ABSTRACT
Prion protein (PRNP) gene is well known for affecting mammal transmissible spongiform encephalopathies (TSE), and is also reported to regulate phenotypic traits (e.g. growth traits) in healthy ruminants. To identify the insertion/deletion (indel) variations of the PRNP gene and evaluate their effects on growth traits, 768 healthy individuals from five sheep breeds located in China and Mongolia were identified and analyzed. Herein, four novel indel polymorphisms, namely, Intron-1-insertion-7bp (I1-7bp), Intron-2-insertion-15bp (I2-15bp), Intron-2-insertion-19bp (I2-19bp), and 3′ UTR-insertion-7bp (3′ UTR-7bp), were found in the sheep PRNP gene. In five analyzed breeds, the minor allelic frequencies (MAF) of the above indels were in the range of 0.008 to 0.986 (I1-7bp), 0.113 to 0.336 (I2-15bp), 0.281 to 0.510 (I2-19bp), and 0.040 to 0.238 (3′ UTR-7bp). Additionally, there were 15 haplotypes and the haplotype ‘II2-15bp-D3’UTR-7bp-DI2-19bp-DI1-7bp’ had the highest frequency, which varied from 0.464 to 0.629 in five breeds. Moreover, association analysis revealed that all novel indel polymorphisms were significantly associated with 13 different growth traits (P < 0.05). Particularly, the influences of I2-15bp on chest width (P = 0.001) in Small Tail Han sheep (ewe), 3′ UTR-7bp on chest circumference (P = 0.003) in Hu sheep, and I2-19bp on tail length (P = 0.001) in Tong sheep, were highly significant (P < 0.01). These findings may be a further step toward the detection of indel-based typing within and across sheep breeds, and of promising target loci for accelerating the progress of marker-assisted selection in sheep breeding.
Introduction
As the vital control gene of fatal prion diseases or transmissible spongiform encephalopathies (TSE), the prion protein (PRNP) gene will always be a focus of ovine research [Citation1–2]. With extensive and thorough studies of PRNP regulating anthropozoonotic scrapie in goat and sheep, outbreaks of scrapie are under control in China and its neighbors (e.g. Mongolia). As well as controlling TSEs, PRNP also has a great impact on the economic performance of healthy livestock, such as the cashmere yield [Citation3], wool thickness [Citation3], and milk yield of goats [Citation4], as well as the waistline [Citation5], body length [Citation5], rump length [Citation6], and weight of cows [Citation5–6].
Recently, the “One Belt and One Road Initiative” was successfully proposed by Chinese paramount leader Xi Jinping. This global strategy focuses on connectivity and cooperation between Eurasian countries in several fields, including husbandry. Under the promotion of the “One Belt and One Road Initiative,” the industrial livestock economy grew dramatically. To reap more economic benefits from the sheep industry, healthy individuals with excellent growth traits were selected through marker-assisted selection (MAS), based on selecting the relevant genetic mutations. Owing to various genetic mutations that regulate scrapie infection and growth traits [Citation3–6], the PRNP gene become a promising target gene when applying MAS to ovine selection and breeding.
Genetics variations of PRNP mostly focus on the coding region of the cellular prion protein (PrPc). There are three exons in the ovine PRNP gene, and the open reading frame (ORF) of the PrPc is located in the third exon. Polymorphisms within the third exon of the ovine PRNP gene at codons 136 [Citation7], 141 [Citation8],146 [Citation9], 154 [Citation10], and 171 [Citation11] are closely associated with variation in the phenotypic expression of scrapie. Other regions of PRNP, either independently or in synergy with the coding region, could also influence susceptibility [Citation12]. With extensive and thorough studies, these pathogenic codons are filtered out by artificial selection (e.g. MAS) in the ovine industry. Under the conditions of scrapie infection, several single nucleotide polymorphisms (SNPs) of the sheep PRNP gene, including mutations of codons 136, 154, and 171, were reported to impact on ewe reproductive performance [Citation13–15], lamb growth traits [Citation16–18], dairy traits [Citation19–20], and the seasonal mobilization of body reserves [Citation21].
Nevertheless, because of the advantages of convenient detection and its remarkable effects, the insertion/deletion (indel) variants have a higher efficiency and wider application than other molecular markers (including SNPs). Compared to SNPs, current research is focused on indel mutations of PRNP impacting TSE infection or phenotypic features in buffalo [Citation22–23], cattle [Citation5, Citation16], and goat [Citation3], but not sheep. For instance, the 14-bp indel variants of the 3′ untranslated region (3′ UTR) within bovine PRNP are significantly associated with body length, body weight, and waistline in Chinese Qinchuan and Xianan cattle [Citation5]. Simultaneously, the 23-bp indel variations within the PRNP promoter and 12-bp indel variations in intron one significantly influence the body length and heart girth of Nanyang cattle and cannon circumference of Ji'an cattle [Citation6]. However, in sheep, there are few reports on indels polymorphisms within the PRNP gene. However, the PRNP genotypes of ewes, such as ARQ/ARQ or ARR/ARQ, are significantly associated with milk traits (milk, protein, and fat yield; and protein and fat content) in Latxa dairy sheep [Citation4]. Further, the mutations in introns and the 3′ UTR are less noted and researched than those in exons. Thus, to elucidate the structural polymorphisms of the ovine PRNP, we screened the indel variants in introns and the 3′ UTR, and further researched polymorphism in Asian sheep breeds.
