ABSTRACT
Abnormal structural changes of the prion protein (PrP) are the cause of prion disease in a wide range of mammals. However, spontaneous infected cases have not been reported in chicken. Genetic variations of the prion protein gene (PRNP) may impact susceptibility to prion disease but have not been investigated thus far. Because an investigation of the chicken PRNP can improve the understanding of characteristics related to resistance to prion disease, research on the chicken PRNP is highly desirable. In this study, we investigated the genetic characteristics of the chicken PRNP gene. For this, we performed direct sequencing in 106 Dekalb White chickens and analyzed the genotype and allele frequencies of chicken PRNP gene. We found two insertion and deletion polymorphisms in the chicken PRNP: c.163_180delAACCCAGGGTACCCCCAT and c.268_269insC. The former is a U2 hexapeptide deletion polymorphism. Of the 106 samples, 13 (12.26%) were insertion homozygotes, 89 (83.96%) were heterozygotes, and 4 (3.77%) were deletion homozygotes in c.163_180delAACCCAGGGTACCCCCAT. In the c.268_269insC polymorphism, 102 (96.23%) were deletion homozygotes, and 4 (3.77%) were heterozygotes. Insertion homozygotes of c.268_269insC were not detected. Two polymorphisms were in perfect linkage disequilibrium (LD) with a D’ value of 1.0, and three haplotypes were identified. Furthermore, PROVEAN evaluates 163_180delAACCCAGGGTACCCCCAT as ‘deleterious’ with a score of – 13.173. Furthermore, single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the PRNP gene were not found in the chicken. To the best of our knowledge, this was the first report on the genetic variations of the chicken PRNP gene.
Introduction
The prion protein (PrP) is coded by the prion protein gene (PRNP), and aberrant structural changes from the cellular form (PrPC) to the scrapie form of the prion protein (PrPSc) causes prion diseases in mammals[Citation1]. Previous studies have tried to identify the cause of prion disease; however, the apparent cause of onset is elusive. To date, spontaneous prion disease in birds has not been reported, and parenteral and oral challenge of prion agent has failed to infected chickens [Citation2,Citation3]. Thus, several studies have attempted to find the feature of the chicken prion protein that leads to prion disease resistance.
In previous studies, it has been reported that homology of the prion protein was very low between mammals and birds; however, important domains, such as signal peptide, tandem repeat domain (octapeptide in mammals and hexapeptide in birds), hydrophobic region and C-terminal globular domain, are conserved[Citation4]. Although amino acids compositions of tandem repeat are different, octapeptide and hexapeptide showed identical copper related functions in mammals and chickens, respectively[Citation5]. In addition, the expression level of mRNA and protein of PRNP was similar in the central nervous system of both species [Citation6–Citation8]. This finding indicates that the major function and expression profile of the prion protein are identical, although several differences existed between mammals and birds. In a recent study, molecular dynamics studies found that the prion protein of chicken has an stable molecular structure analogous to that of humans, which is considered as a susceptible species to prion disease [Citation9]. Because a highly stable molecular structure of the prion protein has been reported in horses, dogs and rabbits, which are classified as resistant animals to prion disease [Citation10–Citation13], there may be another factor that contributes to prion disease resistance in chickens.
The genetic variations of the PRNP gene that can potently impact prion disease susceptibility have not been investigated in chicken. The genetic variations of the PRNP gene are significantly related to prion disease susceptibility in a wide range of hosts and are subdivided into three groups according to the type and location. The first group is non-synonymous single nucleotide polymorphisms (SNPs) and mutations in the C-terminal globular domain of PrPC. SNPs are the most commonly observed form and are detected at codons 129, 180, 200, and 219 in humans [Citation14,Citation15], codons 136, 154, and 171 in sheep [Citation16,Citation17], codons 142, 143, 146, 154, 171, 211, and 222 in goat [Citation18,Citation19], and codons 95 and 96 in deer [Citation20,Citation21]. The second group is insertion/deletion polymorphisms in the promoter region of the PRNP gene. The number of prion disease-affected cattle was correlated with 23-bp insertion/deletion in the promoter region [Citation22]. The third group is octapeptide insertion/deletion polymorphism in the N-terminal octapeptide domain. Octapeptide insertion is significantly associated with human prion disease onset [Citation23]. Because investigation of genetic characteristics of chicken PRNP gene may be important in understanding the functions of PrPC, which is a pre-disease substrate of prion disease, a trial to identify genetic features of chicken PRNP gene is meaningful.
