ABSTRACT
Misfolding of the normally folded prion protein of mammals (PrPC) into infectious fibrils causes a variety of diseases, from scrapie in sheep to chronic wasting disease (CWD) in cervids. The misfolded form of PrPC, termed PrPSc, or in this case PrPCWD, interacts with PrPC to create more PrPCWD. This process is not clearly defined but is affected by the presence and interactions of biotic and abiotic cofactors. These include nucleic acids, lipids, glycosylation, pH, and ionic character. PrPC has been shown to act as a copper-binding protein in vivo, though it also binds to other divalents as well. The significance of this action has not been conclusively elucidated. Previous reports have shown that metal binding sites occur throughout the N-terminal region of PrPC. Other cations like manganese have also been shown to affect PrPC abundance in a transcript-independent fashion. Here, we examined the ability of different divalent cations to influence the stability and in vitro conversion of two variants of PrP from elk (L/M132, 26–234). We find that copper and zinc de-stabilize PrP. We also find that PrP M132 exhibits a greater degree of divalent cation induced destabilization than L132. This supports findings that leucine at position 132 confers resistance to CWD, while M132 is susceptible. However, in vitro conversion of PrP is equally suppressed by either copper or zinc, in both L132 and M132 backgrounds. This report demonstrates the complex importance of ionic character on the PrPC folding pathway selection on the route to PrPSc formation.
Introduction
Chronic Wasting Disease (CWD) is a form of transmissible spongiform encephalopathy (TSE), a unique class of protein-transmissible neurodegenerative diseases [Citation1,Citation2] that affects cervids, including elk, deer, moose, and reindeer. It was first diagnosed in 1978 in captive mule deer and black-tailed deer [Citation3], and for the first time in elk in 1981 in wild-captured adults that had been maintained with other potentially TSE-carrying mammals [Citation4]. Affected elk present with excessive drooling, lowering of the head, teeth-grinding, and excessive, wasting weight loss [Citation4,Citation5]. To date, CWD is the only known prion disease to affect free-ranging wildlife [Citation3,Citation4].
The overall disease outcome from various genetic backgrounds of PrPC in cervids is not fully defined, though controlled experimental studies have shown that higher gene dosages of M132 generally confer susceptibility in deer and elk to CWD, while L132 genes confer relative resistance to CWD. The risk of transmission of CWD to humans appears to be negligible, with no demonstrated cases of CWD in humans to date. Some form of species barrier for CWD transferring to humans is apparent at the molecular level [Citation6]. However, other primates are susceptible in bioassays, either by intracranial or oral transmission route [Citation7]. The ability of PrPCWD to infect other species appears to be heavily dependent on the genotype of host PrPC aligning as closely as possible to PrPCWD. Previous work has shown, for instance, that host encoding S170 confers some level of resistance and N170 confers relative susceptibility [Citation8].
Previous biochemical studies focusing on the structure and in vitro behavior of PrP have focused on proteins derived from humans, mice, or hamsters. However, considering the variety of genetic, cellular, and environmental interactions that can influence a prion disease, it is difficult to make conclusions about one prion disease with data derived from a different species background. There is little in the way of detailed evaluation of cervid PrP that would offer insight into the differential disease outcomes of CWD with different genetic backgrounds. Furthermore, different biochemical properties arise in distinct prion disease strains [Citation9–11]. Therefore, we sought to characterize PrP from Rocky Mountain elk (Cervus elaphus nelsoni), and compare two major, disease-determining variants of the substrate protein, with either a leucine or a methionine at amino acid position 132 (hereafter referred to as L132 and M132, respectively). Previous work has associated a leucine at 132 with relative resistance to CWD and a methionine to relative susceptibility [Citation12,Citation13]. Position 132 is the equivalent of position 129 in humans, which in humans is either a methionine, associated with susceptibility to TSEs, or a valine, associated with a higher relative resistance [Citation14,Citation15]. Other amino acid variants known to influence CWD in cervids occur at positions 95, 96, 169, and 225 [Citation16,Citation17].
CWD has the potential to spread in wild cervid populations. In North America alone, there have been cases of CWD in wild populations in Alberta and Saskatchewan, as far west in the United States as Utah, as far east as upstate New York, and as far south as the Texas/Mexico border [Citation18]. CWD was also identified in South Korea, the result of importing CWD affected elk from North America [Citation19]. Recently, moose and reindeer were identified as having CWD in Norway [Citation20], although the source of the disease in these cases is not known.