Hence, five representative sheep breeds were selected to investigate the indel variants of PRNP. The sartuul sheep (SS), a vital Mongolian domestic sheep breed for meat and wool and reared in Erdenekhairkhan Soum of Zhawkhan Province, has a lack of indel-related research toward it. Additionally, small-tail Han sheep (STHS), Lanzhou fat-tail sheep (LFTS), Tong sheep (TS), and Hu sheep (HS) were the representative indigenous sheep breeds in China. They are characterized by many potential advantages, such as strong endurance [Citation24], rough feedstuff resistance [Citation25], and disease resistance [Citation26]. Particularly, the Hu sheep is one of the most extensively distributed livestock, and its high reproductive capacity can bring huge economic benefits [Citation27]. Owing to the regulation of phenotypic traits by PRNP, and the intron indel polymorphisms of PRNP being limited in Mongolian and Chinese sheep breeds, this study aimed to detect the potential indel loci of the sheep PRNP gene in these representative sheep breeds, and investigate their association with growth traits. These results may provide potential theories for further research on applying MAS to the sheep industry.
Material and methods
Ethics statement
All experiments implemented in this study were approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU), and fully followed local animal welfare guidelines, laws, and policies.
Animal samples and genomic DNA collection
In total, 768 sheep samples, composed of five diverse breeds, sartuul sheep (SS, n = 146) from Zhawkhan Province (Mongolia), Hu sheep (HS, n = 201) from Henan Province (China), small-tail Han sheep (STHS, n = 195) and Lanzhou fat-tail sheep (LFTS, n = 61) from Gansu Province (China), and Tong sheep (TS, n = 165) from Shaanxi Province (China), were used. All of the tested sheep were two to six years old. Health and relationship examinations were performed for individual selection [Citation5–6]. The body measurement traits for all selected individuals [Citation28], including body height (BH), body length (BL), body weight (BW), body back height (BBH), cannon circumference (CaC), chest circumference (ChC), chest width (ChW), head length (HL), sacral height (SH), tail length (TL), and wool length (WL), were measured by the same person with the same standard. Subsequently, according to previously reported descriptions [Citation29–31], the body length index (BLI), chest circumference index (ChCI), chest width index (ChWI), and cannon circumference index (CaCI) were also calculated.
DNA samples in this study were extracted from ear tissues (saved in 70% alcohol at −80°C) and blood leukocytes (frozen at −80°C) by the phenol–chloroform extraction method, according to our previous reports [Citation32–33]. After being assayed by a Nanodrop 1000 Spectrophotometer (Thermo Scientific, Waltham, MA, USA), all DNA samples were diluted to the same standard of 50 ng/μL and were stored at 4 °C temporarily. Additionally, 50 samples from each of the five varieties were mixed in a genomic DNA pool for a polymerase chain reaction (PCR) analysis to determine potential indel loci in the sheep PRNP gene [Citation30].
Novel indel locus discovery and DNA sequencing
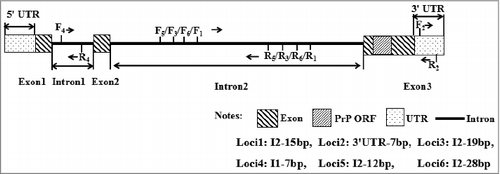
Based on the National Center for Biotechnology Information (NCBI) SNP database (https://www.ncbi.nlm.nih.gov/snp), six potential indel sites were found in the sheep PRNP gene, and all of them were located outside of the PrPc ORF (). Related amplification primers were designed by Primer Premier Software 5.0 (Premier Biosoft International, Palo Alto, CA, USA) to reference the sheep PRNP gene sequence (GenBank No: NC_019470.2)().
Table 1. PCR primer sequences of the sheep PRNP for applification.
The PCR reaction volume and amplification procedure was in accordance with our previous study [Citation34], and the indel loci were separated on 3.5% agarose gels. The products were sequenced only when different genotypes of each pair of primers had appeared [Citation5, Citation32].