The purpose of this study was to investigate genetic characteristics of the chicken PRNP gene. To do so, we performed direct sequencing in 106 Dekalb White chickens and investigated genotype and allele frequencies of the chicken PRNP gene. In addition, we evaluated the impact of polymorphisms on chicken prion protein using PROVEAN.
Results
To investigate the genotype and allele frequencies of chicken PRNP gene polymorphism, we performed PCR to amplify the chicken PRNP gene. Chicken PRNP amplicons were automatically and directly sequenced in 106 Dekalb White chickens using ABI 3730 DNA sequencer. Because tandem repeats of chicken are significantly different from those of mammals, we aligned tandem repeats among mammals and chickens (). Interestingly, we identified only insertion and deletion polymorphisms in the ORF of the chicken PRNP gene. A total two insertion and deletion polymorphisms were identified: c.163_180delAACCAGGGTACCCCCAT and c.268_269insC (). c.163_180delAACCCAGGGTACCCCCAT is a hexapeptide deletion polymorphism. To designate the location of the hexapeptide deletion polymorphisms, we named the tandem repeat from U1 to U7. The identified hexapeptide deletion is located on U2 (). These are the first electropherograms showing a deletion of a single hexapeptide repeat in chicken using an automatic DNA sequencer (). Another polymorphism is c.268_269insC. Interestingly, c.268_269insC entails homozygosity of U2 deletion (data not shown). In addition, the insertion homozygote of c.268_269insC was not detected in this study. Electropherograms of c.268_269insC was first reported in this study (). Furthermore, to validate two PRNP alleles and investigate the haplotype between two polymorphisms in the sample #44, which has heterozygous genotypes at two polymorphisms, the PCR products of chicken PRNP gene from the sample #44 was amplified, subcloned, and sequenced. Only two alleles were observed in sample #44 with WT-WT alleles (clone 1) and deletion-insertion alleles (clone 2) (). Detailed genotype and allele frequencies of these polymorphisms are described in .
Table 1. Genotype and allele frequencies of the PRNP polymorphisms in chickens.
Figure 1. Comparisons of tandem repeat sequence in humans, sheep, goat, cattle and chicken. Tandem repeat sequences were obtained from GenBank at the National Center for Biotechnology Information (NCBI), including those of human (Homo sapiens, BAG32277.1), sheep (Ovis aries, NP_001009481.1), goat (Capra hircus, NP_001301176.1), cattle (Bos taurus, NP_001258555.1) and chicken (Gallus gallus, in this study). Arrows indicate the hexapeptide deletion polymorphism found in this study. Hexapeptide repeat units were named from U1 to U7 in turn. Word spacing and underlines discriminate consecutive tandem repeat units.

Figure 2. Gene map and polymorphism identified in the chicken prion protein gene (PRNP) on chromosome 22. The open reading frame (ORF) is indicated by a shaded block and the 5ʹ and 3ʹ untranslated regions (UTRs) are indicated by white blocks. Horizontal bars with edges indicate the regions sequenced. Arrows indicate the polymorphisms found in this study. The Y-shaped bar indicates the hexapeptide deletion polymorphisms identified in the chicken PRNP gene.
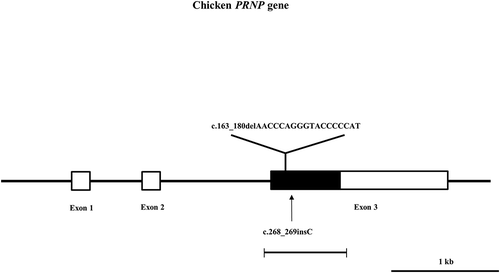
Figure 3. Electropherogram of hexapeptide deletion polymorphism in chicken PRNP gene. Four colors indicate individual bases of DNA sequence using ABI 3730 automatic sequencer (blue: cytosine, red: thymine, black: guanine, green: adenine). Sharps (#) indicate double peaks induced by hexapeptide deletion polymorphism. Arrows indicate the hexapeptide deletion polymorphism (U2) found in this study. WT: wild-type of chicken hexapeptide repeat. Deletion: U2 deletion polymorphisms of chicken hexapeptide repeat.
