Abstract
During brain development, billions of neurons organize into highly specific circuits. To form specific circuits, neurons must build the appropriate types of synapses with appropriate types of synaptic partners while avoiding incorrect partners in a dense cellular environment. Defining the cellular and molecular rules that govern specific circuit formation has significant scientific and clinical relevance because fine scale connectivity defects are thought to underlie many cognitive and psychiatric disorders. Organizing specific neural circuits is an enormously complicated developmental process that requires the concerted action of many molecules, neural activity, and temporal events. This review focuses on one class of molecules postulated to play an important role in target selection and specific synapse formation: the classic cadherins. Cadherins have a well-established role in epithelial cell adhesion, and although it has long been appreciated that most cadherins are expressed in the brain, their role in synaptic specificity is just beginning to be unraveled. Here, we review past and present studies implicating cadherins as active participants in the formation, function, and dysfunction of specific neural circuits and pose some of the major remaining questions.
Abbreviations
| EC | = | extracellular cadherin |
| DG | = | dentate gyrus |
| RGCs | = | retinal ganglion cells |
| DN-cadherin | = | Drosophila N-cadherin |
| SNPs | = | single nucleotide polymorphisms |
| ADHD | = | attention deficit/hyperactivity disorder |
| BC | = | bipolar cell |
| EGF | = | epidermal growth factor |
Introduction
Molecules of the cadherin superfamily are defined by the presence of calcium binding, extracellular cadherin (EC) repeats. Members of the cadherin superfamily include the classic cadherins, protocadherins, desmosomal, Fat, and 7-pass transmembrane cadherins.Citation1 Cadherin superfamily members are conserved across species and most are expressed in the nervous system where they function in several aspects of neural development from neurogenesis and cell migration to synapse formation and plasticity.Citation1-4 By definition the classic cadherins contain precisely 5 extracellular EC repeats (and no other conserved extracellular motifs) and an intracellular catenin-binding domain (). For reference, other cadherin superfamily members have varying numbers of EC repeats, often in combination with other extracellular protein motifs, and do not bind the catenins.Citation3 The classic cadherins have been subject to investigation for many years, yet their role in the nervous system is still unclear. This review focuses on the role of the classic cadherins in target selection and synaptic specificity and aims to unify work spanning several decades starting from early suggestions that cadherins regulate specific circuit development based on their remarkably selective neuronal expression patterns to recent work that directly tests the role of individual cadherins in identified circuits using newly available genetic and imaging tools. For more information regarding the general role of cadherins and cadherin signaling in neural development, synapse formation, and plasticity we refer to several excellent reviews.Citation5-8
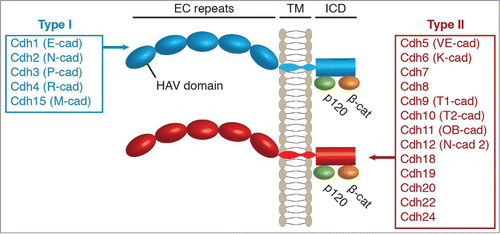
Figure 1. Structure of the classic cadherin protein family in humans. All classic cadherins have 5 extracellular cadherin (EC) repeats, a transmembrane (TM) domain, and an intracellular domain (ICD) that binds p120-catenin and β-catenin. The classic cadherins are sub-divided into Type I and Type II depending on the presence of a histidine-alanine-valine (HAV) motif in the first EC domain. Human Type I and Type II cadherins are indicated as annotated in the HUGO Gene Nomenclature Committee database with common names noted in parentheses.
The Development of Synaptic Specificity
At the turn of the 20th century, Ramón y Cajal famously documented the existence of specific neural connections through his detailed anatomical drawings of neuronal cell types and their orderly and selective axonal projections.Citation9 The term “synaptic specificity” describes the fact that neural connections do not form randomly, but instead are highly organized. Neurons often synapse only with specific types of partner neurons. This requires that presynaptic axons identify correct partners within a complex environment containing a myriad of incorrect cell types. Then neurons must build the appropriate types of synapses relevant to the types of neurons being connected.
The extreme specificity of neural connections is now well established but the molecular mechanisms responsible for generating synaptic specificity are only just beginning to be understood. Many cellular factors contribute to the development of synaptic specificity. These factors include temporal and spatial constraints, genetically specified molecular identities, and neural activity. Moreover, these factors are often inextricably linked. For example, genetically specified molecules can encode spatial constraints on growthCitation10 and neural activity can influence gene expression to provide neurons with a specific molecular identity.Citation11 The role of neural activity in circuit formation has been a particularly active area of investigation since Hubel and Wiesel's classic experiments showing that visual experience shapes the functional connectivity of visual circuits.Citation12 Subsequently, many studies have shown that neural activity mediates the removal of synapses from incorrect targets and the pruning of excessive synapses from correct targets.Citation13 Although activity plays a significant role in circuit refinement, many initial steps in target selection and synapse formation proceed normally in the complete absence of neural activity.Citation14-17 Thus, synaptic specificity is initially mapped out in a large part because neurons are genetically programmed to express unique molecular features allowing them to be identified by other neurons. This idea is rooted in John Langley's early studies on nerve regeneration in cats and was elegantly conceptualized in Roger Sperry's “chemoaffinity hypothesis”.Citation18,19 Sperry predicted that synaptic specificity is based on matching cytochemical tags expressed on pre and postsynaptic neurons. The extreme interpretation of this is that every neuron expresses a unique molecular identity that matches one to one with its correct synaptic partner.
Remarkably, neurons have a unique individual molecular identity that, in vertebrates, is encoded by members of the cadherin superfamily known as the clustered protocadherins. Clustered protocadherins are single-pass transmembrane proteins with 6 extracellular EC domains and intracellular domains with no clear homology or conserved binding domains.Citation20,21 In mice, 58 protocadherin genes are arranged in 3 clusters. Single cell RNA analysis of Purkinje neurons shows that individual neurons express about 1–3 isoforms from each cluster and expression is monoallelic and stochastic.Citation22-24 This means that there are hundreds of thousands of possible combinations, which afford many unique molecular identities.