There is a bevy of data on CWD at the epidemiological level but a relative scarcity of data on the conformational characteristics of monomeric cervid PrPC itself. Considering that, and keeping in mind the unique nature of CWD as the only TSE to circulate in wild populations, we sought to characterize the conformation, stability, and amyloid fibril formation of cervid PrP in response to divalent cation ligands, binding of which is a proposed function of PrP in vivo [Citation21–23]. This is of particular interest after reports that divalents in the diet of cervids affect CWD symptoms and survival time [Citation24]. In this study we purified recombinant mature length (hereafter referred to as rPrP, amino acids 26–234, equivalent to human 23–231) L132 and M132 Rocky mountain elk PrP from E. coli and compared the stability, helical content, and propensity for fibril formation in the presence of a variety of divalent cations. These cations have previously been shown to alter the biochemical characteristics of PrPC from other species backgrounds [Citation25].
Results
Variants of elk PrP at codon 132 are differentially destabilized in the presence of divalent cations
To begin comparing the stabilities of L132 and M132, we monitored by thermal denaturation circular dichroism (CD) on purified, recombinant protein [Citation26,Citation27]. Both proteins have a similar Tm, approximately 67–68°C, in PBS buffer in the absence of salt or divalent cations ( and ). We then performed the same thermal denaturation analysis for both proteins in increasing concentrations of several divalent cations: copper, manganese, calcium, and zinc. All divalent cations used here were in the chloride salt form. These cations have previously been shown to be bound by PrPC in vivo, and to alter the biochemical properties of recombinant PrP in vitro [Citation25,Citation28,Citation29]. The concentration of protein in the experiment was 1.23 µM, so a 1x molar ratio of divalent cation is 1.23 µM, 2x is 2.46µM, 5x is approximately 6 µM, etc.
Figure 1. Determination of Tm of Elk rPrP variants by thermal denaturation circular dichroism spectroscopy. Both rPrP L132 (A) and M132 (B) were melted in either the absence or presence of increasing molar ratios of divalent cations, copper (black circles), manganese (pink squares), calcium (blue triangles), or zinc (purple triangles). In the absence of divalents, both variants have similar melting temperatures – 67.16°C for L132 and 68.08°C for M132. The profile of M132 in the presence of 50x copper did not exhibit a sigmoidal shaped unfolding curve consistent with two state cooperative unfolding and could not be used to determine a Tm and is not included in this figure. In the presence of 120 mM NaCl, L132's Tm was 64.58°C and M132's was 68.01°C.
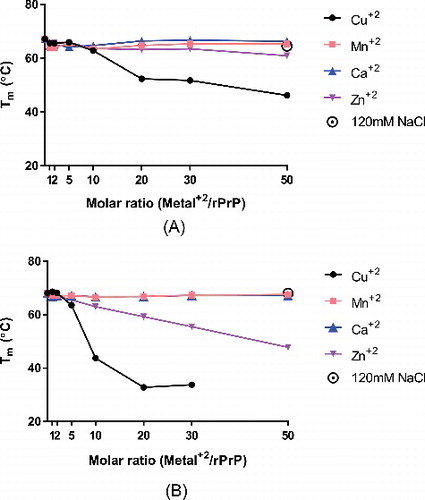
Neither calcium nor manganese altered the melting temperature of either L132 or M132 ( and ). Upwards of 50x molar excess of either divalent cation was unable to perturb the stability of either protein. This is in good agreement with previous results showing that neither calcium nor manganese altered the ability of human rPrP to form fibrils [Citation25].
In contrast, addition of both copper and zinc lowered the Tm of both proteins. However, L132 and M132 had markedly different responses to either cation. L132 maintained an only slightly depressed Tm as concentrations of zinc were increased, dropping only about 6°C in the presence of 50x molar excess zinc. M132 in comparison was far less stable, with its Tm dropping almost 10°C in the presence of 20x molar excess zinc, and 20°C in 50x molar excess.
The trend was similar but even more pronounced when L132 and M132 were incubated with copper. As little as a 10x molar excess of copper ions to M132 protein caused a 25°C reduction of Tm, while a 50x molar excess resulted in data from which a Tm could not be derived because it did not fit a two-state cooperative unfolding model. In contrast, 10x copper only reduced the Tm of L132 by about 5°C. A 50x molar excess of copper reduced L132's Tm to about 46°C, higher than M132's in the presence of as little as 10x molar excess copper.
To control for the possibility that effects of divalent cations are simply altering the osmotic character of the reactions, we also incubated L132 and M132 with 120µM NaCl, which is an equivalent concentration of chloride ions as that in 50x molar excess of divalent copper, zinc, manganese, or calcium. M132's Tm in the presence of this concentration of NaCl was 68.01°C, while L132's was 64.58°C, largely unchanged in comparison to protein in the absence of salts.