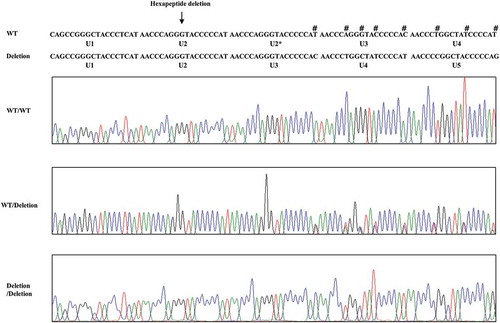
Figure 4. Electropherogram of c.268_269insC polymorphism in chicken PRNP gene. Four colors indicate individual bases of DNA sequence using ABI 3730 automatic sequencer (blue: cytosine, red: thymine, black: guanine, green: adenine). Sanger sequencing was performed by reverse gene-specific primer, as described in the materials and method section. Arrows indicate c.268_269insC polymorphism found in this study. Upper portion: heterozygote of c.268_269insC polymorphism. Lower portion: wild-type of the chicken PRNP gene sequence. WT: wild-type of the chicken PRNP gene sequence. Insertion: c.268_269insC polymorphisms of the chicken PRNP gene sequence.
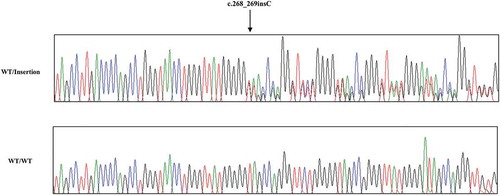
Figure 5. Electropherogram of the chicken PRNP region encompassing hexapeptide deletion and c.268_269insC polymorphisms in sample #44. Clone 1: Electropherogram showed wild-type sequences in hexapeptide deletion and c.268_269insC polymorphisms from the sample #44. Clone 2: The deletion sequence in hexapeptide deletion polymorphism and the insertion sequence at c.268_269insC polymorphism from the sample #44. Arrow indicates c.268_269insC polymorphism found in this study.
To examine if there was a strong linkage disequilibrium (LD) among the 2 polymorphisms, LD (|D’|) was calculated for the chicken PRNP gene. All 2 polymorphisms of chicken PRNP gene were in perfect LD with D’ values 1.0 (). Analysis of the haplotype frequency was carried out in Dekalb White. As shown in , three different haplotypes exist in the chicken PRNP polymorphisms. Among the three haplotypes, the most frequent haplotype is ht1 (54.24%), followed by ht2 (43.87%) and ht3 (1.89%) ().
Table 2. Linkage Disequilibrium (LD) among two polymorphisms of PRNP in chickens.
Table 3. Haplotype frequency of two PRNP polymorphisms in chickens.
Next, to evaluate the impact of insertion/deletion polymorphisms on chicken PrPC, we utilized PROVEAN. PROVEAN can measure the impact of insertion/deletion protein polymorphisms. Surprisingly, PROVEAN estimated hexapeptide deletion as ‘Deleterious’ with a score of −13.173 ().
Table 4. Prediction of protein functional alteration induced by deletion polymorphism of the chicken prion protein using PROVEAN.
Discussion
In previous studies, because PRNP knockout mice showed resistance to prion agents and relatively normal physiology, the function of PrPC was unclear [Citation24,Citation25]. However, Brown et al found that PrPC (23–98 residues) has copper binding capacity and PRNP knockout mice showed significantly lower copper concentrations in the brain among several divalent cations[Citation26]. In addition, these PRNP knockout mice showed reduced copper dependent enzyme reactivity. Thus, these results indicate that PrPC has copper binding ability. Indeed, synthetic octapeptide repeat, which is located on N-terminal domain of the prion protein, confirmed the copper binding domain of PrPC[Citation27]. Among the tandem repeat regions, ‘PHGGGWGQ’ is an octapeptide repeat unit. Specifically, the imidazole ring of histidine is significantly related to the interaction between PrPC and copper [Citation28,Citation29]. In a recent study, neuroprotective α-cleavage event of PrPC significantly influences on Cu(II) coordination at its His111 site [Citation30].