Given that clustered protocadherins are located at synapses,Citation25,26 their discovery generated excitement because it was thought Sperry's one-to-one cytochemical tags allowing neurons to recognize correct synaptic partners had been found. Recent work, however, shows that the unique molecular labels provided by clustered protocadherins have an important but alternative function. They are not directly required for synapse formation between 2 different neurons, but instead mediate self-recognition to prevent dendrites on the same cell from interacting.Citation27,28 Clustered protocadherins are homophilic and their binding mediates repulsion, not adhesion.Citation28 The only dendrites a neuron is likely to contact with a strict homophilic match to its clustered protocadherins are its own dendrites and the repellent signaling causes the dendrites to spread apart from one another. Drosophila neurons use a similar mechanism of dendritic self-avoidance, but it is mediated by a completely different molecular family called Downs Syndrome cell adhesion molecules (DSCAMS).Citation29-31 Thus, studies indicate that neurons use their individual molecular identity to recognize themselves but not their synaptic partners.
Therefore, if not the clustered protocadherins, then what molecules mediate trans-cellular recognition among correct versus incorrect synaptic partners? The answer to this question is not yet solved and is likely to be a very complicated answer. Organizing the brain is an extremely complex process that requires the concerted action of many molecules, neural activity, and temporal events. However, new and increasing evidence suggests that one particularly important molecular family in regulating target recognition and synaptic specificity is the classic cadherins.
The Classic Cadherins
The mammalian classic cadherins, referred to from here as simply “cadherins,” consist of approximately 20 genes that each encode a single-pass transmembrane protein with 5 EC repeats and intracellular binding domains for p120-catenin and β-cateninCitation32 (). Catenins are accepted as the primary intracellular mediators of cadherin signaling although other signaling mechanisms have been reported.Citation5-8 For comparison, other members of the cadherin superfamily, including the protocadherins, do not bind catenins.Citation21 Mammalian cadherins can be further subdivided into type I and type II based on sequence homology in the first EC domain; a domain critical for trans-cellular cadherin-cadherin bindingCitation3,33,34 (). The human genome contains 5 type I and 13 type II cadherinsCitation35 (). Like the clustered protocadherins, type I and type II cadherin binding is largely homophilic,Citation36 however, heterophilic interactions between cadherins of the same subtype have been observed in vitro.Citation37-41
Cadherins are found in dendrites, axons, and growth cones of young neurons. Live imaging studies suggest they preferentially cluster in pre and postsynaptic compartments at nascent synapses and then are maintained at pre and postsynaptic structures as synapses mature.Citation42 Synaptic localization has been shown for several different cadherin molecules by light and electron microscopy in brain sections, cultured neurons, and slice cultures.Citation43-50 More specifically, cadherins are commonly found at puncta adherentia, structures analogous to adherens junctions that are located adjacent to the synaptic active zone,Citation44,45,51 but cadherins have also been shown to associate directly with the postsynaptic density.Citation52 Crystal structures of several cadherin extracellular domains suggest that trans-cellular cadherin interactions are the perfect length (∼40 nm) to span the synaptic cleft.Citation39,53,54 Thus, cadherins are expressed at the right place and time to function throughout synapse development.
Many Cadherins Have Circuit-Specific Expression in the Brain
The cadherins have been postulated to provide a molecular code to guide the formation of specific synaptic connections since the late 1980s and 90s when many cadherin family members were cloned and their expression patterns analyzed.Citation55-61 Although some cadherins, particularly those of the type I class, are widely expressed in the vertebrate brain, most type II cadherins are differentially expressed by subpopulations of neurons. Intriguingly, expression of type II cadherins often follows functional connections within circuits.Citation59,62-64 Takeichi and collegues provided an early demonstration of this when they injected a fluorescent dye in regions of the cortex expressing either cadherin-6 or cadherin-8 and retrograde uptake of the dye resulted in selective labeling of cadherin-6 or cadherin-8-expressing thalamic neurons respectively.Citation62 This indicated that neurons are selectively connected to other neurons that express matching cadherins and suggests that the cadherins themselves may be responsible for guiding the connectivity.
However, the idea that cadherins guide selective connectivity fell out of favor for many years because experiments investigating cadherins in the nervous system failed to provide conclusive evidence to support this role. In hindsight, this likely resulted from several reasons. First, analyses of cadherin function in the nervous system have overwhelmingly centered on the role of N-cadherin (also known as cadherin-2) in cultured neurons.Citation6,8 N-cadherin is a type I cadherin that is broadly expressed by most neurons and, with some exceptions, studies largely suggest N-cadherin has a primary role in general synapse function and plasticity rather than in synaptic specificity. Second, a common approach to study cadherins in the brain has been to simultaneously ablate the function of all cadherins using dominant negative constructs or catenin knockouts.Citation65-67 These first 2 strategies led to important discoveries about the general functions of cadherins in neural development, but by design they do not permit analysis of specificity. Third, overlapping and combinatorial cadherin expression along with potential functional redundancy with other synaptic specificity molecules ensures high fidelity of circuit formation but also means that chances are great that the loss of 1 or 2 cadherin genes will not result in obvious connectivity defects.
Differentially Expressed Cadherins in Synaptic Specificity
Other than the expression data mentioned above, there had been some findings that suggested but did not directly test a role for cadherins in specificity. This includes the finding that overexpression of cadherin-11 increases synapse density in cultured neurons whereas overexpression of either cadherin-13, cadherin-9, or N-cadherin does not.Citation51,65,68 Axons of hippocampal dentate gyrus (DG) neurons grow longer on an N-cadherin substrate compared to a cadherin-8 substrateCitation69 and motor neurons use differential expression of type II cadherins to sort into topographically defined motor pools.Citation63 It was also shown that high frequency stimulation capable of inducing long term potentiation in the medial molecular layer of the hippocampus selectively decreases cadherin-8 from this layer but does not alter N-cadherin expression.Citation70 Together, these observations suggest that different cadherins function distinctly at different types of synapses.