Amplification and genotyping by different detection method
Four novel indel loci (I2-15 bp, 3′ UTR-7 bp, I2-19 bp, I1-7 bp) were detected and the mutation frequencies were also identified. Based on the sample sizes and mutation frequencies of these indel loci, we designed the most efficient pooling strategy to detect all individuals according to the mathematical expectation (ME) [Citation5], multiplex PCR, and traditional detection methods in sheep population. Finally, all the indel loci of the tested individuals were genotyped.
Statistical analyses
The genotype distributions were analyzed according to the Hardy–Weinberg equilibrium using chi-square test. Polymorphism information content (PIC) was calculated by Nei's method implemented in the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx) [Citation35]. The general linear model was used to analyze whether genotypes of different loci and breed affected traits synchronously. Distribution differences for genotypic and allelic frequencies among/between different breeds were implemented with the χ2 test using SPSS (Version 18.0, IBM Corp., Armonk, NY, USA) [Citation36]. Additionally, linkage equilibrium on the populations of four pair alleles and haplotypes of four mutation loci were also analyzed using r2 test and D’ test (http://analysis.bio-x.cn) [Citation37–38]. The independent-samples t-test and analysis of variance (ANOVA) available in SPSS (Version 18.0) [Citation39] were used to explore the associations of the indels of PRNP with several growth traits (e.g. body length [cm]) in different breeds. When necessary, the Bonferroni correction for multiple comparisons was performed. Particularly, the data that did not follow normal distribution and homogeneity of variances were analyzed by the non-parametric (Kruskal–Wallis) test in SPSS (Version 18.0) [Citation36]. Results were considered statistically significant at P < 0.05, and all statistical tests were two-sided.
Results
Result of PCR amplification and genotyping of individuals
The 3.5% gel agarose electrophoretogram revealed four novel indels in the sheep PRNP gene: Intron-1-insertion-7 bp (I1-7 bp), Intron-2-insertion-15 bp (I2-15 bp), Intron-2-insertion-19 bp (I2-19 bp) and 3′ UTR-insertion-7 bp (3′ UTR-7 bp) ()
Figure 2. Amplificationdiagram (a), electrophoresis pattern (b) and sequence diagram of the Intron-2-insertion-15 bp (c1) and 3’UTR-insertion-7 bp (c2) indel variants of sheep PRNP gene.
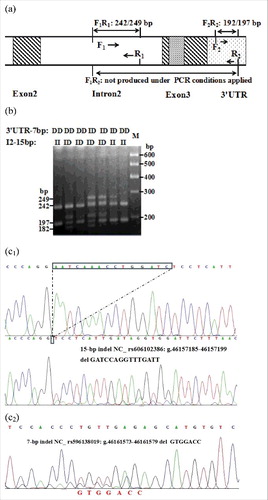
Figure 3. Electrophoresis pattern and sequence diagram of Intron-2-insertion-19 bp indel variants of sheep PRNP gene.
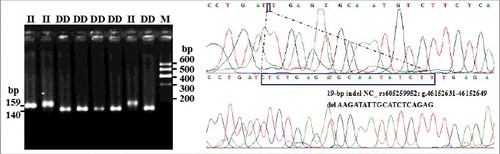
Figure 4. Electrophoresis pattern and sequence diagram of Intron-1-insertion-7 bp indel variants of the PRNP gene in sheep (The insertion sample was genotyped as ID.).
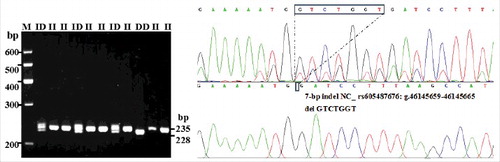
Each indel polymorphism had two or three different genotypes in all individuals: one longer band meant genotype insertion/insertion (II), one shorter band meant genotype deletion/deletion (DD), and two or three (homoduplex) bands meant genotype insertion/deletion (ID). The results of contrasting and analyzing these sequences with BioEdit software (BioEdit, Carlsbad, CA, USA) confirmed those novel indel loci (). The insertion sequences were GTTTACC for 3′ UTR-7 bp, GATCCAGGTTTGATT for I2-15 bp, and GTCTGGT for I1-7 bp. Among the results, only the sequence alignment result for the novel I2-19 bp (AAGATATTGCATCTCAGAG) () was not in concurrence with the predicted sequence data in the NCBI.
Genetic parameters calculation
Frequencies of two alleles and population parameters for each indel locus in the five tested sheep breeds are listed in . In all of the analyzed breeds, the minor allelic frequencies (MAF) of the indels ranged from 0.008 to 0.986 (I1-7 bp), 0.113 to 0.336 (I2-15 bp), 0.281 to 0.510 (I2-19 bp), and 0.040 to 0.238 (3′ UTR-7 bp). The population parameters results showed that the PIC of the indel markers among studied breeds ranged from 0.016 to 0.368. Notably, the I2-19 bp displayed moderate polymorphism in all detected sheep breeds with PIC values ≥ 0.322. Furthermore, some loci were not at Hardy–Weinberg equilibrium (HWE) in several populations (P < 0.05), including the I2-15 bp and 3′ UTR-7 bp loci in LFTS and TS, I2-19 bp locus in STHS and SS, and I1-7 bp locus in TS.
