ABSTRACT
Chronic Wasting Disease (CWD), a well-described transmissible spongiform encephalopathy of the Cervidae family, is associated with the aggregation of an abnormal isoform (PrPCWD) of the naturally occurring host prion protein (PrPC). Variations in the PrP gene (PRNP) have been associated with CWD rate of infection and disease progression. We analysed 568 free-ranging white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties for PRNP polymorphisms. Sampling included 185 CWD-positive, 332 CWD non-detected, and an additional 51 CWD non-detected paired to CWD-positives by sex, age, and harvest location. We found 12 polymorphic sites of which 5 were non-synonymous and resulted in a change in amino acid composition. Thirteen haplotypes were predicted, of which 11 have previously been described. Using logistic regression, consistent with other studies, we found haplotypes C (OR = 0.488, 95% CI = 0.321–0.730, P < 0.001) and F (OR = 0.122, 95% CI = 0.007–0.612, P < 0.05) and diplotype BC (OR = 0.340, 95% CI = 0.154–0.709, P < 0.01) were less likely to be found in deer infected with CWD. As has also been documented in other studies, the presence of a serine at amino acid 96 was less likely to be found in deer infected with CWD (P < 0.001, OR = 0.360 and 95% CI = 0.227–0.556). Identification of PRNP polymorphisms associated with reduced vulnerability to CWD in Michigan deer and their spatial distribution can help managers design surveillance programmesand identify and prioritize areas for CWD management.
Introduction
Chronic Wasting Disease (CWD), a well described, fatal, transmissible spongiform encephalopathy of the Cervidae family, is associated with the aggregation of an abnormal isoform (PrPCWD) of the naturally occurring host prion protein (PrPC) [Citation1–3]. First characterized in 1980 based on clinical and pathological findings in Colorado captive mule deer [Citation2], CWD has since spread within the United States, been found in Canada and Europe, and been detected in imported cervids in Korea [Citation4–7].
CWD is efficiently transmitted both horizontally [Citation8–11] and vertically [Citation12] with effective transmission between cervid species [Citation6,Citation13,Citation14]. Prion shedding can begin during the pre-clinical stage of disease [Citation9,Citation15] through bodily fluids and excreta [Citation9] and shed prions are able to be taken up by vegetation [Citation16] and withstand degradation [Citation17]. Once an animal is infected, CWD is always fatal [Citation3].
CWD prevalence in free-ranging cervid populations has been found to be as high as 35% [Citation18] with population-level impacts seen with prevalence as low as 13% [Citation19–21]. Cervid populations provide not only social and cultural benefits through hunting and viewing, and ecological contributions to biodiversity, they also serve as a financial keystone species for conservation and management making their potential decline of considerable management concern.
Two non-synonymous polymorphisms within the prion gene (PRNP) resulting in changes to amino acids 95 (Q95H) and 96 (G96S), have been most commonly found to be associated with reduced disease susceptibility in white-tailed deer [Citation22–35]. While neither have been shown to provide complete protection from CWD infection, they have been linked to reductions in genotype-specific prevalence rates [Citation26, Citation29–31,Citation36] or increased duration of incubation [Citation37].
CWD was first detected in wild white-tailed deer (Odocoileus virginianus) in Michigan in 2015 through opportunistic passive surveillance, 6 years after the state’s first detection in a captive herd. Since 2015, Michigan has invested in intensive surveillance through localized culling and hunter assisted sampling and CWD has been detected in 9 counties at the time of this study. We examined the current frequency of PRNP polymorphisms among CWD-positive and non-detected deer in 9 CWD-positive Michigan counties, one county in the Upper Peninsula and 8 contiguous counties in central Michigan. We tested for an association between CWD status and PRNP polymorphisms and hypothesized CWD polymorphisms associated with reduced CWD infection are present in Michigan white-tailed deer.