Recent technological advances in neuron and synapse labeling, imaging, and gene manipulation have facilitated more direct and detailed investigations of the role of cadherins in specificity. As a result, studies from the last few years have begun to provide strong evidence that the cadherins play important roles in directing synaptic specificity in the mouse. For example, one study investigated axon targeting of non-image forming retinal ganglion cells (RGCs). Here, the authors observed that among the many neuronal target areas of RGCs, cadherin-6 is selectively expressed by a unique subset of targets used in non-image forming visual processes.Citation71 Furthermore, a subpopulation of RGC axons that expresses cadherins-3 and -6 selectively projects to cadherin-6-expressing targets but not to other nearby RGC target areas. These RGC axons have a variety of projection defects in cadherin-6 knockout mice, often overshooting targets.Citation71 The results suggest cadherin-6 is required for these axons to accurately identify their specific target areas in the brain, likely via cadherin-6 homophilic interactions between this subset of RGC axons and their matching target areas.
Another study in the visual system showed that differentially expressed cadherins are necessary for proper laminar targeting of retinal bipolar cells.Citation72 Experiments with cell-type specific markers revealed that cadherin-8 and cadherin-9 are selectively and specifically expressed in 2 distinct populations of bipolar cells (BCs) that are part of a direction-selective visual circuit. Cadherin-8 is expressed in so called BC2s while cadherin-9 is expressed in BC5s.Citation72 Axons from BC2 and BC5 cells target different synaptic laminae within the inner plexiform layer. Using knockout mice, the authors showed that cadherin-8 and cadherin-9 loss-of-function leads to aberrant laminar targeting and functional connectivity defects only in BC cells that normally express the missing cadherin.Citation72 Furthermore, ectopic overexpression of cadherin-8 in BC5s or cadherin-9 in BC2s is sufficient to redirect axons to the wrong synaptic layer.Citation72 These results provide evidence that differentially expressed type II cadherins play an instructive role in targeting axons to correct laminar targets.
Studies have also examined type-II cadherin mediated specificity at the synaptic level. In the hippocampus, principal cells receive synaptic input from different types of neurons and these different synapses can be identified in cultured hippocampal neurons using unique molecular markers.Citation51 Screening candidate genes for a role in the formation of distinct subtypes of synapses in vitro led to the identification of cadherin-9. Cadherin-9 is strongly expressed in hippocampal DG and CA3 neurons and not by any other types of hippocampal neurons.Citation51,58 Knockdown of cadherin-9 in vitro caused specific reduction of DG-CA3 synapses leaving other hippocampal synapses intact whereas an in vivo knockdown of cadherin-9 resulted in severely altered morphology of DG-CA3 mossy fiber synapses.Citation51 These results suggest that cadherin-9 is selectively required to regulate pre- and postsynaptic development of hippocampal mossy fiber synapses. This is likely mediated via homophilic interactions between DG axons and CA3 dendrites.
In the pontocerebellar circuit, pontine nucleus axons synapse onto granule cells and not with nearby Purkinje neurons. Kuwako et al. showed that cadherin-7 is specifically present in pontine nucleus and granule neurons.Citation73 Knockdown of cadherin-7 in pontine nucleus neurons in vivo causes some pontine nucleus axons to incorrectly invade the Purkinje cell layer. In addition, knockdown of cadherin-7 in vitro causes impaired synapse formation between pontine nucleus and granule neurons.Citation73
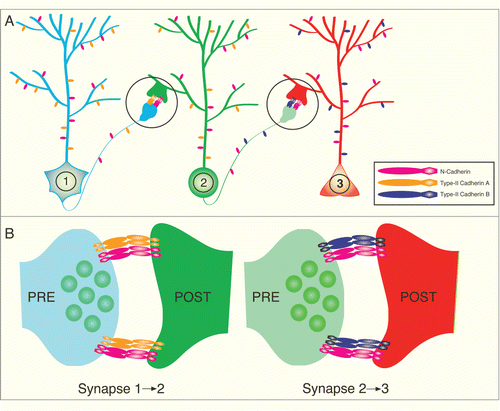
Taken together, data from several studies now supports the hypothesis that cadherins participate in directing the development of synaptic specificity. The primary mechanism used by cadherins to mediate synaptic specificity in the vertebrate brain is likely via differential expression of type II cadherin genes (). This model postulates that when neurons encounter another neuron expressing the same cadherin, synapse formation is more likely to occur than if a neuron encounters another neuron in which cadherin expression does not match. Transcriptional mechanisms that control the differential expression of cadherins is an interesting unsolved question and is an active area of investigation.Citation74 Moreover, it remains unclear whether the actual signal to build a synapse is directly mediated by cadherin intracellular signaling or indirectly mediated by cadherins via increased axo-dendritic adhesion leading to activation of other synaptogenic molecules. Studies on N-cadherin suggest that both possibilities may be correct. On one hand, N-cadherin/catenin signaling can directly recruit synaptic vesicles to the presynapseCitation75,76 and, on the other hand, N-cadherin can indirectly affect synapse formation by enhancing the function of neuroligin, a highly synaptogenic trans-synaptic molecule.Citation77,78
Figure 2. Model of Type II cadherin mediated synaptic specificity. (A) Illustration depicting a 3 neuron circuit with specific connections from neuron 1 to neuron 2 and then neuron 2 to neuron 3. (B) Expanded view of synaptic areas circled in A. Note that all 3 neurons express the Type-I N-cadherin (pink), which is present and required for function of most synapses. However, the neurons express different kinds of Type II cadherins (orange or purple) to restrict synapse formation to select synaptic partners that express the matching Type II cadherin.
Broadly Expressed Cadherins in Synaptic Specificity
The concept that differentially expressed (primarily type II) cadherins regulate synaptic specificity based on locating appropriate partner neurons that express matching cadherins has been technically difficult to test until recently but represents a straightforward mechanism of differential adhesion leading to specific connectivity. Interestingly, studies in Drosophila provide evidence that broadly-expressed cadherins can also regulate synaptic specificity but the mechanisms may be distinct from those used by differentially expressed cadherins in the mammalian brain.
