ABSTRACT
The yeast [PSI+] prion, which is the amyloid form of Sup35, has the unusual property of being cured not only by the inactivation of, but also by the overexpression of Hsp104. Even though this latter observation was made more than two decades ago, the mechanism of curing by Hsp104 overexpression has remained controversial. This question has been investigated in depth by our laboratory by combining live cell imaging of GFP-labeled Sup35 with standard plating assays of yeast overexpressing Hsp104. We will discuss why the curing of [PSI+] by Hsp104 overexpression is not compatible with a mechanism of either inhibition of severing of the prion seeds or asymmetric segregation of the seeds. Instead, our recent data (J. Biol. Chem. 292:8630-8641) indicate that curing is due to dissolution of the prion seeds, which in turn is dependent on the trimming activity of Hsp104. This trimming activity decreases the size of the seeds by dissociating monomers from the fibers, but unlike Hsp104 severing activity, it does not increase the number of prion seeds. Finally, we will discuss the other factors that affect the curing of [PSI+] by Hsp104 overexpression and how these factors may relate to the trimming activity of Hsp104.
KEYWORDS:
It has been more than two decades since it was discovered that unlike other yeast prions, the [PSI+] yeast prion has an unusual property: it is cured both by overexpression of Hsp104 and inactivation of Hsp104 [Citation1]. [PSI+] is the infectious amyloid form of Sup35, a protein that functions in translation termination [Citation2,Citation3]. In yeast, the infectious prion conformation is transmitted from mother to daughter cells as amyloid seeds. Since Hsp104 severs the prion seeds, it is not surprising that inactivating Hsp104 cures yeast prions. However, it seems contradictory that overexpression of Hsp104 cures [PSI+] as well. To cure yeast of the misfolded amyloid form of the prion, all the prion seeds must be eliminated. This can occur by several different mechanisms: (1) inhibition of seed severing (2) asymmetric segregation of the seeds between mother and daughter cells and (3) dissolution of the seeds. Over the years, different studies have supported each of these mechanisms to explain the curing of [PSI+] by Hsp104 overexpression, which highlights the controversy surrounding this question. We will review the data from other laboratories that lead them to propose either the first or second mechanisms for curing and discuss why our results lead us to rule out these mechanisms and instead support a dissolution mechanism for the curing of [PSI+] by Hsp104 overexpression. Finally, questions that remain regarding the curing of [PSI+] by Hsp104 overexpression will be discussed.
Curing by inhibition of severing or asymmetric segregation of the seeds
Winkler et al proposed that overexpression of Hsp104 cures [PSI+] by inhibiting severing of the prion seeds [Citation4]. This proposal was based on their observation that in addition to Hsp104 binding to the Sup35 amyloid conformation in an Ssa1-dependent manner, which produces severing of the prion seeds [Citation5-7], it also binds in an Ssa1-independent manner. Furthermore, when the N-terminal domain of Hsp104 was deleted, the truncated Hsp104 only bound to the misfolded Sup35 conformation in an Ssa1-dependent manner, which may explain the previous observation that [PSI+] is not cured by overexpression of this Hsp104 fragment [Citation8]. Based on these results, they proposed that when Hsp104 is overexpressed, the excess Hsp104 would bind to the Sup35 in an Ssa1-independent manner, and this in turn, would sterically block the Ssa1-dependent binding of Hsp104 that is responsible for the severing of the prion seeds. This model predicts that since severing is inhibited, there should be a pronounced lag prior to curing as the prion seeds are diluted out by cell division similar to that observed when [PSI+] is cured by inhibiting Hsp104 activity with guanidine [Citation9]. However, the curing kinetics of [PSI+] by Hsp104 overexpression does not show this pronounced lag [Citation10-13], as illustrated in . Further evidence against overexpression of Hsp104 cures [PSI+] by inhibiting the severing of the prion seeds is the observation that [PSI+] is cured by overexpression of Hsp104 even when the severing activity of Hsp104 is already inhibited by guanidine [Citation14,Citation15]. Therefore, overexpression of Hsp104 cannot be acting by inhibiting the severing activity of Hsp104.