Table 2. Diversity parameter for indel loci of sheep PRNP gene.
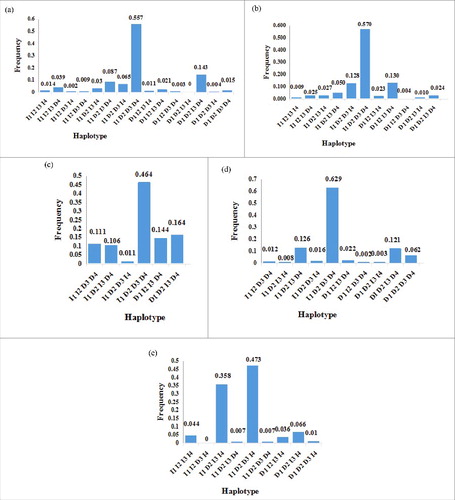
According to the r2 test values(, ), the I2-15 bp and I2-19 bp loci conformed to linkage equilibrium in all five sheep breeds, while the D’ test values indicated adverse consequences (except in LFTS). Concurrently, based on the haplotype analysis of four mutation loci, there were 15 haplotypes, of which ‘II2-15bp-D3’UTR-7bp-DI2-19bp-DI1-7bp’ had the highest frequency (P = 0.569) of occurrence in all tested groups ().
Table 3. Genetic analysis of linkage equilibrium of four pair alleles (D' and r2) on five sheep breeds.
Figure 5. Genetic analysis of linkage equilibrium on different population of four pair alleles in sheep PRNP gene: (a) Lanzhou Fat-Tail sheep, (b) Small Tail Han sheep, (c) Tong sheep, (d) Hu sheep, (e) Sartuul sheep. Loci chosen for hap-analysis: loci1: I2-15 bp, loci2: 3’UTR-7 bp, loci3: I2-19 bp, loci4: I1-7 bp.
Figure 6. Haplotype frequency of four mutation loci within the sheep PRNP gene in different breeds: (a) Small Tail Han sheep, (b) Tong sheep, (c) Lanzhou Fat-Tail sheep, (d) Hu sheep, (e) Sartuul sheep Note: Abscissa represents the haplotype and ordinate means the frequency. Loci chosen for hap-analysis: loci1: I2-15bp, loci2: 3’UTR-7bp, loci3: I2-19bp, loci4: I1-7bp (All those frequency < 0 will be ignored in analysis.).
Novel indel polymorphisms had significant relationships with growth traits.
Owing to the excellent economic performance of tested sheep breeds, we investigated the association between the novel indel of the PRNP gene and sheep growth traits. Each indel locus had significant relationships with growth traits(), such as cannon circumference and chest circumference, in STHS, TS, and HS. For example, the mutations of 3′ UTR-7 bp significantly influenced the ChCI of STHS (ram), WL of TS (ewe), and ChC and ChCI of HS (ewe). Particularly, the influences of I2-15 bp on the chest width (P = 0.001) in STHS (ewe), 3′ UTR-7 bp on chest circumference (P = 0.003) in HS, and I2-19 bp on tail length (P = 0.001) in TS, were highly significant (P < 0.01).
Table 4. Relationship between the different indel locus of PRNP gene and growth related traits in different breeds (LSMa ± SE).
Furthermore, the results of the intersubjectivity effect test on genotype and breed-affecting traits revealed that the influence of I2-15 bp on CaCI (P = 0.034), and 3′ UTR-7 bp on ChC (P = 1.122e-4) and ChCI (P = 1.234e-9), were significantly correlated among varieties. Nevertheless, interactions between genotypes and breeds were non-existent.
Discussion
In the PubMed database, there are thousands of studies on PRNP regulating fatal TSEs, while studies on PRNP affecting production performance are in the hundreds. Apart from regulating disease, the genetic mutations of PRNP also affect production traits in healthy ruminants, such as the dairy traits in goats [Citation3] and sheep [Citation4, Citation19–20]. Simultaneously, codon polymorphisms within the sheep PRNP gene influence ewe reproductive performance [Citation13–15], lamb growth traits [Citation16–18], and the seasonal mobilization of body reserves [Citation21]. Hence, the process of economic sheep breeding could be accelerated through PRNP polymorphism loci selection. Nevertheless, existing studies on the associated polymorphism of the ovine PRNP gene are mostly focused on SNP or haplotype mutations, while there are few studies on the indel mutation of this gene [Citation2, Citation15, Citation18, Citation40–41], especially in sheep. Hence, to our knowledge, this is the first study to identify four novel indels within the sheep PRNP in Chinese and Mongolian typical sheep breeds, as well as explore the genetic diversities and effects.