To further probe this phenomena, we repeated the titration of divalent cations in L132 and M132, but instead examined the cations' effect on the protein's CD spectra. PrPC is a largely α-helical protein, so its CD spectra is a classical “double-minima” spectra, with two minima at approximately 210 nm and 225 nm [Citation30,Citation31]. Perturbation of that secondary structure by either divalent cations or denaturants results in loss of the characteristic α-helix spectra and frequently converting to a single minimum at 215 nm, indicating a change in secondary structure consistent with reports of increased β-sheet content mixed with some residual helical structure30,32,33.
In similar fashion to the thermal denaturation experiments, increasing amounts of copper, manganese, calcium, and zinc were titrated into L132 or M132 containing no additional salts, and the resultant spectra were compared ( and ). Manganese and calcium, as observed in the thermal denaturation assays, did not have any discernable effect on either L132 or M132's far UV CD spectra, even at 50x molar excess.
Figure 2. Circular dichroism spectra of Elk rPrP variants in presence and absence of increasing molar ratios of divalent cations. The far UV CD spectra of both L132 (A) and M132 (B) was determined incubated in PBS alone (solid green line), and in 1x (dotted line), 5x (dashed line), and 50x (solid line) molar ratios of copper (black), manganese (pink), and zinc (purple). Angle of deflection data was converted to molar ellipticity.
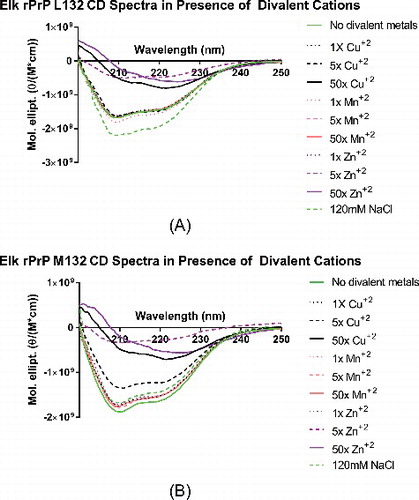
The result was quite different when L132 and M132 were incubated with copper or zinc. L132, when incubated with copper, retained its secondary structure in 1x or 5x molar ratios. In 50x copper, however, L132's secondary structure had converted from a classic helical structure to one more suggestive of β-sheet [Citation32]. Zinc, however, much more readily perturbed L132's structure, with as little as 5x molar ratio disrupting the native CD spectra.
M132 was, once again, less stable than L132 in the face of the same challenge. The protein retained its structure in 1x copper, but that structure was severely reduced in 5x copper, and entirely disrupted in 50x. In zinc, 5x and 50x were also sufficient to disrupt the helical state. It appears that while M132 is less able to retain its structure when incubated with divalent cations, both L132 and M132's structure are more easily disrupted with zinc than copper, despite a more profound effect on Tm in the presence of copper than zinc, when probed by thermal denaturation.
We also measured L132 and M132's spectra in the presence of 120mM NaCl, and found that both were largely unchanged in comparison to the protein's spectra in the absence of salt (, dotted green line). This control demonstrates that the effects seen here are specifically a result of interaction with copper and zinc ions in solution, and not simply a result of changing osmotic character of the solution.
Divalent cations have similar effects on RT-QuIC conversion, regardless of substrate genotype
To investigate whether the differential responses of L132 and M132 to challenge by divalent cations had an effect on their ability to form fibrils, we performed real-time, quaking-induced conversion (RT-QuIC) assays in the presence or absence of divalent cations [Citation34,Citation35]. These assays track the fluorescence of Thioflavin T (ThT). ThT is a fluorophore whose emission increases as it is incorporated into growing fibrils [Citation34,Citation36]. We used rPrP, either L132 or M132, as a substrate and a normalized amount of PrPCWD, sourced from elk brain homogenate (see Methods) as a template. Note that seed genotype is matched to substrate genotype, so L132 rPrP is incubated with brain homogenate of LL homozygous elk, and M132 rPrP with MM homozygous homogenate. The relative concentration of PrPCWD was ascertained by ELISA, and the results were used to normalize the amount of PrPCWD added to all reactions. Reactions containing substrate rPrP but not seeded with brain homogenate did not convert to fibril (data not shown).