Figure 1. Comparison of the kinetics of curing [PSI+] by inactivation of Hsp104 and overexpression of Hsp104. The rate of curing of the L2888[PSI+] variant was measured after inactivating Hsp104 with 5 mM guanidine (open circles) or overexpressing Hsp104 from the GAL1 promoter (closed circles).
![Figure 1. Comparison of the kinetics of curing [PSI+] by inactivation of Hsp104 and overexpression of Hsp104. The rate of curing of the L2888[PSI+] variant was measured after inactivating Hsp104 with 5 mM guanidine (open circles) or overexpressing Hsp104 from the GAL1 promoter (closed circles).](/cms/asset/6a2d6388-f779-4f8a-9744-c5bc06a2347d/kprn_a_1412911_f0001_b.gif)
An alternative mechanism for the curing of [PSI+] by Hsp104 overexpression is asymmetric segregation of the prion seeds, a mechanism strongly supported by the Tuite laboratory [Citation15,Citation16]. In this mechanism, there is reduced transmission of seeds to the daughter cells during cell division, which, in turn, results in faster curing of the daughter cells relative to the mother cells. This can be achieved by increasing the size of or aggregating the seeds. In fact, an increase in the size of the seeds was suggested based on semi-denaturing gels of denatured Sup35 prepared from [PSI+] yeast overexpressing Hsp104. These gels showed that Hsp104 overexpression reduced the mobility of Sup35 from [PSI+] yeast, suggesting an increase in size of the SDS-resistant aggregate [Citation17]. Further evidence for an asymmetric model of segregation comes from the Tuite laboratory who observed that during the curing of [PSI+] by Hsp104 overexpression, the mother cells had a significantly greater number of seeds than the daughter cells at 5.7 generations when the yeast were about 50% cured [Citation15].
On the other hand, the following facts do not fit with an asymmetric model of curing of [PSI+] by Hsp104 overexpression. First, in contrast to the earlier study mentioned above using semi-denaturing gels [Citation17], the Tuite laboratory did not find a change in mobility of the SDS-resistant Sup35 aggregates in [PSI+] yeast overexpressing Hsp104 [Citation15]. Second, it is not clear what the SDS-resistant aggregates represent on the semi-denaturing gels; i.e. whether they are the intact prion seeds themselves or just small amyloid polymers from the seeds that we will refer to as the core of the fiber. Third, the Tuite laboratory did not observe asymmetric seed segregation at 3.6 generations when the yeast were about 30% cured [Citation15]. Fourth, even if there were a slight asymmetric bias in the transmission of the seeds, this may not result in significantly faster curing of the daughter cells relative to the mother cells because one seed remaining in the daughter cell is sufficient to generate a [PSI+] colony. Fifth and most importantly, our previous study determined that daughter cells and mother cells cure at the same rate in [PSI+] yeast overexpressing Hsp104 when fluorescence-activated cell sorting (FACS) was used to separate mother cells from their progeny or to separate the newborn cells (with a birth scar) from the old cells (with numerous bud scars) [Citation12]. The plating assay showed no significant difference in the extent of curing among the sorted populations. Therefore, these results do not support a mechanism of asymmetric segregation for the curing of [PSI+] by Hsp104 overexpression.
Curing by dissolution
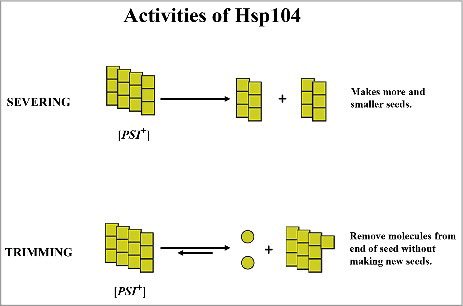
A dissolution model in which Hsp104 causes the loss of seeds by dissociating monomers from the misfolded amyloid form of Sup35 is consistent with our data [Citation12,Citation13]. A dissolution model for curing [PSI+] by Hsp104 overexpression was originally proposed by Paushkin et al [Citation18], who observed an increase in the soluble pool of Sup35 in [PSI+] yeast overexpressing Hsp104. Our data supporting a dissolution model are largely based on the imaging of GFP-labeled Sup35 in [PSI+] yeast, which revealed that in a process, which we called trimming, Hsp104 reduces the size of the seeds without increasing their number. In contrast, the severing activity of Hsp104 decreases the size of the seeds, but also increases their number (see model in ). The trimming activity of Hsp104 first became evident when we examined the curing of [PSI+] by guanidine inactivation of Hsp104 using yeast expressing GFP-labeled Sup35 [Citation19]. Imaging showed that the fluorescent foci were no longer detectable in yeast even though plating assays showed that the yeast were still [PSI+]. The decrease in size of the foci was confirmed by measuring the diffusional mobility of GFP-labeled Sup35 [Citation19]. Importantly, the foci again became detectable after stress if the yeast were still [PSI+], but not in the cured [psi−] cells [Citation20]. This shows that the trimming reaction is reversible; the Sup35 monomers both dissociate and rebind to the fibers. Therefore, trimming of the prion seeds is detected when the rate of dissociation of the monomers is faster than their rate of rebinding.