The Drosophila genome has only 3 classic cadherins; E-cadherin, N-cadherin, and the less understood N-cadherin2. Notably, the sequences of these cadherins are significantly different from their vertebrate counterparts.Citation3,79 Drosophila N-cadherin (DN-cadherin) has 15 EC repeats, a fly classic cadherin box region, and several EGF-like repeat regions.Citation3,79 Nonetheless, like vertebrate N-cadherin, DN-cadherin is widely expressed in most neurons and plays many roles in neural development including synaptic specificity. How does DN-cadherin regulate synaptic specificity when it is ubiquitously expressed and does not provide a target-specific cue? One mechanism is that DN-cadherin expression is temporally regulated. It is known that Drosophila axons from the R8 class of photoreceptor cells terminate in the medulla layer known as M3. In contrast, R7 photoreceptor axons terminate in the more distal layer M6. All the axons and target cells express DN-cadherin, but it was shown that differential expression of a zinc-finger transcription factor drives high levels of DN-cadherin expression earlier in R8 cells compared to R7 cells.Citation80 DN-cadherin is thought to cause R8 axons to stop growing and adhere at the peak of DN-cadherin expression that happens to occur when axons arrive at the M3 layer.Citation80 In contrast, R7 axons have lower adhesion and continue growing to the M6 layer.Citation80 Thus, although eventually most neurons express DN-cadherin, they express it at different time points in development when the axons are encountering different synaptic partners. A similar mechanism could also occur in mammalian neurons. In cultured rat and mouse neurons, N-cadherin has different functions depending on the age of the neurons. Blocking cadherin function in neurons prior to synapse maturation has strong effects on synapse density whereas blocking cadherin function after synapse formation does not affect synapse density.Citation81 In addition, although N-cadherin is broadly expressed in the mammalian cortex, it is enriched in specific brain regions at different developmental time points. In particular, the enrichment of N-cadherin in cortical layer IV in early postnatal animals may play a role in directing thalamacortical axons precisely to this layer during development.Citation82
In addition to tightly controlled temporal expression of N-cadherin, it is possible that broadly expressed cadherins regulate specificity through differential subcellular localization or regulated surface trafficking. This remains to be conclusively tested, but there is correlative evidence to support each possibility. For example, as rodent neurons mature, N-cadherin is retained at excitatory synapses and excluded from inhibitory synapses.Citation45,46 Conversely, E-cadherin localizes to and is required for inhibitory synapse formation.Citation45,83 It is possible that the selective retention of N- and E-cadherin at excitatory vs. inhibitory synapses is primarily mediated by other synaptic specificity molecules like the neuroligins. Different neuroligin genes are well known to have a strong bias toward excitatory (neuroligin-1) versus inhibitory (neuroligin-2) synapses and cadherins have been shown to associate with neuroligins via intracellular synaptic scaffolds.Citation84-86 Although this idea remains to be directly tested, it emphasizes the complex nature of synaptic specificity, which is likely mediated by a network of synaptic molecules. A second example of regulated surface trafficking of cadherins was shown in Drosophila. Photoreceptor cell targeting is disrupted in Rab6 (a GTPase) and Ric-1 homolog (Rich, a Rab6 binding protein) mutants.Citation87 Rab6 regulates vesicle transport and causes a specific reduction of DN-cadherin expression at synapses suggesting that regulated trafficking of cadherins is another way that broadly-expressed N-cadherin can influence synaptic specificity.Citation87 One mechanism that does not appear to play a major role in cadherin-directed synaptic specificity is alternative splicing. DN-cadherin has up to 12 isoforms, but the isoforms do not have biochemical differences and isoform diversity is not required for R7/R8 photoreceptor targeting.Citation88,89 Vertebrate cadherins have few splice forms and evidence suggests that the classic cadherin gene family diversified by gene duplication events rather than alternative splicing mechanisms.Citation3
Combinatorial Coding Increases the Selectivity and Fidelity of Neural Circuits
There are billions of neurons in the human brain that must assemble and organize into countless numbers of specific circuits and microcircuits. One question that often arises about the cadherins is how such a relatively small number of cadherin molecules could possibly accomplish this task? Each cadherin is independently transcribed from an individual gene locus and expression studies indicate most neuronal cell types express more than one cadherin. Thus, from a purely mathematic perspective there are hundreds of thousands of unique combinations that the 18 classic cadherins or just the 13 classic type II cadherins can generate. This level of diversity is not necessarily even needed given that a developing brain goes through many stages prior to synapse formation. By the time an axon has been guided to the correct area where it will make a synapse, it is faced with a much simpler problem of choosing between a few cell types in the local area. For this task, the diversity of the classic cadherins is more than sufficient to identify specific target cell populations and construct different types of synapses. It is important to distinguish that although cadherins may theoretically be capable of providing a unique cellular identity, there is little evidence for this and we favor a model in which the cadherins provide a population identity. This is based on in situ and expression studies showing that cadherin expression levels vary substantially between different cell types but not between individual neurons of the same type.Citation58,90 How does a neuron choose between one cell and another cell of the same type? This aspect of specificity is likely to occur independent of the cadherins and although the mechanism is not known, timing, spatial constraints, randomization, and neural activity may all play a role in refining circuits at this level.