Similar to the two sets of circular dichroism assays, neither calcium nor manganese had an effect on fibril formation in either L132- or M132-background assays (, , , ). 1x, 5x, and 50x molar excess of either divalent cation was not sufficient to either prolong the lag time of conversion from monomer to fibril or suppress fibril formation at all. We do note that there was a slightly shorter lag time for M132 fibrils in the presence of manganese, though it is unclear if this is of biological significance. There was also a mild enhancement of maximum ThT fluorescence in M132 fibrils when incubated with either manganese or calcium, though raw ThT fluorescence seems to vary from protein to protein (see the difference between both minimum and maximum ThT fluorescence between L132 and M132), so we do not ascribe significance to this either. Raw fluorescence also appears to be affected by the type of brain homogenate used as seed [Citation9]
Figure 3. Real-time quaking induced conversion (RT-QuIC) of Elk rPrP L132 in presence and absence of increasing molar ratios of divalent cations. Monomeric L132 (A-D) was used as substrate and seeded with brain homogenate from homozygous L132 elk infected with CWD. Relative amounts of PrPCWD were determined by ELISA and equal amounts of seed are used as described in the Methods. Conversion of monomers into fibrils monitored by increased Thioflavin T (ThT) fluorescence (arbitrary units) as a function of cycle number are shown either in the absence (green line) or presence of increasing concentrations of divalent cations. L132 incubated with 1x (dotted line), 5x (dashed line), or 50x (solid line) molar ratios of (A) copper (black), (B) manganese (pink), (C) calcium (blue), or (D) zinc (purple). Under the conditions of the experiment, unseeded controls did not convert and, for clarity, are not shown.
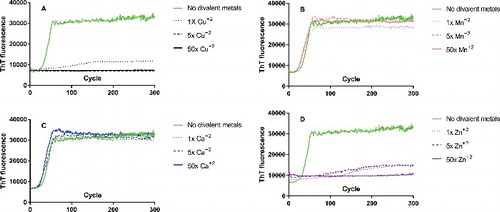
Figure 4. Real-time quaking induced conversion (RT-QuIC) of Elk rPrP M132 in presence and absence of increasing molar ratios of divalent cations. Monomeric M132 (A-D) was used as substrate and seeded with brain homogenate from homozygous M132 elk infected with CWD. Relative amounts of PrPCWD were determined by ELISA and equal amounts of seed are used in both (see Methods). Conversion of monomers into fibrils monitored by increased Thioflavin T (ThT) fluorescence (arbitrary units) as a function of cycle number are shown either in the absence (green line) or presence of increasing concentrations of divalent cations. M132 incubated with 1x (dotted line), 5x (dashed line), or 50x (solid line) molar ratios of (A) copper (black), (B) manganese (pink), (C) calcium (blue), or (D) zinc (purple). Under the conditions of the experiment, unseeded controls did not convert and, for clarity, are not shown.
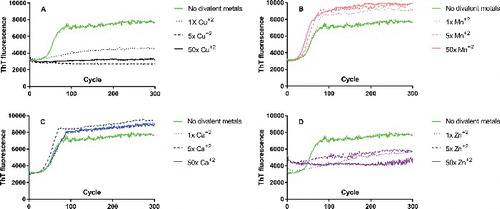
Both copper and zinc had profound effects on fibril formation when tracked via ThT fluorescence (, , , ). Both L132 and M132 conversion into fibrils were completely blocked by as little as a 1x equimolar concentration of either copper or zinc. Even more remarkably, the effects of copper and zinc do not appear to be different between L132 and M132; neither was more or less susceptible to suppression, they appear to be equally suppressed. In addition, we are confident that suppression of fibril formation is a bona fide effect of copper and zinc ions, instead of a contribution to ionic character from Cl- ions, as the presence of millimolar quantities of NaCl mean that the addition of micromolar amounts of divalent cations does not meaningfully change the ionic character of the reaction.
Discussion
Reports in the literature with regard to the effects of divalent cations on PrP vary from prion protein to prion protein, and the assay used to measure the effect. Copper is shown to have a profound effect. Zinc has been shown to reduce fibril conversion efficiency in vitro, though less so than copper [Citation25,Citation28]. Zinc has also been shown to have no effect on rPrP folding in one type of refolding assay, where copper and manganese did have an effect [Citation29]. Nickel was also shown to have no effect on refolding, though rPrP has been shown to bind nickel in vitro. [Citation37] The same group in [Citationref. 27] found a different type of refolding assay – one that probed proteinase K resistance after refolding in the presence of various divalents – where manganese didn't have an effect but nickel did. At the same time, previous work has shown that while zinc can physically associate with rPrP, it is bound secondarily to copper in a complex interplay of molar ratios [Citation23]. In general, loss of helical content upon addition of copper and zinc, with little to no effect from manganese or calcium, in elk PrPC appears to be consistent with results reported for PrPC sourced from other species backgrounds (see for example [Citation21,Citation29,Citation38].
In the NMR-determined structure of elk rPrP (residues 121–231), the overall secondary structure of the protein is consistent with structures determined for other mammalian species [Citation39]. However, one variation is a stable loop formed by residues 166–175, where in other species this loop is not stable. Residue 132 forms part of the N-terminal ribbon that sits just across from this loop in space [Citation39]. It is possible that because of where it is located, residue 132's position may contribute to different stabilities in the overall protein.
