 6:24178-91; PMID:26172304; http://dx.doi.org/10.18632/oncotarget.4680
- Iguchi Y, Eid L, Parent M, Soucy G, Bareil C, Riku Y, Kawai K, Takagi S, Yoshida M, Katsuno M, et al. Exosome secretion is a key pathway for clearance of pathological TDP-43. Brain 2016; 139:3187-201; PMID:27679482; http://dx.doi.org/10.1093/brain/aww237
- Feiler MS, Strobel B, Freischmidt A, Helferich AM, Kappel J, Brewer BM, Li D, Thal DR, Walther P, Ludolph AC, et al. TDP-43 is intercellularly transmitted across axon terminals. J Cell Biol 2015; 211:897-911; PMID:26598621; http://dx.doi.org/10.1083/jcb.201504057
- Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 2013; 4:2980; PMID:24356509; http://dx.doi.org/10.1038/ncomms3980