Interestingly, just as was observed for the curing of [PSI+] in guanidine, overexpression of Hsp104 produced a loss of detectable foci in yeast that plating assays showed were still [PSI+] [Citation12]. Furthermore, the foci were again readily detected after the cells were stressed. This is illustrated in , which shows confocal images of maximized Z-stacks of GFP-labeled Sup35 in [PSI+] yeast before and after induction of Hsp104 overexpression. The images show a loss of detectable foci in most cells even though plating assays showed the yeast are still 80% [PSI+]. After stressing the yeast by starvation, the foci again became visible in most of the cells. These results suggest that overexpression of Hsp104 increases the trimming of the prion seeds, which in turn causes a reduction in their size as well as an increase in the soluble pool of Sup35 monomers. Consistent with our in vivo observations, in vitro studies support a dissolution model; these studies showed that the Sup35 amyloid is dissolved by high concentrations of Hsp104. [Citation21,Citation22] This dissolution did not occur with high concentrations of the N-terminal deletion fragment of Hsp104, [Citation22] a fragment that does not cure [PSI+] when overexpressed [Citation8].
Figure 3. Trimming of the GFP-labeled Sup35 foci in [PSI+] cells by overexpression of Hsp104. 1074 [PSI+] yeast, which expresses GFP-labeled Sup35 from the chromosomal SUP35 locus, was imaged prior to Hsp104 overexpression (panel a). Hsp104 was overexpressed for 1 generation using the tetracycline inducible expression system.[Citation12] Images were obtained before stress (panel b) and after stress (panel c). Maximized images of confocal Z-stacks obtained under the identical settings.
![Figure 3. Trimming of the GFP-labeled Sup35 foci in [PSI+] cells by overexpression of Hsp104. 1074 [PSI+] yeast, which expresses GFP-labeled Sup35 from the chromosomal SUP35 locus, was imaged prior to Hsp104 overexpression (panel a). Hsp104 was overexpressed for 1 generation using the tetracycline inducible expression system.[Citation12] Images were obtained before stress (panel b) and after stress (panel c). Maximized images of confocal Z-stacks obtained under the identical settings.](/cms/asset/9417c5b1-54a3-49ea-a7d9-6d59a5c7fb6d/kprn_a_1412911_f0003_b.gif)
The relationship between Hsp104 trimming activity and curing of [PSI+] by Hsp104 overexpression
What factors affect trimming of the prion seeds by Hsp104 and therefore, in turn, affect [PSI+] curing by Hsp104 overexpression? One factor that affects trimming of the prion seeds is the protein Ssa1, a member of the Hsp70 family. Overexpression of Ssa1 inhibits trimming of the prion seeds by Hsp104 [Citation19], and as shown previously, reduces the rate of [PSI+] curing by Hsp104 overexpression [Citation23]. Another factor that affects trimming is the [PSI+] variant. Weak [PSI+] variants have fewer seeds, more soluble Sup35 monomers and different biophysical properties than strong variants. When Hsp104 was overexpressed, we observed faster trimming of the prion seeds of a weak [PSI+] variant than of a strong [PSI+] variant [Citation13], and consistent with this observation, Hsp104 overexpression cured the weak variants at a faster rate than the strong variants [Citation24]. Trimming is also affected by the properties of Hsp104, itself. Our model predicts that a mutation of Hsp104 that prevents trimming should also prevent curing of [PSI+] by overexpression of this mutant Hsp104. We determined that this was indeed the case for two different point mutants of Hsp104, Hsp104(D184S) and Hsp104(T160M) [Citation12]. In addition, overexpression of Hsp104 orthologs from Schizosaccharomyces pombe and Candida albicans did not show detectable trimming of the prion seeds of the strong [PSI+] variants and consistent with this observation, overexpression of these Hsp104 orthologs did not cure the strong [PSI+] variants [Citation13]. In contrast to what occurred with the strong [PSI+] variants, these Hsp104 orthologs trimmed the prion seeds of the weak [PSI+] variants and therefore, as expected, overexpression of these Hsp104 orthologs also cured the weak [PSI+] variants.