Although the cadherins may be capable of providing molecular diversity to wire the entire brain, the most realistic answer is that they likely do not – at least not on their own. As mentioned throughout this review, it is almost certain that the cadherins function together with other cell adhesion, axon guidance, and synaptogenic molecules on the neuronal cell surface. Interactions between cadherins and other cell adhesion systems can take several forms. First, cadherins act redundantly with other adhesion systems whereby both sets of molecules signal to accomplish the same task. For example, DN-cadherin and a 7-pass transmembrane cadherin Flamingo act redundantly to polarize Drosophila photoreceptor growth cones toward correct synaptic partners.Citation91 Cadherins can also act in a complementary push-pull manner with other cell surface proteins. In this case, the cadherins are usually the positive adhesive cues promoting appropriate synapse formation while repellent cues push an axon or dendrite away from incorrect synaptic partners. An example of this type of mechanism was reported in the Drosophila medulla where DN-cadherin mediates adhesion between an axon and its correct synaptic target zone while Sema/Plexin signaling provides a simultaneous repellent signal to keep the axon out of nearby incorrect synaptic areas.Citation92
Cadherins also affect synaptic specificity through their ability to modulate synaptogenic signals. In most cases, cadherins are necessary, but not sufficient, to induce de novo synapse formation.Citation51,65,Citation81,93-95 However, recent work showed that N-cadherin modulates recruitment of neuroligin-1. Previous studies found an indirect biochemical link between neuroligins and N-cadherin via S-SCAM and β-cateninCitation84-86 and more recent results from 2 different groups show that N-cadherin is necessary to cluster neuroligin-1 at synaptic sites.Citation77,78 Thus, overexpression of neuroligin-1 is synaptogenic, but only in the presence of N-cadherin.Citation77,78 Together, these 2 cell adhesion systems promote the clustering of synaptic vesicles and increase the probability of synaptic vesicle release.Citation77,78 As discussed in the previous sections, almost all individual neurons (certainly all cortical and hippocampal neurons) also express other classic cadherins in addition to N-cadherin. An unanswered question is whether other classic cadherins also synergize with synaptogenic molecules like neuroligin? One intriguing possibility is that different cadherins may synergize with different cell adhesion molecules to organize distinct classes of synapses. Future studies are needed to understand how specific members of the classic cadherin family interact with neuroligins and other synaptogenic cues.
Combinatorial coding among the cadherins themselves and with other synaptic molecules is expected to dramatically increase the fidelity of neural circuit formation. It also is likely a major reason why direct evidence showing that cadherins regulate synaptic specificity has been so sparse. In the hippocampus, each class of excitatory neuron expresses 4–5 different cadherins.Citation58,90 This provides each cell type with a unique extracellular identity but there is enough overlap among them that the global loss or overexpression of any one cadherin may do little to significantly alter the molecular adhesion or identity of synaptic partners. Depleting all cadherin signaling by knocking out cadherin signaling molecules reveals synaptic defectsCitation67,86 but also obscures any analysis of specificity. One experimental paradigm that may overcome this conundrum is to sparsely deplete individual cadherins from neurons using viral transfections, in utero electroporation, or conditional genetic techniques. As recently shown for neuroligin, sparse deletion can put individual cells at a competitive disadvantage and reveal the function of a molecule when global changes in gene expression do not.Citation96
Cadherins, Synaptic Specificity, and Neurodevelopmental Disorders
The cadherins are emerging as important regulators of synapse form and function. Therefore, it is not surprising that cadherin genes are implicated in the etiology of a variety of neurological disorders. Supporting a role for cadherins in synaptic specificity, alterations in different cadherin genes are associated with distinct neurological disorders in humans. Genetic deletions or copy number variations of cadherin-8 and the GPI-linked cadherin-13 were identified in patients with autism and learning disability.Citation97,98 Single nucleotide polymorphisms (SNPs) within the cadherin-13 gene associate with attention deficit/hyperactivity disorder (ADHD) and alcoholism and a SNP between cadherin-9 and -10 associates with autism.Citation99-101 In addition, a chromosomal rearrangement involving cadherin-9 was identified in a family with non-syndromic mental retardation.Citation102 Finally, analysis of a monozygotic twin pair discordant for schizophrenia identified a genetic deletion between cadherins-12 and -18.Citation103
So far, at least 2 of these genetic associations have led to follow up studies supporting their role in disease. First, in a study of over 7,000 participants, the high-risk autism SNP between cadherin-9 and cadherin-10 correlates with lower communication traits in the general population.Citation104 This interesting finding suggests isolated genetic aberrations are important, but need to accumulate or undergo environmental stresses before behavioral impairments are severe enough to reach functional and clinical significance.Citation105 Second, in a cohort of 238 children, the presence of the ADHD-associated SNP in cadherin-13 was found to specifically correlate with a defect in verbal working memory, but not with spatial working memory.Citation106 This finding provides the first glimpse that cadherins may also regulate circuit specificity in humans. It suggests that cadherin-13 may be required for the formation of specific circuits involved with verbal working memory but not for circuits specific to spatial working memory or other cognitive functions. It is important to note that most of these studies identified SNPs or deletions in non-coding genomic regions associated with a cadherin. These alterations may be located in regulatory regions affecting expression of the nearby cadherin gene, but more studies are required to determine whether or not expression of individual cadherins are actually affected by these genomic alterations.
Conclusions and Future Directions
Experiments over the last 10 years provide increasing evidence that cadherins are important mediators in the formation of specific neural circuits, however, many questions remain. One major unresolved issue is the nature of cadherin heterogeneity. While it is likely all cadherins undergo strong homophilic interactions, in vitro binding assays indicate that some cadherins form heterophilic interactions with other cadherin family members.Citation37-41 Moreover, cadherins are differentially expressed throughout the brain, but not in an exclusive manner. Most neurons express multiple cadherins yet little is known about how adhesion is affected when single cells express multiple cadherins. Do all cadherins have to match for binding to occur, or just a few? Does adhesion depend on the expression levels of each cadherin? Do different cadherins segregate to different subcellular compartments? Our understanding of how multiple classic cadherins function within the context of a single cell is very limited. We do not yet know if different cadherins segregate to different synapses or if they are all present at every synapse made by a single cell.