However, this well conserved octapeptide repeat sequence was found only in mammals. Chicken has a significantly different form of tandem repeat, called hexapeptide repeat. Due to this significantly different composition of amino acids of the tandem repeat, we performed sequence alignment and compared sequence differences of tandem repeat domains (). In previous studies, the repeat unit of chicken hexapeptide repeat is controversial [Citation31,Citation32]. Here, we suggested a novel tandem repeat unit based on polymorphisms of the chicken PRNP gene: U1 to U7 (). Interestingly, heterozygosity of hexapeptide deletion genotype frequencies was found in 89 (83.96%) of 106 Dekalb White chickens. Because octapeptide in/del polymorphisms in mammals are rare, highly polymorphic hexapeptide polymorphism in chicken PRNP gene are noticeable. Thus, the heterozygosity of the hexapeptide deletion genotype does not significantly affect survival of chicken. In fact, in a previous study, it was reported that two amino acids, histidine and tyrosine, provide copper binding ability of the chicken prion protein. Since chicken has twice the number of amino acids in the tandem repeat unit compared mammals (mammals: histidine, chicken: histidine, tyrosine), it was presumed that the impact of a single deletion of hexapeptide seems to be relatively low and does not act as selection pressure [Citation33]. In addition, we did not find SNPs in the chicken PRNP gene. Since the PRNP gene is highly polymorphic in mammals and several disease-associated SNPs were identified [Citation34–Citation37], it is noteworthy that no SNP was found in the chicken PRNP gene. Since the chicken samples have been collected from an animal farm, it is possible that whole genetic diversity of chicken isn’t reflected in this study. Thus, future studies will be necessary to identify SNPs of the chicken PRNP gene in large groups and other breeds of the chicken.
Lastly, we assessed the impact of hexapeptide deletion polymorphism on the protein. For this, we used PROVEAN to evaluate the impact of c.163_180delAACCCAGGGTACCCCCAT. PROVEAN measured this deletion protein polymorphism as ‘Deleterious’ with a score of −13.173. This score indicates that hexapeptide deletion may make impact prion protein function. In a recent in vitro functional study, because it was reported that the number of hexapeptide repeat units is directly proportional to copper binding ability [Citation32], the heterozygotes or homozygotes of U2 deletion found in this study demonstrated decreased copper binding ability. An exact estimate of copper binding ability according to hexapeptide genotype is highly desirable in the future.
Overall, our study suggested three main points. First, we investigated chicken PRNP gene polymorphisms and found only in/del polymorphisms. Second, we suggested a novel tandem repeat unit of chicken hexapeptide repeat based on polymorphisms. Third, we evaluated the impact of deletion of hexapeptide polymorphisms in silico. To the best of our knowledge, this was the first report of chicken PRNP gene polymorphisms.
Materials and methods
Ethics statement
A total of 106 Dekalb White (3 weeks old) chickens were obtained from an animal farm in South Korea. All experimental processes performed in the present study approved according to regulations of the Animal Care and Use Committee of Chonbuk National University (IACUC Number: CBNU 2017–0030).
DNA extraction
Genomic DNA was isolated from 20 mg peripheral tissue using the LaboPass Tissue Genomic DNA Isolation Kit (Cosmo Genetech CO., Ltd., Korea) following the manufacturer’s instructions.
Genetic analysis
Polymerase chain reaction (PCR) was carried out with gene-specific sense and antisense primers as follows: chicken PRNP-F (TGGGATGATGCTTGATTTCGGT) and chicken PRNP-R (ATCCCTGTCACGCTCCAGAA) using S-1000 Thermal Cycler (Bio-Rad Laboratories, USA). The PCR reagents contained 25 pmol of each primer, 5 μl of 10 × Taq DNA polymerase buffer, 1 µl of 10 mM dNTPs and 2.5 units of Taq DNA polymerase (Promega, USA). The PCR conditions were as follows: 94°C for 2 min to denature, and 35 cycles of 94°C for 45 sec, 63°C for 45 sec, 72°C for 1 min 30 sec, and then 1 cycle of 72°C for 10 min to extend the reaction. A 5 μl aliquot of the PCR product was analyzed by electrophoresis on a 1.2% agarose gel stained with EcoDyeTM DNA Staining Solution (BIOFACT, KOREA) to confirm the target band size (978 bp). The PCR product (sample # 44) was subcloned into pGEM-T easy vector (Promega, USA). The recombinant plasmid pGEM-T easy vector was transformed into the E. coli TOP 10 competent cells (Thermo Fisher Scientific, USA). The purified PCR products and the positive clones were performed with Sanger DNA sequencing using ABI 3730 automatic sequencer with Taq Dideoxy Terminator Cycle Sequencing Kit (ABI, USA). The sequencing result was processed by Finch TV software (Geospiza Inc, Seattle, USA), and genotyping of the chicken PRNP gene was performed. The haplotypes of two polymorphisms in the chicken PRNP gene were analyzed by SNP Analyzer 1.2A (http://snp.istech21.com/snpanalyzer/1.2A/). We also examined Lewontin’s D’ (|D’|) to investigate LD among these two polymorphisms by SNP Analyzer 1.2A.