The difference in the ability of these Hsp104 orthologs to cure the weak and strong [PSI+] variants reconciles previous studies on the overall ability of the Hsp104 orthologs to cure [PSI+]. The Tuite laboratory showed that overexpression of Hsp104 from C. albicans cures [PSI+] [Citation25], whereas no curing was observed with overexpression of Hsp104 from S. pombe [Citation26,Citation27]. In the former study, the variants used must have been weaker than the strong [PSI+] variants used in our study, whereas in the latter study, relatively strong [PSI+] variants must have been used.
As our dissolution model predicts, overexpression of Hsp104 does not cure [PSI+] in the absence of net trimming. However, there were several conditions in which we observed trimming and either no curing or very slow curing by Hsp104 overexpression. One condition was low levels of S. cerevisiae Hsp104 overexpression in strong [PSI+] variants. There was a large amount of partial trimming in about half the population, while no obvious trimming occurred in the remaining population.[Citation13] This variability in trimming was first observed several generations after the induction of Hsp104 overexpression and then persisted for 20 generations at which time the yeast were still >95% [PSI+] based on plating assays. The variable extent of trimming in these cells is probably due to a variable number of Hsp104 expressing centromeric plasmids, which could vary from one to three plasmids per cell [Citation28]. As for the observation that, even after 20 generations, there is only partial trimming of the seeds in some of the cells, this must be due to the reversibility of Sup35 dissociation from the seeds. If the Sup35 monomers could not rebind to the seeds, even very slow trimming of the seeds would go to completion and most of the cells would cure over the 20-generation time span.
Another situation where we observed trimming by Hsp104, but no concomitant curing, was curing in a mutant strain defective in proteasome assembly; here there was trimming of the prion seeds when Hsp104 was overexpressed, but curing was very much delayed relative to this trimming [Citation12]. Although it is not clear why inhibiting proteasome activity reduces the rate of [PSI+] curing by Hsp104 overexpression, these results are consistent with the reduction in the rate of [PSI+] curing by Hsp104 overexpression caused by the deletion of genes encoding either the ubiquitin-conjugating enzyme, Ubc4, or the ubiquitin-recycling enzyme, Ubp6 [Citation29]. Conversely, raising the ubiquitin levels in the cell was found to increase the rate of curing [PSI+] by Hsp104 overexpression [Citation10]. These data suggest that at least under certain conditions, e.g. with strong [PSI+] variants, trimming alone is not sufficient for curing. It has been suggested that certain [PSI+] variants have a core of small amyloid polymers, which have been identified on semi-denaturing gels and visualized by electron microscopy [Citation17,Citation30]. It may be that trimming dissociates Sup35 monomers from the prion fibers, but the core must be digested by the proteasome to ultimately cure [PSI+] as depicted in the model shown in .
Figure 4. Model of the curing of [PSI+] by Hsp104 overexpression. Curing occurs in a 2-step process with first trimming of the amyloid fiber followed by proteolysis of the amyloid core, consisting of small amyloid polymers. The rate of trimming (step 1) is increased by Hsp104 overexpression and decreased by Ssa1 overexpression. As for other factors that inhibit curing of [PSI+] by Hsp104 overexpression, including expressing Sis1 without its dimerization domain or deleting the following proteins: Sti1, Ssbs, or members of the Hsp90 family, it has yet to be determined which step is affected in this model.