Equally important for understanding the role of different cadherins in synaptic specificity is to understand how they signal. Is specificity governed exclusively by adhesive extracellular matching or are distinct intracellular signaling mechanisms required? By definition, all classic cadherins bind the catenins and catenins are critical for synapse formation, but some cadherins have been shown to interact directly and indirectly with other synaptic proteins including glutamate receptors (AMPA, NMDA, and kainate type), scaffolding proteins (AKAP79/150, GRIP), and signaling molecules (Vangl2, p38 MAPK).Citation95,107-111 Nonetheless, the precise signaling mechanisms used by cadherins during the process of synaptic specificity remain unknown. Another unresolved question is at what stage in synapse development are cadherins required? Are they activated at the moment of axo-dendritic contact or are they recruited some time later? This answer is likely to be complex because previous work suggests the role of the cadherins changes as neurons mature.Citation81
In conclusion, although cadherin and cadherin-signaling has been extensively studied since the first cadherin was cloned in 1980 (resulting in tens of thousands of published manuscripts), there is still much more to learn about the role of cadherins in the brain. Now that cadherins are becoming established as important molecules in the development of synaptic specificity, future experiments can be aimed at elucidating the specialized mechanisms used by cadherins to wire the brain during neural development.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Joris DeWit and Anne Martin for comments on the manuscript.
Funding
This work was supported by grants from the Whitehall, Edward Mallinckrodt Jr., and Sloan foundations (M.E.W).
References
- Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: diversity in form and function. J Cell Sci 2001; 114:629-41; PMID:11171368
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci 2007; 8:11-20; PMID:17133224
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members1. J Mol Biol 2000; 299:551-72; PMID:10835267; http://dx.doi.org/10.1006/jmbi.2000.3777
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell 2010; 143:343-53; PMID:21029858; http://dx.doi.org/10.1016/j.cell.2010.10.009
- Tai C-Y, Kim SA, Schuman EM. Cadherins and synaptic plasticity. Curr Opin Cell Biol 2008; 20:567-75; PMID:18602471; http://dx.doi.org/10.1016/j.ceb.2008.06.003
- Arikkath J, Reichardt LF. Cadherins and catenins at synapses: roles in synaptogenesis and synaptic plasticity. Trends Neurosci 2008; 31:487-94; PMID:18684518; http://dx.doi.org/10.1016/j.tins.2008.07.001
- Hirano SS, Takeichi MM. Cadherins in brain morphogenesis and wiring. Physiol Rev 2012; 92:597-634; PMID:22535893; http://dx.doi.org/10.1152/physrev.00014.2011
- Brigidi GS, Bamji SX. Cadherin-catenin adhesion complexes at the synapse. Curr Opin Neurobiol 2011; 21:208-14; PMID:21255999; http://dx.doi.org/10.1016/j.conb.2010.12.004
- Sotelo C. Viewing the brain through the master hand of Ramón y Cajal. Nat Rev Neurosci 2003; 4:71-7; PMID:12511863
- Vrieseling E, Arber S. Target-induced transcriptional control of dendritic patterning and connectivity in motor neurons by the ETS gene Pea3. Cell 2006; 127:1439-52; PMID:17190606
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell 2006; 127:1057-69; PMID:17129788; http://dx.doi.org/10.1016/j.cell.2006.10.031
- Espinosa JS, Stryker MP. Development and plasticity of the primary visual cortex. Neuron 2012; 75:230-49; PMID:22841309; http://dx.doi.org/10.1016/j.neuron.2012.06.009
- Kano M, Hashimoto K. Synapse elimination in the central nervous system. Curr Opin Neurobiol 2009; 19:154-61; PMID:19481442; http://dx.doi.org/10.1016/j.conb.2009.05.002
- Verhage M, Maia AS, Plomp JJ, Brussaard AB, Heeroma JH, Vermeer H, Toonen RF, Hammer RE, van den TK, Berg, et al. Synaptic assembly of the brain in the absence of neurotransmitter secretion. Science 2000; 287:864-9; PMID:10657302
- Nevin LM, Taylor MR, Baier H. Hardwiring of fine synaptic layers in the zebrafish visual pathway. Neural Dev 2008; 3:36; PMID:19087349; http://dx.doi.org/10.1186/1749-8104-3-36
- Godinho L, Mumm JS, Williams PR, Schroeter EH, Koerber A, Park SW, Leach SD, Wong ROL. Targeting of amacrine cell neurites to appropriate synaptic laminae in the developing zebrafish retina. Development 2005; 132:5069-79; PMID:16258076; http://dx.doi.org/10.1242/dev.02075
- Mumm JS, Williams PR, Godinho L, Koerber A, Pittman AJ, Roeser T, Chien C-B, Baier H, Wong ROL. In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron 2006; 52:609-21; PMID:17114046; http://dx.doi.org/10.1016/j.neuron.2006.10.004
- Langley JN. Note on regeneration of Pr ๆ-ganglionic fibres of the sympathetic. J Physiol (Lond) 1895; 18:280-4; PMID:16992254
- Sperry RW. Chemoaffinity in the orderly growth of nerve fiber patterns and connections. Proc Natl Acad Sci USA 1963; 50:703-10; PMID:14077501
- Kohmura N, Senzaki K, Hamada S, Kai N, Yasuda R, Watanabe M, Ishii H, Yasuda M, Mishina M, Yagi T. Diversity revealed by a novel family of cadherins expressed in neurons at a synaptic complex. Neuron 1998; 20:1137-51; PMID:9655502; http://dx.doi.org/10.1016/S0896-6273(00)80495-X
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 1999; 97:779-90; PMID:10380929; http://dx.doi.org/10.1016/S0092-8674(00)80789-8