It is also possible that the difference in reaction to excess copper and zinc in our circular dichroism assays is due to the difference in how PrP coordinates these two ions. The octarepeat region of PrP binds copper ions in a 1:1 ratio, one ion bound for each repeat, with additional coordination possible at residues H96 and H111 (in human PrPC numbering) [Citation23]. PrP binds zinc, in contrast, in a 1:4 ratio, with all four octarepeats needed to coordinate one zinc ion [Citation23]. Our data shows that monomeric M132 is less stable than L132. If the loss of structure resulting from interaction with divalents is exacerbated by the methionine substitution's effect on a structural feature such as the stable loop in residues 166–175, this would explain M132's disproportionate loss of stability in the presence of divalent cations. On the other hand, both copper and zinc have been shown to mediate a dramatic N-to-C-terminus interaction that serves apparently to stabilize the whole protein [Citation40,Citation41]. How this whole-protein stabilization corresponds with our results showing remarkable destabilization of secondary structure by both copper and zinc is unclear.
There are several fundamental disconnects at the heart of our data as presented here. The L132 genotype is generally associated with longer incubation times and relative resistance to CWD, while M132 is associated with shorter incubation times and susceptibility. It makes sense, then, that rPrP-M132 is less able to retain its secondary structure in comparison to L132, lending credence to the idea that M132 easily transitions to non-native structure. However, in RT-QuIC, L132 not only converts from monomer to fibril with a shorter lag time than M132, but both M132 and L132 fibril formation is suppressed equally in the presence of copper or zinc.
This is consistent with findings from studies on prions occurring in yeasts. That work showed that the formation of fibrils involved a complex interplay between template and substrate genotypes, as well as the chemical environment present, which resulted in more transmissible prions aggregating slower than expected in vitro, and less transmissible prions aggregating more quickly [Citation42].
There are other possible explanations as well. The first is that divalent cations can affect the secondary structure of both monomeric and oligomeric PrP. If that is the case, then the effects of divalent cations on fibril formation in RT-QuIC likely result from their interaction with PrPCWD, which is present in molar quantities orders of magnitude smaller than that of monomeric protein or divalent cations. So, PrPCWD is likely saturated with divalents in our RT-QuIC assays. Previous work has shown that some divalent cations, including copper and zinc, but not manganese, can act to stabilize oligomeric structures of PrP [Citation43]. Stabilization of fibrils by divalents may be unaffected by genotype at position 132, and then might result in equal suppression of formation of fibrils in our RT-QuIC results. Furthermore, research on amyloid-β has shown that oligomers formed by the same monomeric protein can have different metal binding properties, depending on the conditions under which the oligomers were formed [Citation44–46]. Taking this into consideration, it is possible that oligomers formed by L132 may have different metal binding properties than those formed by M132 that might not be in concordance with binding properties of the monomer. It also stands to reason that since the concentration range of divalent metals that affect secondary structure of rPrP differs depending on the assay – for instance, equimolar Cu+2 has no effect on melting temperature or far-UV CD spectra but completely suppresses in vitro fibril formation – that there are other unconsidered factors beyond the scope of this investigation at play.
Furthermore, that L132 more readily forms fibrils in RT-QuIC than M132 suggests that differential disease outcomes stemming from the two genotypes is unrelated to the constituent protein's propensity for fibril formation in vitro. Rather, we suggest that while individual monomers may have less propensity to follow prion-specific folding pathways, once fibrils are formed by M132, those fibrils may themselves be less stable and prone to fragmentation in comparison to L132 fibrils, resulting in shorter incubation times in vivo. These hypotheses will become easier to study as more and more bioassay samples became available for testing of the stability of bona fide CWD fibrils.
Methods
Protein purification
Mature length Rocky Mountain elk PrP (amino acids 26–234, Uniprot identifier P67986, EMBL accession number AAC12860.2) with either leucine or methionine at position 132 was synthesized and cloned by Genscript (Piscataway, NJ) between the NdeI and EcoRI sites in the plasmid pET28a. Protein was expressed and purified from E. coli inclusion bodies as described previously [Citation37].
Briefly, plasmid encoding the relevant protein was transformed into E. coli BL21 DE3 (expression in Rosetta DE3 was not found to alter expression levels). Cultures were grown at 37°C to A600 ∼ = 0.6-0.8, induced with IPTG, and allowed to incubate with shaking overnight again at 37°C. Cultures were harvested the following morning and resuspended in lysis buffer (20 mM Tris pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 2mM EDTA). A protease inhibitor tablet (Roche, cat. #04693159001) was added, followed by addition of lysozyme to 10 µg/mL. Cells were incubated on ice for 10–15 minutes and then further disrupted by sonication (three cycles of sonication, consisting of one minute at 18W followed by resting for one minute).