Results
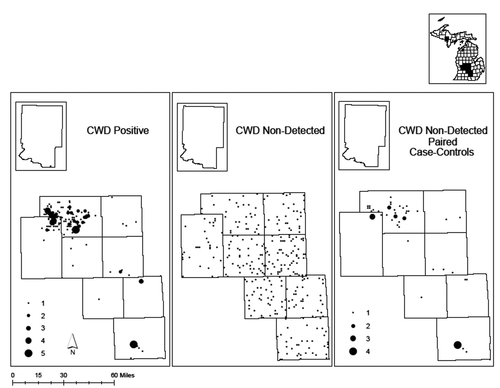
PRNP sequences were determined for 568 free-ranging white-tailed deer from 9 CWD-positive Michigan counties. Of these samples, 185 were CWD-positive, 332 were CWD non-detected, and an additional 51 CWD non-detected were paired to CWD-positives to control for sex, age, and harvest location (). Within the analysed 625bp region of the PRNP gene, we detected 12 single nucleotide polymorphisms (SNPs), 9 of which had been previously reported [Citation22, Citation29, Citation33, Citation36, Citation38–41]. Of the 12 SNPs, 5 were non-synonymous, resulting in a change to the amino acid sequence (). BLAST and literature searches indicated that 589A/G, 642 G/A, and 643 C/A had not previously been reported. Full associated sequences have been deposited in GenBank under accession numbers MZ913400 – MZ913401.
Table 1. Thirteen haplotypes and associated single nucleotide polymorphisms (SNP) of the PRNP gene from 568 white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties. Bold text indicated non-synonymous SNPs. Asterisks indicate previously unreported SNPs
Figure 1. Distribution of sampled (a) chronic wasting disease (CWD) positive, (b) CWD non-detected, and (c) CWD non-detected paired control free-ranging white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties
Thirteen haplotypes were predicted from the 12 SNPs, 11 of which have previously been described [Citation22, Citation40, Citation41]. Of the 13 haplotypes, B was most common (n = 368) and was used as the reference in logistic regression. Haplotypes J and MI-1 were found only in CWD non-detected deer, precluding them from analysis. Haplotypes C (OR = 0.488, 95% CI = 0.321–0.730, P < 0.001) and F (OR = 0.122 and 95% CI = 0.007–0.612, P < 0.05) were less likely to be found in deer infected with CWD ().
Table 2. PRNP haplotype frequency (f) and count for chronic wasting disease positive (+) and non-detected (-) white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties. Odds ratios and 95% confidence intervals are shown for significant variables (P < 0.05) determined by logistic regression against the most frequent haplotype, B. Asterisks indicate haplotypes found in only positive or non-detected deer precluding them from analysis. Bolding indicates previously unreported haplotypes
We identified 49 diplotypes with AB being the most common (n = 89) and used this as the reference in logistic regression. Twenty-four diplotypes were found only in positive or non-detected deer, precluding them from analysis. Of the remaining 25 diplotypes, BC (OR = 0.340 and 95% CI = 0.154–0.709, P < 0.01) was less likely to be found in deer infected with CWD ().
Table 3. PRNP diplotype frequency (f) and count for chronic wasting disease positive (+) and non-detected (-) white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties. Odds ratios and 95% confidence intervals are shown for significant variables (P < 0.05) determined by logistic regression against the most frequent diplotype, AB. Asterisks indicate diplotypes found in only positive or non-detected deer precluding them from analysis
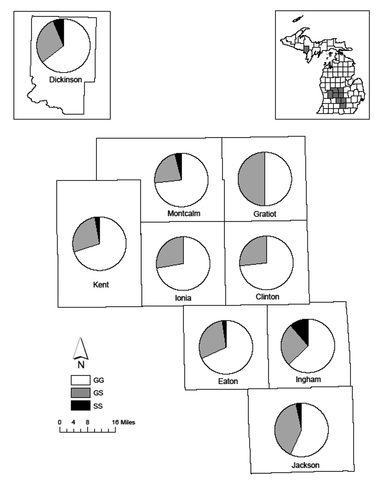
Three genotypes at aa96 were observed; aa96GG was most common (n = 387) and was used as the reference in logistic regression. aa96GS was less likely to be found in deer infected with CWD (OR = 0.360 and 95% CI = 0.227–0.556, P < 0.001; ; ); however, we did not detect a reduced likelihood of infection for homozygous individuals (aa96SS). Two genotypes at aa95 were observed, aa95QQ was most common (n = 552). No significant associations were seen for genotype at aa95 and CWD infection.
Table 4. PRNP genotype frequency (f) and count at codons 95 and 96 for chronic wasting disease positive (+) and non-detected (-) white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties
Figure 2. Proportion of PRNP amino acid 96 genotypes of white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties
Among the case-controlled samples, the presence of one C haplotype or one serine at aa96 was confirmed to be associated with reduced CWD infection by 0.191 (95% CI = 0.065–0.555, P < 0.01) and 0.182 (95% CI = 0.063–0.528, P < 0.01), respectively. As with the full dataset comparison, no evidence for protection was seen in homozygous CC or aa96SS individuals.