- Esumi S, Kakazu N, Taguchi Y, Hirayama T, Sasaki A, Hirabayashi T, Koide T, Kitsukawa T, Hamada S, Yagi T. Monoallelic yet combinatorial expression of variable exons of the protocadherin-α gene cluster in single neurons. Nat Genet 2005; 37:171-6; PMID:15640798; http://dx.doi.org/10.1038/ng1500
- Kaneko R, Kato H, Kawamura Y, Esumi S, Hirayama T, Hirabayashi T, Yagi T. Allelic gene regulation of Pcdh- and Pcdh- clusters involving both monoallelic and biallelic expression in single purkinje cells. J Biol Chem 2006; 281:30551-60; PMID:16893882; http://dx.doi.org/10.1074/jbc.M605677200
- Hirano K, Kaneko R, Izawa T, Kawaguchi M, Kitsukawa T, Yagi T. Single-neuron diversity generated by Protocadherin-β cluster in mouse central and peripheral nervous systems. Front Mol Neurosci 2012; 5; PMID:22969705; http://dx.doi.org/10.3389/fnmol.2012.00090
- Phillips GR, Tanaka H, Frank M, Elste A, Fidler L, Benson DL, Colman DR. γ-Protocadherins are targeted to subsets of synapses and intracellular organelles in neurons. J Neurosci 2003; 23:5096-104; PMID:12832533
- Junghans D, Heidenreich M, Hack I, Taylor V, Frotscher M, Kemler R. Postsynaptic and differential localization to neuronal subtypes of protocadherin β16 in the mammalian central nervous system. Eur J Neurosci 2008; 27:559-71; PMID:18279309; http://dx.doi.org/10.1111/j.1460-9568.2008.06052.x
- Lefebvre JL, Kostadinov D, Chen WV, Maniatis T, Sanes JR. Protocadherins mediate dendritic self-avoidance in the mammalian nervous system. Nature 2012; 488:517-21; PMID:22842903; http://dx.doi.org/10.1038/nature11305
- Chen WV, Maniatis T. Clustered protocadherins. Development 2013; 140:3297-302; PMID:23900538; http://dx.doi.org/10.1242/dev.090621
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB. Dendrite self-avoidance is controlled by Dscam. Cell 2007; 129:593-604; PMID:17482551; http://dx.doi.org/10.1016/j.cell.2007.04.013
- Soba P, Zhu S, Emoto K, Younger S, Yang S-J, Yu H-H, Lee T, Jan LY, Jan Y-N. Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 2007; 54:403-16; PMID:17481394; http://dx.doi.org/10.1016/j.neuron.2007.03.029
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW. DSCAM and DSCAML1 function in self-avoidance in multiple cell types in the developing mouse retina. Neuron 2009; 64:484-97; PMID:19945391; http://dx.doi.org/10.1016/j.neuron.2009.09.027
- Hulpiau P, van Roy F. Molecular evolution of the cadherin superfamily. Int J Biochem Cell Biol 2009; 41:349-69; PMID:18848899; http://dx.doi.org/10.1016/j.biocel.2008.09.027
- Nose A, Tsuji K, Takeichi M. Localization of specificity determining sites in cadherin cell adhesion molecules. Cell 1990; 61:147-55; PMID:2317870; http://dx.doi.org/10.1016/0092-8674(90)90222-Z
- Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grübel G, Legrand JF, Als-Nielsen J, Colman DR, Hendrickson WA. Structural basis of cell-cell adhesion by cadherins. Nature 1995; 374:327-37; PMID:7885471; http://dx.doi.org/10.1038/374327a0
- Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids… 2012; 41:D545-52; PMID:23161694
- Takeichi M. The cadherins: cell-cell adhesion molecules controlling animal morphogenesis. Development 1988; 102:639-55; PMID:3048970
- Ounkomol C, Yamada S, Heinrich V. Single-cell adhesion tests against functionalized microspheres arrayed on AFM cantilevers confirm heterophilic E- and N-cadherin binding. Biophys J 2010; 99:L100-2; PMID:21156120; http://dx.doi.org/10.1016/j.bpj.2010.11.013
- Volk T, Cohen O, Geiger B. Formation of heterotypic adherens-type junctions between L-CAM-containing liver cells and A-CAM-containing lens cells. Cell 1987; 50:987-94; PMID:3621349; http://dx.doi.org/10.1016/0092-8674(87)90525-3
- Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, et al. Type II cadherin ectodomain structures: implications for classical cadherin specificity. Cell 2006; 124:1255-68; PMID:16564015; http://dx.doi.org/10.1016/j.cell.2005.12.046
- Shimoyama Y, Tsujimoto G, Kitajima M, Natori M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem J 2000; 349:159-67; PMID:10861224; http://dx.doi.org/10.1042/0264-6021:3490159
- Prakasam AK, Maruthamuthu V, Leckband DE. Similarities between heterophilic and homophilic cadherin adhesion. Proc Natl Acad Sci USA 2006; 103:15434-9; PMID:17023539
- Jontes JD. In vivo trafficking and targeting of N-cadherin to nascent presynaptic terminals. J Neurosci 2004; 24:9027-34; PMID:15483121; http://dx.doi.org/10.1523/JNEUROSCI.5399-04.2004
- Yamagata M, Herman JP, Sanes JR. Lamina-specific expression of adhesion molecules in developing chick optic tectum. J Neurosci 1995; 15:4556-71; PMID:7790923
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J Cell Biol 1996; 135:767-79; PMID:8909549; http://dx.doi.org/10.1083/jcb.135.3.767
- Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron 1996; 17:423-34; PMID:8816706; http://dx.doi.org/10.1016/S0896-6273(00)80175-0
- Benson DL, Tanaka H. N-cadherin redistribution during synaptogenesis in hippocampal neurons. J Neurosci 1998; 18:6892-904; PMID:9712659
- Bartelt-Kirbach B, Langer-Fischer K, Golenhofen N. Different regulation of N-cadherin and cadherin-11 in rat hippocampus. Cell Commun Adhes 2010; 17:75-82; PMID:21250828; http://dx.doi.org/10.3109/15419061.2010.549977