Inclusion bodies were harvested by centrifugation, discarding the supernatant, and resuspending the pellet in high detergent buffer (20 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100). Lysate was again centrifuged and the pellet resuspended in high salt buffer (20 mM Tris pH 7.4, 2M NaCl). Lysate was once again centrifuged and resuspended in urea-containing buffer (20 mM Tris pH 7.4, 150 mM NaCl, 2M urea). Finally, protein was solubilized by centrifugation and resuspension in guanidinium hydrochloride buffer (10mM Tris pH 8.0, 100mM Na2HPO4, 6M GdHCl, 0.5% Triton X-100, 10mM β-ME). Protein was allowed to rock at room temperature for one hour and then rocked overnight at 4°C. The following morning the lysate was clarified by centrifugation and filtered through a 0.2µm PES filter (Thermo Fisher, cat. #564-0020). During that time a Ni+2 column was prepared and equilibrated in the same GdHCl buffer that the lysate was in. After clarification and filtering, the lysate was applied to the Ni+2 column. The column was rinsed with three column volumes of wash buffer (8M urea, 100mM Na2HP04, 10mM Tris pH 8.0, 10mM β-ME), and protein was eluted with 15mL of elution buffer (8M urea, 10mM Tris pH 8.0, 0.5M imidazole, 10mM β-ME). Fractions containing protein were selected and dialyzed overnight into urea buffer lacking reducing agent to re-form PrP's disulfide bond (8M urea, 100mM Na2HP04, 10mM Tris pH 8.0). Then, protein was dialyzed into potassium phosphate buffer (10mM K2HPO4, 10mM KH2PO4) for two hours, placed into fresh potassium phosphate buffer, and dialyzed overnight. In the morning, misfolded precipitate was pelleted and soluble protein aliquoted and stored at 4°C.
Circular dichroism
Analysis was performed on a Jasco J-815 spectrophotometer. To track changes in secondary structure, 1.23µM rPrP in potassium phosphate buffer (6.15mM K2HPO4 + 3.85mM KH2HPO4) was incubated on ice with increasing amounts of the indicated divalent cation prepared from the chloride salt (CuCl2, CaCl2, MnCl2, ZnCl2)for 30 minutes. Spectra were taken in a 1cm path-length cuvette with data recorded every 0.1nm from 260nm to 200nm, averaged over eight scans.
For thermal denaturation analysis, samples were prepared in the same fashion. Circular dichroism signal was monitored at 222nm from 20°C to 95°C, sampled every 3°C with a ramp rate of 1°C/min. Thermodynamic parameters describing the curves were determined by fitting the data to equations provided in reference [Citation47].
Real time quaking-induced conversion (RT-QuIC)
Assays were conducted with a reaction volume of 100µL in the well of a black, anti-reflective 96-well plate. Each 100µL well contained a reaction mixture of 6µM rPrP, 400mM NaCl, 10mM potassium phosphate buffer, 100µM thioflavin T, and the specified concentration of divalent cation prepared from the chloride salt (CuCl2, CaCl2, MnCl2, ZnCl2). To the seeded wells, PrPCWD derived from brain homogenate prepared from elk infected with CWD was added. Archived samples from a previously published experimental transmission study addressing oral transmissibility of CWD were used to seed the RT-QuiC experiments. Details of the MM and LL brain homogenates used as seed are available in ref. [Citation13] The relative amount of PrPCWD in each brain homogenate was ascertained by ELISA (data not shown), and each assay, regardless of substrate genotype, received the same amount of PrPCWD template based on the ELISA for that sample. To unseeded control wells, extra ddH20 was added to make up the total volume to 100µL. Parameters of the plate reader used for RT-QuIC were the same as that described previously [Citation34]. Plates were placed in a BMG Fluostar Omega (Cary, NC) plate reader for 72–96 hours, at 42°C. Plates were shaken for one minute (700 rpm, double orbital) and rested for one minute, with measurements taken every 15 minutes (450nm excitation, 480nm emission, 20 flashes per well, manual gain of 2000, 20 µs integration time).
Disclaimer
Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Trudy Tatum and Semakaleng Lebepe-Mazur for excellent technical support.