The ratio of haplotypes C and F, diplotype BC, and genotype aa96GS (associated with reduced susceptibility) relative to non-protective haplotypes, diplotypes and genotypes, respectively, were compared among the nine studied counties. Pairwise comparisons using Fisher’s exact tests failed to detect significant differences among counties in the distribution of protective genetic types after p-values were corrected for multiple comparisons.
Discussion
This is the first examination of PRNP variation for a wild white-tailed deer population in Michigan. We established baseline frequencies of PRNP genotypes, haplotypes, and diplotypes in nine known CWD-positive counties. We found aa96GS, haplotypes C and F, and diplotype BC to be less frequent in CWD-positive deer, consistent with other studies [Citation22, Citation24–27, Citation29–35]. The results of our analyses of paired samples controlling for potential confounding variables of age, sex, and harvest location further reinforce the finding of an association between haplotype C and the presence of a serine at aa96 with reduced vulnerability to CWD. While the C haplotype and aa96S were less frequent in CWD-positive deer, we did not find evidence to support a reduced vulnerability to CWD for homozygotes. Previous work has also failed to detect a reduced likelihood of infection among aa96SS individuals [Citation22]. In the current study, this could be indicative of a biological process due to strain type, or an artefact of the low prevalence of aa96SS reducing our power to detect an effect. To account for strain-type differences, these results should be used to target aa96GG, aa96GS, and aa96SS CWD-positive individuals for inclusion in strain-type assessments. And with increased sampling over time we may produce a greater proportion of aa96SS individuals for evaluation.
Annual apparent CWD prevalence between 2015 and 2019 varied across the 9 positive counties with the highest prevalence of 1.95% seen in Kent county in 2019 (). We found no statistically significant differences in PRNP genotype or haplotype frequencies across these counties suggesting that, within the current known distribution of CWD in Michigan, infection vulnerability based on PRNP is relatively homogeneous. Given that deer in some counties in Michigan seem to have higher CWD prevalence than others, it will be of interest to monitor the potential selective impacts of CWD across these areas. While no PRNP types have been associated with complete resistance, the presence of aa96S has been associated with slower disease progression and longer survival post-infection [Citation30, Citation31]. Longer survival may provide deer with aa96S a selective advantange leading to changes in PRNP frequencies in wild populations over time [Citation27]. Our data present the current localized prevalence of G96S (28.7%) as similar to studies in white-tailed deer in Wyoming [Citation38] (20%), but higher than those in Illinois [Citation33] (13.8%), and northern Illinois and southern Wisconsin [Citation22] (11%). Our characterization of PRNP frequencies, presumably relatively early in the disease’s occurrence in Michigan, provides a baseline for monitoring selective effects of CWD on PRNP frequencies and white-tailed deer population characteristics over time and should be used in disease modelling efforts to map risk and rate of spread.
Table 5. Apparent prevalence of chronic wasting disease in white-tailed deer (Odocoileus virginianus) from 9 CWD-positive Michigan counties. Number in parentheses corresponds to total number of animals tested for the year in the given county
It is important to note some possible limitations to our study that point towards the need for future investigation. This assessment was a snapshot of polymorphisms restricted to a 625bp region from deer in a relatively small geographic area. Future work to monitor frequencies of haplotype C, aa96S, and any new informative polymorphisms outside of the 625bp region will help inform disease impact, possible selection within the population, and target regions for special management attention. Additional assessments of genetic connectivity among deer in the CWD-positive regions would also inform delineation of management areas.
Surveillance is currently being used as the leading indicator to inform CWD management in wild deer populations, and while beneficial, surveillance is costly, limited in scope, and is not in itself a management tool. As CWD detections continue to increase the areas under surveillance, the use of regionally specific data to allocate testing efforts and funding will be pivotal for success. Identification of PRNP polymorphisms associated with reduced vulnerability to CWD and their spatial distribution and prevalence may help managers design surveillance programmes to identify and prioritize areas for CWD management when partnered with movement data and anticipated deposition of prions onto the landscape over time.
Materials and methods
Sampling
Medial retropharyngeal lymph nodes were collected from white-tailed deer by Michigan Department of Natural Resource staff during routine disease surveillance between April 2015 and January 2020 from 9 CWD-positive Michigan counties. Sex, harvest location, and age, as assessed by tooth wear and replacement, were collected from all sampled deer. Samples were stored at −20°C or −80°C until prepared for DNA extraction.