The genetic diversities of ovine PRNP, especially the exon polymorphisms, have been addressed thoroughly for decades, whereas the magnitude of intron and UTR mutation are often neglected. Herein, indel mutations in introns and the 3′ UTR were detected, and all were insertion mutations. According to the MAF value, the mutant frequencies of 3′ UTR-7 bp in HS and I1-7 bp in LFTS were low (MAF value < 0.01) while the four mutations in other species were high. Additionally, genotypes of the same indel loci were distributed differently among the five sheep breeds. This may be because the tested sheep were scattered across different geographic areas [Citation25–27]; environmental factors may have led to the differences in mutant frequencies and distribution. Moreover, most indels had moderate polymorphism (PIC value ≥ 0.3) in the Chinese sheep breeds. But this was not the case in Sartuul sheep, which may result in a lower degree of selection and breeding. Thus, I2-19 bp, with its moderate polymorphism (PIC value = 0.375) in Sartuul, could be crucial to research into improving the performance of this sheep breed. Furthermore, our results also found that some mutation loci (e.g. I1-7 bp in TS) were not at HWE in several tested breeds, which might be because of the small sample group, as well as long-term artificial selection in breeding. Because the causal mutation could be better captured by haplotype-based methods, it is important to note that the indel polymorphisms, whether they influence the phenotypic traits or disease, could be considered for haplotype investigation to elucidate the relationship between indels and the specific traits [Citation42].
Because all of the all analyzed sheep were healthy, and previously reported pathogenic codons (such as codons 136, 141, 146, 154, and 171) were not located in the studied indels, the present study cannot provide results on the relationship between these indels and scrapie susceptibility. As well as being considered a pathogenic control, PRNP has also become an emerging target gene for cancer therapies, because PrPc participate in the processes of intercellular junctions, tumor growth, and metastasis [Citation43–44]. Further cancer investigations revealed that PrPc contribute toward the self-renewal of embryonic, tissue-specific, and cancer stem cells [Citation44], which provides a potential basis for researching the PRNP regulation of animal phenotypic traits. Since PRNP polymorphisms reportedly affect the economic traits of healthy sheep [Citation45], phenotypic records were used to determine the relationship between indels and growth performance in this study.
In the present study, the association analyses confirm that four loci were significantly correlated with growth traits. Several associations were highly significant. Furthermore, individuals with homozygous mutation genotypes (II) often have better body trait measurement values than other genotypes. For instance, STHS rams, with genotype II in the I2-15 bp mutation, have a larger ChW than individuals with genotype ID or DD. Previous studies have indicated that mutations of basic sequences may alter mRNA stability, processing, and maturation, thereby affecting the allelic expression and co-translational folding pathway [Citation46]. Predicting that the binding sites of transcription factors might exist in the PRNP indel loci, the expression of other growth-related genes would be influenced by bonding with miRNA [Citation47]. For another, indel polymorphism in the 3′ UTR could modulate traits or disease susceptibility via a microRNA-mediated post-transcriptional mechanism or elements of DNA functions [Citation48]. Interaction between genes was also inferred as a hypothetical factor contributing to this correlation, for PRNP genotypes associated with the prion-like protein doppel (PRND) [Citation49], and polymorphic PRND had an impact on sheep growth traits [Citation34]. Although the polymorphisms of PRNP affecting growth traits in sheep had been identified [Citation50–51], the specific regulatory mechanisms and relationships between polymorphisms of PRND and mutations of PRNP still need to be further researched.
These findings may be a further step toward the detection of indel-based typing within and across sheep breeds, as well as the detection of potentially useful DNA markers for the selection of high-quality individuals with MAS for sheep breeding.
Disclosure of potential conflicts of interest
The authors certify that there is no conflict of interest with any financial organization regarding the material discussed in the manuscript.
Financial disclosure statements
The study was funded by the National Natural Science Foundation of China (No.31660642), Natural Science Foundation of Gansu Province (No.1610RJZA103), and Central Special Funds for Basic Research in Universities Operating Expenses of “An Excellent and Three Special” Discipline Construction (No.31920170170). We greatly thanked the staffs of TS Elite Reservation Farm (Baishui county, Shannxi Province), Ruilin Sci-Tech Culture and Breeding Limit Company (Yongjing county, Gansu Province), Shanshan Agriculture and Animal Husbandry Sci-tech Company (Mengjin county, Henan Province), and Zhawkhan Tsum Sureg Farm, (Tsagann Khairkhan Sum, Zhawkhan Province, Mongolia) for collecting samples.