Evaluation impact of insertion/deletion to chicken PrPc
PROVEAN assesses the impact of polymorphisms by building up and comparing the clusters of related sequences and predicting the score. If the final score is below −2.5, protein variants are predicted to be ‘neutral’; if the final score is above −2.5, protein variants are predicted as ‘deleterious’ (http://provean.jcvi.org/seq_submit.php).
Abbreviations
| LD | = | linkage disequilibrium |
| ORF | = | open reading frame |
| PRNP | = | prion protein gene |
| PrP | = | prion protein |
| PrPC | = | cellular form of prion protein |
| PrPSc | = | scrapie form of the prion protein |
| SNP | = | single nucleotide polymorphism |
| UTRs | = | untranslated regions |
Acknowledgments
We wish to thank DVM Yun-Hwan Yeo for isolating tissue and blood samples of Chickens.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. PMID: 9811807.
- Matthews D, Cooke BC. The potential for transmissible spongiform encephalopathies in non-ruminant livestock and fish. Rev Sci Tech. 2003;22: 283–296. PMID: 12793786.
- Moore J, Hawkins SA, Austin AR, et al. Studies of the transmissibility of the agent of bovine spongiform encephalopathy to the domestic chicken. BMC Res Notes. 2011;4:501. PMID: 22093239.
- Wopfner F, Weidenhöfer G, Schneider R, et al. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol. 1999;289:1163–1178. PMID: 10373359.
- Redecke L, Meyer-Klaucke W, Koker M, et al. Comparative analysis of the human and chicken prion protein copper binding regions at pH 6.5. J Biol Chem. 2005;280:13987–13992. PMID: 15684434.
- Atoji Y, Ishiguro N. Distribution of the cellular prion protein in the central nervous system of the chicken. J Chem Neuroanat. 2009;38:292–301. PMID: 19751818.
- Liu T, Zwingman T, Li R, et al. Differential expression of cellular prion protein in mouse brain as detected with multiple anti-PrP monoclonal antibodies. Brain Res. 2001;896: 118–129. PMID: 11277980.
- Diaz-San Segundo F, Salguero FJ, de Avila A, et al. Distribution of the cellular prion protein (PrPC) in brains of livestock and domesticated species. Acta Neuropathol. 2006;112:587–595. PMID: 16957924.
- Ji HF, Zhang HY. A comparative molecular dynamics study on thermostability of human and chicken prion proteins. Biochem Biophys Res Commun. 2007;359:790–794. PMID: 17560545.
- Zhang J. The structural stability of wild-type horse prion protein. J Biomol Struct Dyn. 2011;29:369–377. PMID: 21875155.
- Zhang J. Comparison studies of the structural stability of rabbit prion protein with human and mouse prion proteins. J Theor Biol. 2011;269:88–95. PMID: 20970434.
- Zhang J, Liu DD. Molecular dynamics studies on the structural stability of wild-type dog prion protein. J Biomol Struct Dyn. 2011;28:861–869. PMID: 21469747.
- Qing LL, Zhao H, Liu LL. Progress on low susceptibility mechanisms of transmissible spongiform encephalopathies. Dongwuxue Yanjiu. 2014;35:436–445. PMID: 25297084.
- Jeong BH, Lee KH, Kim NH, et al. Association of sporadic Creutzfeldt-Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics. 2005;6:229–232. PMID: 16217673.
- Jeong BH, Nam JH, Lee YJ, et al. Polymorphisms of the prion protein gene (PRNP) in a Korean population. J Hum Genet. 2004;49:319–324. PMID: 15148589.
- Groschup MH, Lacroux C, Buschmann A, et al. Classic scrapie in sheep with the ARR/ARR prion genotype in Germany and France. Emerg Infect Dis. 2007;13:1201–1207. PMID: 17953092.