Additional information
Funding
References
- Prusiner SB.. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216(4542):136–44. doi:10.1126/science.6801762
- Prusiner SB. The prion diseases. Brain Pathol. 1998;8(3):499–513. doi:10.1111/j.1750-3639.1998.tb00171.x
- Williams ES, Young S. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis. 1980;16(1):89–98. doi:10.7589/0090-3558-16.1.89
- Williams ES, Young S. Spongiform encephalopathy of Rocky Mountain elk. J Wildl Dis. 1982;18(4):465–71. doi:10.7589/0090-3558-18.4.465
- Spraker TR, Miller MW, Williams ES, et al. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J Wildl Dis. 1997;33(1):1–6. doi:10.7589/0090-3558-33.1.1
- Raymond GJ, Bossers A, Raymond LD, et al. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 2000;19(17):4425–30. doi:10.1093/emboj/19.17.4425
- Kurt TD, Sigurdson CJ. Cross-species transmission of CWD prions. Prion. 2016;10(1):83–91. doi:10.1080/19336896.2015.1118603
- Kurt TD, Telling GC, Zabel MD, et al. Trans-species amplification of PrP(CWD) and correlation with rigid loop 170N. Virology. 2009;387(1):235–43. doi:10.1016/j.virol.2009.02.025
- Hwang S, Greenlee JJ, Nicholson EM. Use of bovine recombinant prion protein and real-time quaking-induced conversion to detect cattle transmissible mink encephalopathy prions and discriminate classical and atypical L- and H-Type bovine spongiform encephalopathy. PLoS One. 2017;12(2):e0172391. doi:10.1371/journal.pone.0172391
- Moore SJ, Smith JD, Greenlee MH, et al. Comparison of Two US Sheep Scrapie Isolates Supports Identification as Separate Strains. Vet Pathol. 2016;53(6):1187–1196. doi:10.1177/0300985816629712
- Moore SJ, West Greenlee MH, Smith JD, et al. A Comparison of Classical and H-Type Bovine Spongiform Encephalopathy Associated with E211K Prion Protein Polymorphism in Wild-Type and EK211 Cattle Following Intracranial Inoculation. Front Vet Sci. 2016;3:78. doi:10.3389/fvets.2016.00078
- Hamir AN, Gidlewski T, Spraker TR, et al. Preliminary observations of genetic susceptibility of elk (Cervus elaphus nelsoni) to chronic wasting disease by experimental oral inoculation. J Vet Diagn Invest. 2006;18(1):110–4. doi:10.1177/104063870601800118
- O'Rourke KI, Spraker TR, Zhuang D, et al. Elk with a long incubation prion disease phenotype have a unique PrPd profile. Neuroreport. 2007;18(18):1935–8. doi:10.1097/WNR.0b013e3282f1ca2f
- Asante EA, Linehan JM, Gowland I, et al. Dissociation of pathological and molecular phenotype of variant Creutzfeldt-Jakob disease in transgenic human prion protein 129 heterozygous mice. Proc Natl Acad Sci U S A. 2006;103(28):10759–64. doi:10.1073/pnas.0604292103
- Wadsworth JD, Asante EA, Desbruslais M, et al. Human prion protein with valine 129 prevents expression of variant CJD phenotype. Science. 2004;306(5702):1793–6. doi:10.1126/science.1103932
- Meade-White K, Race B, Trifilo M, et al. Resistance to chronic wasting disease in transgenic mice expressing a naturally occurring allelic variant of deer prion protein. J Virol. 2007;81(9):4533–9. doi:10.1128/JVI.02762-06
- Mitchell GB, Sigurdson CJ, O'Rourke KI, et al. Experimental oral transmission of chronic wasting disease to reindeer (Rangifer tarandus tarandus). PLoS One. 2012;7(6):e39055. doi:10.1371/journal.pone.0039055
- USGS National Wildlife Health Center. Map of chronic wasting disease in North America. <http://www.nwhc.usgs.gov/disease_information/chronic_wasting_disease/>.
- Lee YH, Sohn HJ, Kim MJ, et al. Strain characterization of the Korean CWD cases in 2001 and 2004; J Vet Med Sci. 2013;75(1):95–8. doi:10.1292/jvms.12-0077
- Benestad SL, Mitchell G, Simmons M, et al. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res. 2016;47(1):88. doi:10.1186/s13567-016-0375-4
- Stockel J, Safar J, Wallace AC, et al. Prion protein selectively binds copper(II) ions. Biochemistry. 1998;37(20):7185–93. doi:10.1021/bi972827k
- Walter ED, Stevens DJ, Spevacek AR, et al. Copper binding extrinsic to the octarepeat region in the prion protein. Curr Protein Pept Sci. 2009;10(5):529–35. doi:10.2174/138920309789352056
- Walter ED, Stevens DJ, Visconte MP, et al. The prion protein is a combined zinc and copper binding protein: Zn2+ alters the distribution of Cu2+ coordination modes. J Am Chem Soc. 2007;129(50):15440–1. doi:10.1021/ja077146j
- Nichols TA, Spraker TR, Gidlewski T, et al. Dietary magnesium and copper affect survival time and neuroinflammation in chronic wasting disease. Prion. 2016;10(3):228–50. doi:10.1080/19336896.2016.1181249
- Bocharova OV, Breydo L, Salnikov VV, et al. Copper(II) inhibits in vitro conversion of prion protein into amyloid fibrils. Biochemistry. 2005;44(18):6776–87. doi:10.1021/bi050251q
- Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1(6):2527–35. doi:10.1038/nprot.2006.204
- Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1 Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants Biochemistry. 1988;27(21):8063–8.