![Figure 4. Model of the curing of [PSI+] by Hsp104 overexpression. Curing occurs in a 2-step process with first trimming of the amyloid fiber followed by proteolysis of the amyloid core, consisting of small amyloid polymers. The rate of trimming (step 1) is increased by Hsp104 overexpression and decreased by Ssa1 overexpression. As for other factors that inhibit curing of [PSI+] by Hsp104 overexpression, including expressing Sis1 without its dimerization domain or deleting the following proteins: Sti1, Ssbs, or members of the Hsp90 family, it has yet to be determined which step is affected in this model.](/cms/asset/b150b6f3-38e8-43bc-b55c-b6e632a88b1d/kprn_a_1412911_f0004_c.jpg)
Additional factors that affect curing of [PSI+] by Hsp104 overexpression
There are other chaperones and cochaperones that affect the rate of [PSI+] curing by Hsp104 overexpression, but their effect on trimming has not yet been determined. Overexpression of Ssb1, a member of the Hsp70 family, increased the rate of curing and conversely, deleting both members of the Ssb family decreased the rate of [PSI+] curing by Hsp104 overexpression [Citation31]. Since it has been suggested that the Ssb family of proteins compete with the Ssa family of proteins in binding to prion fibers [Citation32], and overexpressing Ssa1 inhibits trimming of the prions seeds [Citation19], overexpressing Ssb1 should presumably increase trimming, but this has yet to be determined. Sis1, a member of the Hsp40 family, also plays an important role in [PSI+] curing by Hsp104 overexpression as shown by the fact that deletion of its dimerization domain causes a marked inhibition of [PSI+] curing [Citation33]. However, since Sis1 affects proteasome degradation [Citation34,Citation35], it is not clear whether the effect of Sis1 on the curing [PSI+] by Hsp104 overexpression is due to altering trimming or proteasome degradation. In addition, Sti1 and Cpr7, two tetratricopeptide cochaperones that bind to both Hsp70 and Hsp90, also function in [PSI+] curing by Hsp104 overexpression [Citation10,Citation11]. Specifically, deletion of CPR7 caused a modest inhibition in the rate of [PSI+] curing while deletion of STI1 caused a marked inhibition in this rate. Interestingly, these two cochaperones were identified in a suppressor screen of Ssa1-21, a point mutant of Ssa1 that causes mitotic instability of [PSI+] [Citation5]. This implies that the mitotic instability of [PSI+] caused by Ssa1–21 may be related to the curing of [PSI+] by Hsp104 overexpression [Citation10].
To add to the complex interactions involved in curing of [PSI+] by Hsp104 overexpression, Hsc80 and Hsp82, members of the Hsp90 family also affect curing of [PSI+] by Hsp104 overexpression; the rate of curing was decreased by genomic deletions of either HSP82 or HSC82 [Citation10]. It is not clear whether Hsp90 affects trimming or whether Hsp90 has an indirect effect on curing through its role in the assembly and maintenance of the 26 proteasome [Citation36]. Therefore, members of the Hsp40, Hsp70, and Hsp90 families all affect curing of [PSI+] by Hsp104 overexpression, but further work will be required to determine if they do so by affecting trimming or through some other mechanism.
In conclusion, most of the experimental evidence now supports a dissolution model for the curing of [PSI+] by Hsp104 overexpression in which the dissolution is caused by the trimming activity of Hsp104. Structural studies suggest that in addition to the N-terminal domain of Hsp104 [Citation8], there is a region in the middle domain of Sup35 that is necessary for the curing of [PSI+] by Hsp104 overexpression, but not for propagation of [PSI+] [Citation37]. Perhaps the N-terminal domain of Hsp104 interacts with this region of Sup35 to trim the prion seeds. So far, the [PSI+] prion is the only yeast prion that has been shown to be trimmed by Hsp104. Since Sup35 is an essential protein, it