Abbreviations
| BBH | = | body back height |
| BH | = | body height |
| BL | = | body length |
| BLI | = | body length index |
| BW | = | body weight |
| CaC | = | cannon circumference |
| CaCI | = | cannon circumference index |
| ChC | = | chest circumference |
| ChCI | = | chest circumference index |
| ChW | = | chest width |
| ChWI | = | chest width index |
| DD | = | deletion/deletion |
| He | = | heterozygosity |
| HL | = | head length |
| Ho | = | homozygosity |
| HS | = | Hu sheep |
| HWE | = | Hardy-Weinberg equilibrium |
| ID | = | insertion/deletion |
| II | = | insertion/insertion |
| indel | = | insertion/deletion |
| I1-7 bp | = | Intron-1-insertion-7 bp |
| I2-15 bp | = | Intron-2-insertion-15 bp |
| I2-19 bp | = | Intron-2-insertion-19 bp |
| LFTS | = | Lanzhou Fat-Tail sheep |
| MAS | = | application of marker assisted selection |
| ME | = | mathematical expectation |
| ORF | = | open reading frame |
| PCR | = | polymerase chain reaction |
| PIC | = | Polymorphism information content |
| PRND | = | prion-related doppel gene |
| PRNP, | = | Prion protein gene |
| PrPc | = | prion protein |
| SH | = | sacral height |
| SNPs | = | single nucleotide polymorphisms |
| SS | = | Sartuul sheep |
| STHS | = | Small Tail Han sheep |
| TL | = | tail length |
| TS | = | Tong sheep |
| TSE | = | transmissible spongiform encephalopathies |
| WL | = | wool length. |
| 3′ UTR | = | 3′ untranslated region |
| 3′ UTR-7 bp | = | 3′ UTR-insertion-7 bp |
Additional information
Funding
References
- Houston F, Goldmann W, Foster J, et al. Comparative susceptibility of sheep of different origins, breeds and PRNP genotypes to challenge with bovine spongiform encephalopathy and scrapie. PLoS One. 2015;10(11):e0143251. doi:10.1371/journal.pone.0143251.
- Stepanek O, Horin P. Genetic diversity of the prion protein gene (PRNP) coding sequence in Czech sheep and evaluation of the national breeding programme for resistance to scrapie in the Czech Republic. Animal Genetics. 2017;58:111–121.
- Lan XY, Zhao HY, Li ZJ, et al. A novel 28 bp insertion-deletion polymorphism within goat PRNP gene and its association with production traits in Chinese native breeds. Genome. 2012;55(7):547–552. doi:10.1139/g2012-040.
- Vitezica ZG, Beltran de Heredia I, Ugarte E. Short communication: Analysis of association between the prion protein (PRNP) locus and milk traits in Latxa dairy sheep. J Dairy Sci. 2013;96(9):6079–6083. doi:10.3168/jds.2013-6570.
- Yang Q, Zhang SH, Liu LL, et al. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: an example for evaluating 14 bp insertion/deletion (indel) within the bovine PRNP gene. Prion. 2016;10:409–419. doi:10.1080/19336896.2016.1211593.
- Yang Q, Zhang SH, Liu LL, et al. The evaluation of 23-bp and 12-bp insertion /deletion within the PRNP gene and their effects on growth traits in healthy Chinese native cattle breeds. J Applied Animal Res. 2017;(2):1–7. doi:10.1080/09712119.2017.1348950.
- de Andrade CP, de Almeida LL, de Castro LA, et al. Development of a real-time polymerase chain reaction assay for single nucleotide polymorphism genotyping codons 136, 154, and 171 of the prnp gene and application to Brazilian sheep herds. J Veterinary Diagnostic Invest. 2013;25(1):120–124. doi:10.1177/1040638712471343.
- Konold T, Phelan LJ, Donnachie BR, et al. Codon 141 polymorphisms of the ovine prion protein gene affect the phenotype of classical scrapie transmitted from goats to sheep. BMC Veterinary Res. 2017;13(1):122. doi:10.1186/s12917-017-1036-1.
- Papasavva-Stylianou P, Simmons MM, Ortiz-Pelaez A, et al. The effect of polymorphisms at codon 146 of the goat PRNP gene on susceptibility to challenge with classical scrapie by different routes. J Virol. 2017;91(22):e01142–17. doi:10.1128/JVI.01142-17.
- Seabury CM, Derr JN. Identification of a novel ovine PrP polymorphism and scrapie-resistant genotypes for St. Croix White and a related composite breed. Cytogenetic Genome Res. 2003;102:85–88. doi:10.1159/000075730.
- Zabavnik J, Cotman M, Juntes P, et al. A decade of using small-to-medium throughput allele discrimination assay to determine prion protein gene (Prnp) genotypes in sheep in Slovenia. J Veterinary Diagnostic Invest. 2017. doi:10.1177/1040638717723946.