Subsampling within each county for this study represented: 1) CWD-positive deer; 2) CWD non-detected deer; and 3) and additional CWD non-detected paired controls. Sampling aimed to obtain three individuals from unique sections (2.6 km2) in each township (93 km2) for CWD non-detected animals. To control for factors known to be associated with CWD infection probability, paired controls were identified for a subset of CWD-positive deer by matching a CWD-positive deer to a CWD non-detected deer of the same age, sex, and harvest location. Due to the already small sample size for paired controls, we were unable to control for background relatedness as done previously [Citation42].
Samples were collected within a short period of time that led us to assume relatively similar exposure to CWD between paired case-controls and CWD-positive and CWD non-detected deer.
CWD diagnosis
All animals were tested for CWD using a USDA approved enzyme-linked immunosorbent assay to detect PrPCWD at either the Michigan (East Lansing, MI) or Wisconsin (Madison, WI) Veterinary Diagnostic Laboratory. Confirmation by immunohistochemistry was done by the diagnostic laboratories or by the National Veterinary Services Laboratory (Ames, IA). Sampling did not allow for the assessment of disease stage in different tissue types; however, the use of lymph tissue, where PrPCWD deposition first occurs, reduced the chance that false negatives might impact these results [Citation23].
Prnp sequence analysis
Genomic DNA was isolated from lymph node tissue using Qiagen DNeasy Blood and Tissue Kits (Qiagen Inc., Valencia, CA) following manufacturer’s guidelines with a final elution volume of 200uL in Buffer AE.
The PRNP gene was amplified using a primer pair specific for the functional gene (223 5ʹ-acaccctctttattttgcag-3ʹ and 224 5ʹ-agaagataatgaaaacaggaag-3ʹ) [Citation36]. PCR amplicons were purified using ExoSAP-IT (Applied Biosystems, Foster City, CA) and products were sequenced using the Big Dye Terminator system (Applied Biosystems, Foster City, CA). Sequence products were purified using ethanol/EDTA precipitation and resolved on an ABI 3500.
Sequences were visualized and edited in SEQUENCHER (Gene Codes Corporation, Ann Arbor, MI). Re-sequencing was done until regions of variability were confirmed three times. Haplotypes were generated from unphased sequences using DNA Sequence Polymorphism 5.10.01 (Rozas et al., Universitat de Barcelona). Markov chain Monte Carlo (MCMC) samples were taken from a minimum of 1,000 iterations, with a discarded burn-in of 100 iterations. Previously published haplotype sequences [Citation22, Citation43] were uploaded from NCBI and a local BLAST was run to match phased sequences to published haplotypes.
Phased sequences were translated in SEQUENCHER to their amino acid composition for final reporting.
Statistical analyses
We used logistic regression to identify associations between CWD status and haplotype, diplotype, and aa95 and aa96 genotypes. Chronic wasting disease status was a binomial variable with CWD-positive deer coded as 1 and non-detected deer coded as 0. Genetic data were treated as categorical variables. The most common genetic type was used as the reference type in each analysis. Genetic types significantly associated with CWD status were those with P-values ≤ 0.05. Odds ratios (ORs) and associated 95% confidence intervals were also calculated. Odds ratios with 95% confidence intervals that did not include one were considered significant. Genetic types with significant ORs less than one were interpreted as exhibiting reduced susceptibility to CWD.
To further explore associations between CWD status and genetic type while controlling for other factors that might affect CWD status (eg, age, sex, location), conditional logistic regression was used to identify associations between CWD status and genetic types for matched case-control pairs. The lesser number of available pairs (n = 51) limited the analyses we could conduct. Based on findings from the analyses described above as well as previous studies, we tested for associations between CWD status and presence of a C haplotype, CC genotype, and presence of at least one serine at aa96 using the clogit function in the survival package [Citation44] in R [Citation45] (version 3.6.1). We coded CWD status as described above. We did not assess haplotype F as only one available deer pair had a F haplotype. Significance was interpreted as described above.
We assessed differences in the frequency of presumably protective haplotypes, diplotypes, and genotypes among the 9 counties where CWD had been identified using Fisher’s exact tests.