is tempting to speculate that the trimming activity of Hsp104 may have evolved to maintain a steady-state concentration of Sup35 monomers that is required for cellular viability. Curing of [PSI+] by Hsp104 overexpression may then just be a consequence of the inherent trimming activity of Hsp104. Interestingly, recent studies have shown that some [PSI+] variants are cured by either very low levels of Hsp104 overexpression or even endogenous Hsp104 levels. [Citation13,Citation38] Finally, there are still the unanswered questions as to whether weak and strong [PSI+] variants show the same chaperone dependence for curing, whether trimming alone can cure some [PSI+] variants, and in a related question, what is the role of the various chaperones as well as the proteasome in curing [PSI+] by Hsp104 overexpression. Until we can answer these questions, the complete mechanism of [PSI+] curing by Hsp104 overexpression will remain a puzzle with only clues as to how the various pieces of this puzzle fit together.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Chernoff YO, Lindquist SL, Ono B, et al. Role of the chaperone protein hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 1995;268(5212):880–884
- Patino MM, Liu JJ, Glover JR, et al. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273(5275):622–626. doi:10.1126/science.273.5275.622
- Stansfield I, Jones KM, Kushnirov VV, et al. The products of the sup45 (erf1) and sup35 genes interact to mediate translation termination in saccharomyces cerevisiae. EMBO J. 1995;14(17):4365–4373
- Winkler J, Tyedmers J, Bukau B, et al. Hsp70 targets hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198(3):387–404. doi:10.1083/jcb.201201074
- Jung G, Jones G, Wegrzyn RD, et al. A role for cytosolic hsp70 in yeast [[psi+] prion propagation and [psi+] as a cellular stress. Genetics. 2000;156(2):559–570
- Tipton KA, Verges KJ, Weissman JS. In vivo monitoring of the prion replication cycle reveals a critical role for sis1 in delivering substrates to hsp104. Mol Cell. 2008;32(4):584–591
- Lee J, Kim JH, Biter AB, et al. Heat shock protein (hsp) 70 is an activator of the hsp104 motor. Proc Natl Acad Sci U S A. 2013;110(21):8513–8518. doi:10.1073/pnas.1217988110
- Hung G-C, Masison DC. N-terminal domain of yeast hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by hsp104 overexpression. Genetics. 2006;173(2):611–620. doi:10.1534/genetics.106.056820
- Ness F, Ferreira P, Cox BS, et al. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol Cell Biol. 2002;22(15):5593–5605. doi:10.1128/MCB.22.15.5593-5605.2002
- Reidy M, Masison DC. Sti1 regulation of hsp70 and hsp90 is critical for curing of saccharomyces cerevisiae [psi+] prions by hsp104. Mol Cell Biol. 2010;30(14):3542–3552
- Moosavi B, Wongwigkarn J, Tuite MF. Hsp70/hsp90 co-chaperones are required for efficient hsp104-mediated elimination of the yeast [psi+] prion but not for prion propagation. Yeast. 2010;27(3):167–179
- Park YN, Zhao X, Yim YI, et al. Hsp104 overexpression cures saccharomyces cerevisiae [psi+] by causing dissolution of the prion seeds. Eukaryot Cell. 2014;13(5):635–647. doi:10.1128/EC.00300-13
- Zhao X, Rodriguez R, Silberman RE, et al. Heat shock protein 104 (hsp104)-mediated curing of [psi+] yeast prions depends on both [psi+] conformation and the properties of the hsp104 homologs. J Biol Chem. 2017;292(21):8630–8641. doi:10.1074/jbc.M116.770719
- Zhao X, Park YN, Todor H, et al. Sequestration of sup35 by aggregates of huntingtin fragments causes toxicity of [psi+] yeast. J Biol Chem. 2012;287(28):23346–23355. doi:10.1074/jbc.M111.287748
- Ness F, Cox BS, Wongwigkarn J, et al. Over-expression of the molecular chaperone hsp104 in saccharomyces cerevisiae results in the malpartition of [psi+] propagons. Mol Microbiol. 2017;104(1):125–143. doi:10.1111/mmi.13617
- Cox B, Tuite M. The life of [psi]. Curr Genet. 2017;:. doi:10.1007/s00294-017-0714-7.