- Saunders GC, Cawthraw S, Mountjoy SJ, et al. Ovine PRNP untranslated region and promoter haplotype diversity. J General Virol. 2009;90:1289–1293. doi:10.1099/vir.0.007997-0.
- Ponz R, Tejedor MT, Monteagudo LV, et al. Scrapie resistance alleles are not associated with lower prolificity in Rasa Aragonesa sheep. Res Veterinary Sci. 2006;81:37–39. doi:10.1016/j.rvsc.2005.10.001.
- Casellas J, Caja G, Bach R, et al. Association analyses between the prion protein locus and reproductive and lamb weight traits in Ripollesa sheep. J Animal Sci. 2007;85:592–597. doi:10.2527/jas.2006-308.
- Guan F, Pan L, Li J, et al. Polymorphisms of the prion protein gene and their effects on litter size and risk evaluation for scrapie in Chinese Hu sheep. Virus Genes. 2011;43(1):147–152. doi:10.1007/s11262-011-0609-5.
- Isler BJ, Freking BA, Thallman RM, et al. Evaluation of associations between prion haplotypes and growth, carcass, and meat quality traits in a Dorset x Romanov sheep population. J Animal Sci. 2006;84:783–788. doi:10.2527/2006.844783x.
- Sawalha RM, Brotherstone S, Man WYN, et al. Associations of polymorphisms of the ovine prion protein gene with growth, carcass, and computerized tomography traits in Scottish Blackface lambs. J Animal Sci. 2007;85:632–640. doi:10.2527/jas.2006-372.
- Sawalha RM, Villanueva B, Brotherstone S, et al. Prediction of prion protein genotype and association of this genotype with lamb performance traits of Suffolk sheep. Am Soc Animal Sci. 2010;88(2):428–434.
- Álvarez L, Gutiérrez-Gil B, San Primitivo F, et al. Influence of prion protein genotypes on milk production traits in Spanish Churra sheep. J Dairy Sci. 2006;89:1784–1791. doi:10.3168/jds.S0022-0302(06)72247-0.
- Psifidi A, Basdagianni Z, Dovas CI, et al. Characterization of the PRNP gene locus in Chios dairy sheep and its association with milk production and reproduction traits. Animal Genetics. 2011;42(4):406–414. doi:10.1111/j.1365-2052.2010.02159.x.
- Sawalha RM, Brotherstone S, Lambe NR, et al. Association of the prion protein gene with individual tissue weights in Scottish Blackface sheep. J Animal Sci. 2008;86:1737–1746. doi:10.2527/jas.2007-0650.
- Zhao H, Du Y, Chen SM, et al. The prion protein gene polymorphisms associated with bovine spongiform encephalopathy susceptibility differ significantly between cattle and buffalo. Infect Genetics Evolution. 2015;36:531–538. doi:10.1016/j.meegid.2015.08.031.
- Yaman Y, Karadağ O, Ün C. Investigation of the prion protein gene (PRNP) polymorphisms in Anatolian, Murrah, and crossbred water buffaloes (Bubalus bubalis). Tropical Animal Health Production. 2017;49(2):427–430. doi:10.1007/s11250-016-1185-4.
- Xu XC, Li BB, Wei X, et al. Differential expression of peroxisome proliferator-activated receptor γ, fatty acid synthase, and hormone-sensitive lipase in fat-tailed and thin-tailed sheep breeds. Genetics Mol Res. 2015;14(4):15624–15633. doi:10.4238/2015.December.1.14.
- Cheng X, Zhao SG, Yue Y, et al. Comparative analysis of the liver tissue transcriptomes of Mongolian and Lanzhou fat-tailed sheep. Genetics Mol Res. 2016;15(2): e15028572. doi:10.4238/gmr.15028572.
- Sun W, Chang H, Yang ZP, et al. Hussein, Analysis on the origin and phylogenetic status of Tong Sheep using 12 blood protein and nonprotein markers. J Genetics Genomics. 2007;34(12):1097–1105. doi:10.1016/S1673-8527(07)60125-8.
- Sun LW, Guo YX, Fan YX, et al. Metabolic profiling of stages of healthy pregnancy in Hu sheep using nuclear magnetic resonance (NMR). Theriogenology. 2017;92:121–128. doi:10.1016/j.theriogenology.2017.01.025.
- Zhao HD, He S, Zhu YJ, et al. A novel 29 bp insertion/deletion (indel) variant of the LHX3 gene and its influence on growth traits in four sheep breeds of various fecundity. Archives Animal Breeding. 2017;60:79–85. doi:10.5194/aab-60-79-2017.
- Lan XY, Pan CY, Chen H, et al. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Ruminant Res. 2007;73(1):8–12. doi:10.1016/j.smallrumres.2006.10.009.
- Lan XY, Zhao HY, Li ZJ, et al. Exploring the novel genetic variant of PITX1 gene and its effect on milk performance in dairy goats. J Integrative Agriculture. 2013;12(1):118–126. doi:10.1016/S2095-3119(13)60212-9.