Acknowledgments
The authors would like to acknowledge the following for their contributions in preparing this manuscript: Tyler Petroelje, Sarah Mayhew, Ansley Rowland, Joe Darish, Samantha Ludlam, and Jessica Clark.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Williams ES. Chronic wasting disease. Vet Pathol. 2005;42:530–549.
- Williams ES, Young S. Chronic wasting disease of captive mule deer: a spongiform encephalopathy. J Wildl Dis. 1980;16:89–98.
- Williams ES, Young S. Spongiform encephalopathies in Cervidae. Rev Sci Tech. 1992;11:551–567.
- Sohn HJ, Kim JH, Choi KS, et al. A case of chronic wasting disease in an elk imported to Korea from Canada. J Vet Med Sci. 2002;64:855–858.
- Kim TY, Shon HJ, Joo YS, et al. Additional cases of chronic wasting disease in imported deer in Korea. J Vet Med Sci. 2005;67:753–759.
- Benestad SL, Mitchell G, Simmons M, et al. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res. 2016;47:88.
- United States Geological Survey. Distribution of chronic wasting disease in North America [Internet]; 2020. Accessed 7 September 2020. Available from: https://www.usgs.gov/media/images/distribution-chronic-wasting-disease-north-america-0
- Haley NJ, Seelig DM, Zabel MD, et al. Detection of CWD prions in urine and saliva of deer by transgenic mouse bioassay. PLoS One. 2009;4:e4848.
- Mathiason CK, Hays SA, Powers J, et al. Infectious prions in pre-clinical deer and transmission of chronic wasting disease solely by environmental exposure. PLoS One. 2009;4:e5916.
- Pritzkow S, Morales R, Lyon A, et al. Efficient prion disease transmission through common environmental materials. J Biol Chem. 2018;293:3363–3373.
- Saunders SE, Bartelt-Hunt SL, Bartz JC. Prions in the environment: occurrence, fate and mitigation. Prion. 2008;2:162–169.
- Nalls AV, McNulty E, Powers J, et al. Mother to offspring transmission of chronic wasting disease in Reeves’ Muntjac deer. PLoS One. 2013;8:e71844.
- Baeten LA, Powers BE, Jewell JE, et al. A natural case of chronic wasting disease in a free-ranging moose (Alces alces shirasi). J Wildl Dis. 2007;43(2):309–314.
- Spraker TR, Miller MW, Williams ES, et al. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus vircinianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J Wildl Dis. 1997;33:1–6.
- Tamgüney G, Miller MW, Wolfe LL, et al. Asymptomatic deer excrete infectious prions in feces. Nature. 2009;761:529–532.
- Pritzkow S, Morales R, Moda F, et al. Grass plants bind, retain, uptake and transport infectious prions. Cell Reprod. 2015;11:1168–1175.
- Wyckoff AC, Kane S, Lockwood K, et al. Clay components in soil dictate environmental stability and bioavailability of cervid prions in mice. Front Microbiol. 2016;7:1885.
- Wisconsin Department of Natural Resources. CWD prevalence in Wisconsin [Internet]; 2020. Accessed 13 July 2020. Available from: https://dnr.wi.gov/topic/wildlifehabitat/prevalence.html
- Monello RJ, Powers JG, Hobbs NT, et al. Survival and population growth of a free-ranging elk population with a long history of exposure to chronic wasting disease. J Wildl Manage. 2014;78:214–223.
- DeVivo MT, Edmunds DR, Kauffman MJ, et al. Endemic chronic wasting disease causes mule deer population decline in Wyoming. PLoS One. 2017;12:e0186512.
- Edmunds DR, Kauffman MJ, Schumaker BA, et al. Chronic wasting disease drives population decline of white-tailed deer. PLoS One. 2016;11:e0161127.
- Brandt AL, Kelly AC, Green ML, et al. Prion protein gene sequence and chronic wasting disease susceptibility in white-tailed deer (Odocoileus virginianus). Prion. 2015;9:449–462.
- Fox KA, Jewell JE, Williams ES, et al. Patterns of PrPCWD accumulation during the course of chronic wasting disease infection in orally inoculated mule deer (Odocoileus hemionus). J Gen Virol. 2006;87:3451–3461.
- Wilson GA, Nakada SM, Bollinger TK, et al. Polymorphisms at the PRNP gene influence susceptibility to chronic wasting disease in two species of deer (Odocoileus Spp.) in Western Canada. J Toxicol Environ Heal Part A. 2009;72:1025–1029.