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, et al. Yeast [psi+] prion aggregates are formed by small sup35 polymers fragmented by hsp104. J Biol Chem. 2003;278(49):49636–49643. doi:10.1074/jbc.M307996200
- Paushkin SV, Kushnirov VV, Smirnov VN, et al. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the sup35-encoded polypeptide chain release factor. EMBO J. 1996;15(12):3127–3134
- Park YN, Morales D, Rubinson EH, et al. Differences in the curing of [psi+] prion by various methods of hsp104 inactivation. PLoS One. 2012;7(6):e37692. doi:10.1371/journal.pone.0037692
- Greene LE, Park YN, Masison DC, et al. Application of gfp-labeling to study prions in yeast. Protein Pept Lett. 2009;16(6):635–641. doi:10.2174/092986609788490221
- Shorter J, Lindquist S. Destruction or potentiation of different prions catalyzed by similar hsp104 remodeling activities. Mol Cell. 2006;23(3):425–438. doi:10.1016/j.molcel.2006.05.042
- Sweeny EA, Jackrel ME, Go MS, et al. The hsp104 n-terminal domain enables disaggregase plasticity and potentiation. Mol Cell. 2015;57(5):836–849. doi:10.1016/j.molcel.2014.12.021
- Newnam GP, Wegrzyn RD, Lindquist SL, et al. Antagonistic interactions between yeast chaperones hsp104 and hsp70 in prion curing. Mol Cell Biol. 1999;19(2):1325–1333. doi:10.1128/MCB.19.2.1325
- Derkatch IL, Chernoff YO, Kushnirov VV, et al. Genesis and variability of [psi] prion factors in saccharomyces cerevisiae. Genetics. 1996;144(4):1375–1386
- Zenthon JF, Ness F, Cox B, et al. The [psi+] prion of saccharomyces cerevisiae can be propagated by an hsp104 orthologue from candida albicans. Eukaryot Cell. 2006;5(2):217–225. doi:10.1128/EC.5.2.217-225.2006
- Senechal P, Arseneault G, Leroux A, et al. The schizosaccharomyces pombe hsp104 disaggregase is unable to propagate the [psi] prion. PLoS One. 2009;4(9):e6939. doi:10.1371/journal.pone.0006939
- Reidy M, Sharma R, Masison DC. Schizosaccharomyces pombe disaggregation machinery chaperones support saccharomyces cerevisiae growth and prion propagation. Eukaryot Cell. 2013;12(5):739–745. doi:10.1128/EC.00301-12
- Jordan BE, Mount RC, Hadfield C. Determination of plasmid copy number in yeast. Methods Mol Biol. 1996;53:193–203
- Allen KD, Chernova TA, Tennant EP, et al. Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem. 2007;282(5):3004–3013. doi:10.1074/jbc.M609597200
- Bagriantsev SN, Gracheva EO, Richmond JE, et al. Variant-specific [psi+] infection is transmitted by sup35 polymers within [psi+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19(6):2433–2443. doi:10.1091/mbc.E08-01-0078
- Chernoff YO, Newnam GP, Kumar J, et al. Evidence for a protein mutator in yeast: Role of the hsp70-related chaperone ssb in formation, stability, and toxicity of the [psi] prion. Mol Cell Biol. 1999;19(12):8103–8112. doi:10.1128/MCB.19.12.8103
- Chernova TA, Wilkinson KD, Chernoff YO. Prions, chaperones, and proteostasis in yeast. Cold Spring Harbor perspectives in biology. 2017;9(2):a023663. doi:10.1101/cshperspect.a023663
- Kirkland PA, Reidy M, Masison DC. Functions of yeast hsp40 chaperone sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics. 2011;188(3):565–577. doi:10.1534/genetics.111.129460
- Park SH, Kukushkin Y, Gupta R, et al. Polyq proteins interfere with nuclear degradation of cytosolic proteins by sequestering the sis1p chaperone. Cell. 2013;154(1):134–145. doi:10.1016/j.cell.2013.06.003
- Summers DW, Wolfe KJ, Ren HY, et al. The type ii hsp40 sis1 cooperates with hsp70 and the e3 ligase ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8(1):e52099. doi:10.1371/journal.pone.0052099
- Imai J, Maruya M, Yashiroda H, et al. The molecular chaperone hsp90 plays a role in the assembly and maintenance of the 26s proteasome. EMBO J. 2003;22(14):3557–3567. doi:10.1093/emboj/cdg349
- Helsen CW, Glover JR. Insight into molecular basis of curing of [psi+] prion by overexpression of 104-kda heat shock protein (hsp104). J Biol Chem. 2012;287(1):542–556. doi:10.1074/jbc.M111.302869
- Gorkovskiy A, Reidy M, Masison DC, Wickner RB. Hsp104 disaggregase at normal levels cures many [psi+] prion variants in a process promoted by sti1p, hsp90, and sis1p. Proc Natl Acad Sci U S A. 2017;114(21):E4193–E4202. doi:10.1073/pnas.1704016114