- Bossers A, Schreuder BE, Muileman IH, et al. PrP genotype contributes to determining survival times of sheep with natural scrapie. J Gen Virol. 1996;77:2669–2673. PMID: 8887505.
- Goldmann W, Martin T, Foster J, et al. Novel polymorphisms in the caprine PrP gene: a codon 142 mutation associated with scrapie incubation period. J Gen Virol. 1996;77:2885–2891. PMID: 8922485.
- Babar ME, Abdullah M, Nadeem A, et al. Prion protein gene polymorphisms in four goat breeds of Pakistan. Mol Biol Rep. 2009;36:141–144. PMID: 17934795.
- Johnson CJ, Herbst A, Duque-Velasquez C, et al. Prion protein polymorphisms affect chronic wasting disease progression. PloS One. 2011;6:e17450. PMID: 21445256.
- Brandt AL, Kelly AC, Green ML, et al. Prion protein gene sequence and chronic wasting disease susceptibility in white-tailed deer (Odocoileus virginianus). Prion. 2015;9:449–462. PMID: 26634768.
- Jeong BH, Jin HT, Carp RI, et al. Bovine spongiform encephalopathy (BSE)-associated polymorphisms of the prion protein (PRNP) gene in Korean native cattle. Anim Genet. 2013;44:356–357. PMID: 23134411.
- Jansen C, Van Swieten JC, Capellari S, et al. Inherited Creutzfeldt-Jakob disease in a Dutch patient with a novel five octapeptide repeat insertion and unusual cerebellar morphology. J Neurol Neurosurg Psychiatry. 2009;80:1386–1389. PMID: 19917818.
- Sailer A, Bueler H, Fischer M, et al. No propagation of prions in mice devoid of PrP. Cell. 1994;77: 967–968. PMID: 7912659.
- Weissmann C, Bueler H, Fischer M, et al. PrP-deficient mice are resistant to scrapie. Ann N Y Acad Sci. 1994;724: 235–240. PMID: 8030944.
- Brown DR, Qin K, Herms JW, et al. Nature. 1997;390:684–687. PMID: 9414160.
- Jackson GS, Murray I, Hosszu LL, et al. Location and properties of metal-binding sites on the human prion protein. Proc Natl Acad Sci U S A. 2001;98:8531–8535. PMID: 11438695.
- Miura T, Hori-I A, Mototani H, et al. Raman spectroscopic study on the copper(II) binding mode of prion octapeptide and its pH dependence. Biochemistry. 1999;38:11560–11569. PMID: 10471308.
- Samorodnitsky D, Nicholson EM. Differential effects of divalent cations on elk prion protein fibril formation and stability. Prion. 2018;12:63–71. PMID: 29310497.
- Sánchez-López C, Fernández CO, Quintanar L. Neuroprotective alpha-cleavage of the human prion protein significantly impacts Cu(ii) coordination at its His111 site. Dalton Trans. 2018 Feb 8. PMID: 29417110. DOI:10.1039/c7dt03400h
- Di Natale G, Pappalardo G, Milardi D, et al. Membrane interactions and conformational preferences of human and avian prion N-terminal tandem repeats: the role of copper(II) ions, pH, and membrane mimicking environments. J Phys Chem B. 2010;114:13830–13838. PMID: 20936829.
- Stanczak P, Luczkowski M, Juszczyk P, et al. Interactions of Cu2+ ions with chicken prion tandem repeats. Dalton Trans. 2004;14:2102–2107. PMID: 15249945.
- Pietropaolo A, Muccioli L, Zannoni C, et al. Unveiling the role of histidine and tyrosine residues on the conformation of the avian prion hexarepeat domain. J Phys Chem B. 2008;112:5182–5188. PMID: 18386869.
- Kim YC, Jeong BH. Lack of germline mutation at codon 211 of the prion protein gene (PRNP) in Korean native cattle. Acta Vet Hung. 2017;65:147–152. PMID: 28244340.
- Jeong BH, Kim YS. Genetic studies in human prion diseases. J Korean Med Sci. 2014;29:623–632. PMID: 24851016.
- Sander P, Hamann H, Drogemuller C, et al. Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 2005;280:37408–37414. PMID: 16141216.
- Belt PB, Muileman IH, Schreuder BE, et al. Identification of five allelic variants of the sheep PrP gene and their association with natural scrapie. J Gen Virol. 1995;76:509–517. PMID: 7897344.