- Robinson SJ, Samuel MD, Lopez DL, et al. The walk is never random: subtle landscape effects shape gene flow in a continuous white-tailed deer population in the Midwestern United States. Mol Ecol. 2012;21:4190–4205.
- Robinson SJ, Samuel MD, O’Rourke KI, et al. The role of genetics in chronic wasting disease of North American cervids. Prion. 2012;6:153–162.
- Robinson SJ, Samuel MD, Johnson CJ, et al. Emerging prion disease drives host selection in a wildlife population. Ecol Soc Am. 2012;22:1050–1059.
- Jewell JE, Conner MM, Wolfe LL, et al. Low frequency of PrP genotype 225SF among free-ranging mule deer (Odocoileus hemionus) with chronic wasting disease. J Gen Virol. 2005;86:2127–2134.
- Johnson C, Johnson J, Clayton M, et al. Prion protein gene heterogeneity in free-ranging white-tailed deer within the chronic wasting disease affected region of Wisconsin. J Wildl Dis. 2003;39:576–581.
- Johnson C, Johnson J, Vanderloo JP, et al. Prion protein polymorphisms in white-tailed deer influence susceptibility to chronic wasting disease. J Gen Virol. 2006;87:2109–2114.
- Johnson CJ, Herbst A, Duque-Velasquez C, et al. Prion protein polymorphisms affect chronic wasting disease progression. PLoS One. 2011;6:e17450.
- Keane DP, Barr DJ, Bochsler PN, et al. Chronic wasting disease in a Wisconsin white-tailed deer farm. J Vet Diagn Investig. 2008;20:698–703.
- Kelly AC, Mateus-Pinilla NE, Diffendorfer J, et al. Prion sequence polymorphisms and chronic wasting disease resistance in Illinois white-tailed deer (Odocoileus virginianus). Prion. 2008;2:28–36.
- Miller WL, Walter WD. Spatial heterogeneity of prion gene polymorphisms in an area recently infected by chronic wasting disease. Prion. 2019;13:65–76.
- O’Rourke KI, Besser TE, Miller MW, et al. PrP genotypes of captive and free-ranging Rocky Mountain elk (Cervus elaphus nelsoni) with chronic wasting disease. J Gen Virol. 1999;80:2765–2769.
- O’Rourke KI, Spraker TR, Hamburg LK, et al. Polymorphisms in the prion precursor functional gene but not the pseudogene are associated with susceptibility to chronic wasting disease in white-tailed deer. J Gen Virol. 2004;85:1339–1346.
- Denkers ND, Hoover CE, Davenport KA, et al. Very low oral exposure to prions of brain or saliva origin can transmit chronic wasting disease. PLoS One. 2020;15:1–17.
- Heaton MP, Leymaster KA, Freking BA, et al. Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer. Mamm Genome. 2003;14:765–777.
- Raymond GJ. Evidence of a molecular barrier limiting susceptibility of humans, cattle and sheep to chronic wasting disease. EMBO J. 2000;19:4425–4430.
- Perrin-Stowe TIN, Ishida Y, Terrill EE, et al. Prion Protein Gene (PRNP) sequences suggest differing vulnerability to chronic wasting disease for Florida Key deer (Odocoileus virginianus clavium) and Columbian white-tailed deer (O. v. leucurus). J Hered. 2020;111:564–572.
- Vázquez-Miranda H, Zink RM. Geographic distribution of chronic wasting disease resistant alleles in Nebraska, with comments on the evolution of resistance. J Fish Wildl Manag. 2020;11:46–55.
- Blanchong JA, Heisey DM, Scribner KT, et al. Genetic susceptibility to chronic wasting disease in free-ranging white-tailed deer: complement component C1q and Prnp polymorphisms. Infect Genet Evol. 2009;9:1329–1335.
- Brandt AL, Green ML, Ishida Y, et al. Influence of the geographic distribution of prion protein gene sequence variation on patterns of chronic wasting disease spread in white-tailed deer (Odocoileus virginianus). Prion. 2018;12:204–215.
- Therneau T. A Package for Survival Analysis in R. R package version 2.37-4; 2013. Accessed 6 September 2021. Available from: http://CRAN.R-project.org/package=survival.
- R Core Team. R: a language and environment for statistical computing. R Foundation for statistical computing. Vienna Austria; 2018. Accessed 11 May 2020. Available from: http://www.R-project.org/.