- Jia WC, Wu XF, Li XC, et al. Novel genetic variants associated with mRNA expression of signal transducer and activator of transcription 3 (STAT3) gene significantly affected goat growth traits. Small Ruminant Res. 2015;129:25–36. doi:10.1016/j.smallrumres.2015.05.014.
- Zhang SH, Sun K, Bian YN, Zhao Q, Wang Z, Ji CN, Li CT. Developmental validation of an X-Insertion/Deletion polymorphism panel and application in HAN population of China. Scientific Reports. 2015;5:18336. doi:10.1038/srep18336.
- Zhang XY, Wu XF, Jia WC, et al. Novel nucleotide variations, haplotypes structure and associations with growth related traits of goat at motif-binding factor (ATBF1) gene. Asian-Australasian J Animal Sci. 2015;28(10):1394–1406. doi:10.5713/ajas.14.0860.
- Li J, Zhu XC, Ma L, Xu HW, et al. Detection of a new 20bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion. 2017;11(2):143–150. doi:10.1080/19336896.2017.1300740.
- Czarnik U, Grzybowski G, Zabolewicz T, et al. Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish red cattle, Polish White-backed cattle and European bison (Bison bonasus L., 1758). Genetika. 2009;45(4):519–525.
- Pan CY, Wu CY, Jia WC, et al. A critical functional missense mutation (H173R) in the bovine PROP1 gene significantly affects growth traits in cattle. Gene. 2013;531(2):398–402. doi:10.1016/j.gene.2013.09.002.
- Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19(4):519–523. doi:10.1038/cr.2009.33.
- Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi:10.1038/sj.cr.7290272.
- Li Y, Wang K, Jiang YZ, et al. 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inhibits human ovarian cancer cell proliferation. Cell Oncol (Dordr). 2014;37(6):429–437. doi:10.1007/s13402-014-0206-4.
- Pritchard TC, Cahalan CM, Ap Dewi I. Association between PrP genotypes and performance traits in a Welsh mountain flock. Animal. 2008;2(10):1421–1426. doi:10.1017/S1751731108002747.
- Moore RC, Boulton K, Bishop SC. Associations of PrP genotype with lamb production traits in three commercial breeds of British hill sheep. Animal. 2009;3(3):336–346. doi:10.1017/S1751731108003637.
- Huo ZX, Luo XG, Zhan XN, et al. Genetic analysis of indel markers in three loci associated with Parkinson's disease. PLoS One. 2017;12(9):e0184269. doi:10.1371/journal.pone.0184269.
- Rousset M, Leturque A, Thenet S. The nucleo-junctional interplay of the cellular prion protein: A new partner in cancer-related signaling pathways? Prion. 2016;10(2):143–152. doi:10.1080/19336896.2016.1163457.
- Santos TG, Lopes MH, Martins VR. Targeting prion protein interactions in cancer. Prion. 2015;9(3):165–173. doi:10.1080/19336896.2015.1027855.
- Alexander BM, Stobart RH, Russell WC, et al. The incidence of genotypes at codon 171 of the prion protein gene (PRNP) in five breeds of sheep and production traits of ewes associated with those genotypes. J Animal Sci. 2005;83(2):455–459. doi:10.2527/2005.832455x.
- Komar AA. Genetics. SNPs, silent but not invisible. Science. 2007;315:466–467. doi:10.1126/science.1138239.
- Hou JX, An XP, Song YX, et al. Two mutations in the caprine MTHFR 3’ UTR regulated by microRNAs are associated with milk production traits. PLoS One. 2015;10(7):e0133015. doi:10.1371/journal.pone.0133015.
- Peletto S, Bertolini S, Maniaci MG, et al. Association of an indel polymorphism in the 3’UTR of the caprine SPRN gene with the scrapie positivity in the central nervous system. J General Virol. 2012;93:1620–1623. doi:10.1099/vir.0.041400-0.
- Mesquita P, Garcia V, Marques MR, et al. The prion-related protein (testis-specific) gene (PRNT) is highly polymorphic in Portuguese sheep. Animal Genet. 2015;47:128–132. doi:10.1111/age.12380.
- Vitezica ZG, Moreno CR, Lantier F, Lantier I, Schibler L, Roig A, François D, Bouix J, Allain D, Brunel JC, Barillet F, Elsen JM. Quantitative trait loci linked to PRNP gene controlling health and production traits in INRA 401 sheep. Genet Sel Evol. 2007;39:421–430. doi:10.1186/1297-9686-39-4-421.
- Sweeney T, Hanrahan JP. The evidence of associations between prion protein genotype and production, reproduction, and health traits in sheep. Veterinary Res. 2008;39:28. doi:10.1051/vetres:2008004.