- Bocharova OV, Breydo L, Parfenov AS, et al. In vitro conversion of full-length mammalian prion protein produces amyloid form with physical properties of PrP(Sc). J Mol Biol. 2005;346(2):645–59. doi:10.1016/j.jmb.2004.11.068
- Brown DR, Hafiz F, Glasssmith LL, et al. Consequences of manganese replacement of copper for prion protein function and proteinase resistance. EMBO J. 2000;19(6):1180–6. doi:10.1093/emboj/19.6.1180
- Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1(6):2876–90. doi:10.1038/nprot.2006.202
- Greenfield NJ. Determination of the folding of proteins as a function of denaturants, osmolytes or ligands using circular dichroism. Nat Protoc. 2006;1(6):2733–41.
- Johnson WC, Jr.. Protein secondary structure and circular dichroism: A practical guide. Proteins. 1990;7(3):205–14. doi:10.1002/prot.340070302
- Greenfield NJ.. Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal Biochem. 1996;235(1):1–10. doi:10.1006/abio.1996.0084
- Wilham JM, Orru CD, Bessen RA, et al. Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 2010;6(12):e1001217. doi:10.1371/journal.ppat.1001217
- Vascellari S, Orru CD, Hughson AG, et al. Prion seeding activities of mouse scrapie strains with divergent PrPSc protease sensitivities and amyloid plaque content using RT-QuIC and eQuIC. PLoS One. 2012;7(11):e48969. doi:10.1371/journal.pone.0048969
- Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 2010;1804(7):1405–12. doi:10.1016/j.bbapap.2010.04.001
- Vrentas CE, Onstot S, Nicholson EM. A comparative analysis of rapid methods for purification and refolding of recombinant bovine prion protein. Protein Expr Purif. 2012;82(2):380–8. doi:10.1016/j.pep.2012.02.008
- Zhu F, Davies P, Thompsett AR, et al. Raman optical activity and circular dichroism reveal dramatic differences in the influence of divalent copper and manganese ions on prion protein folding. Biochemistry. 2008;47(8):2510–7. doi:10.1021/bi7022893
- Gossert AD, Bonjour S, Lysek DA, et al. Prion protein NMR structures of elk and of mouse/elk hybrids. Proc Natl Acad Sci U S A. 2005;102(3):646–50. doi:10.1073/pnas.0409008102
- Spevacek AR, Evans EG, Miller JL, et al. Zinc drives a tertiary fold in the prion protein with familial disease mutation sites at the interface. Structure. 2013;21(2):236–46. doi:10.1016/j.str.2012.12.002
- Evans EG, Pushie MJ, Markham KA, et al. Interaction between Prion Protein's Copper-Bound Octarepeat Domain and a Charged C-Terminal Pocket Suggests a Mechanism for N-Terminal Regulation. Structure. 2016;24(7):1057–67. doi:10.1016/j.str.2016.04.017
- Sharma A, Bruce KL, Chen B, et al. Contributions of the Prion Protein Sequence, Strain, and Environment to the Species Barrier. J Biol Chem. 2016;291(3):1277–88. doi:10.1074/jbc.M115.684100
- Gonzalez-Iglesias R, Pajares MA, Ocal C, et al. Prion protein interaction with glycosaminoglycan occurs with the formation of oligomeric complexes stabilized by Cu(II) bridges. J Mol Biol. 2002;319(2):527–40. doi:10.1016/S0022-2836(02)00341-8
- Miller Y, Ma B, Nussinov R. Zinc ions promote Alzheimer Abeta aggregation via population shift of polymorphic states. Proc Natl Acad Sci U S A. 2010;107(21):9490–5. doi:10.1073/pnas.0913114107
- Miller Y. MB, Nussinov R.. Metal binding sites in amyloid oligomers: Complexes and mechanisms. Coordination Chemistry Reviews. 2012;256(19-20):7. doi:10.1016/j.ccr.2011.12.022
- Parthasarathy S, Long F, Miller Y, et al. Molecular-level examination of Cu2+ binding structure for amyloid fibrils of 40-residue Alzheimer's beta by solid-state NMR spectroscopy. J Am Chem Soc. 2011;133(10):3390–400. doi:10.1021/ja1072178
- Swint L, Robertson AD. Thermodynamics of unfolding for turkey ovomucoid third domain: thermal and chemical denaturation. Protein Sci. 1993;2(12):2037–49. doi:10.1002/pro.5560021205