- Suzuki SC, Furue H, Koga K, Jiang N, Nohmi M, Shimazaki Y, Katoh-Fukui Y, Yokoyama M, Yoshimura M, Takeichi M. Cadherin-8 is required for the first relay synapses to receive functional inputs from primary sensory afferents for cold sensation. J Neurosci 2007; 27:3466-76; PMID:17392463; http://dx.doi.org/10.1523/JNEUROSCI.0243-07.2007
- Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron 2000; 28:245-59; PMID:11086998; http://dx.doi.org/10.1016/S0896-6273(00)00100-8
- Manabe T. Loss of cadherin-11 adhesion receptor enhances plastic changes in hippocampal synapses and modifies behavioral responses. MolCell Neurosci 2000; 15:534-46; PMID:10860580; http://dx.doi.org/10.1006/mcne.2000.0849
- Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, et al. Cadherin-9 regulates synapse-specific differentiation in the developing hippocampus. Neuron 2011; 71:640-55; PMID:21867881; http://dx.doi.org/10.1016/j.neuron.2011.06.019
- Bayés A, van de Lagemaat LN, Collins MO, Croning MDR, Whittle IR, Choudhary JS, Grant SGN. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat Neurosci 2011; 14:19-21; PMID:21170055; http://dx.doi.org/10.1038/nn.2719
- Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, Shapiro L. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 2002; 296:1308-13; PMID:11964443; http://dx.doi.org/10.1126/science.1071559
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G. The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins. Structure 2011; 19:244-56; PMID:21300292; http://dx.doi.org/10.1016/j.str.2010.11.016
- Miskevich F, Zhu Y, Ranscht B, Sanes JR. Expression of multiple cadherins and catenins in the chick optic tectum. Mol Cell Neurosci 1998; 12:240-55; PMID:9828089; http://dx.doi.org/10.1006/mcne.1998.0718
- Hatta K, Takagi S, Fujisawa H, Takeichi M. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev Biol 1987; 120:215-27; PMID:3817290; http://dx.doi.org/10.1016/0012-1606(87)90119-9
- Suzuki S, Sano K, Tanihara H. Diversity of the cadherin family: evidence for eight new cadherins in nervous tissue. Cell Regul 1991; 2:261-70; PMID:2059658
- Bekirov IH, Needleman LA, Zhang W, Benson DL. Identification and localization of multiple classic cadherins in developing rat limbic system. Neuroscience 2002; 115:213-27; PMID:12401335; http://dx.doi.org/10.1016/S0306-4522(02)00375-5
- Inoue T, Tanaka T, Suzuki SC, Takeichi M. Cadherin-6 in the developing mouse brain: expression along restricted connection systems and synaptic localization suggest a potential role in neuronal circuitry. Dev Dyn 1998; 211:338-51; PMID:9566953; http://dx.doi.org/10.1002/(SICI)1097-0177(199804)211:4<338::AID-AJA5>3.0.CO;2-I
- Arndt K, Nakagawa S, Takeichi M, Redies C. Cadherin-defined segments and parasagittal cell ribbons in the developing chicken cerebellum. Mol Cell Neurosci 1998; 10:211-28; PMID:9618214; http://dx.doi.org/10.1006/mcne.1998.0665
- Yoon MS, Puelles L, Redies C. Formation of cadherin-expressing brain nuclei in diencephalic alar plate divisions. J Comp Neurol 2000; 427:461-80; PMID:11183875; http://dx.doi.org/10.1002/(SICI)1096-9861(20000612)421:4<461::AID-CNE2>3.0.CO;2-M
- Suzuki SC, Inoue T, Kimura Y, Tanaka T, Takeichi M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol Cell Neurosci 1997; 9:433-47; PMID:9361280; http://dx.doi.org/10.1006/mcne.1997.0626
- Price SR, De Marco Garcia NV, Ranscht B, Jessell TM. Regulation of motor neuron pool sorting by differential expression of type II cadherins. Cell 2002; 109:205-16; PMID:12007407; http://dx.doi.org/10.1016/S0092-8674(02)00695-5
- Korematsu K, Redies C. Expression of cadherin-8 mRNA in the developing mouse central nervous system. J Comp Neurol 1997; 387:291-306; PMID:9336230; http://dx.doi.org/10.1002/(SICI)1096-9861(19971020)387:2<291::AID-CNE10>3.0.CO;2-Y
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron 2002; 35:77-89; PMID:12123610; http://dx.doi.org/10.1016/S0896-6273(02)00748-1
- Demireva EY, Shapiro LS, Jessell TM, Zampieri N. Motor neuron position and topographic order imposed by β- and γ-catenin activities. Cell 2011; 147:641-52; PMID:22036570; http://dx.doi.org/10.1016/j.cell.2011.09.037
- Arikkath J, Peng I-F, Ng YG, Israely I, Liu X, Ullian EM, Reichardt LF. Delta-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J Neurosci 2009; 29:5435-42; PMID:19403811; http://dx.doi.org/10.1523/JNEUROSCI.0835-09.2009
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron 2007; 53:217-32; PMID:17224404; http://dx.doi.org/10.1016/j.neuron.2006.12.012
- Bekirov IH, Nagy V, Svoronos A, Huntley GW, Benson DL. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus 2008; 18:349-63; PMID:18064706; http://dx.doi.org/10.1002/hipo.20395
- Huntley GW, Elste AM, Patil SB, Bozdagi O, Benson DL, Steward O. Synaptic loss and retention of different classic cadherins with LTP-associated synaptic structural remodeling in vivo. Hippocampus 2010; 22:17-28; PMID:20848607; http://dx.doi.org/10.1002/hipo.20859
- Osterhout JA, Josten N, Yamada J, Pan F, Wu S-W, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, et al. Cadherin-6 mediates axon-target matching in a non-image-forming visual circuit. Neuron 2011; 71:632-9; PMID:21867880; http://dx.doi.org/10.1016/j.neuron.2011.07.006
- Duan X, Krishnaswamy A, la Huerta De I, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell 2014; 158:793-807; PMID:25126785; http://dx.doi.org/10.1016/j.cell.2014.06.047
- Kuwako K-I, Nishimoto Y, Kawase S, Okano HJ, Okano H. Cadherin-7 regulates mossy fiber connectivity in the cerebellum. Cell Rep 2014; 9:311-23; PMID:25284782; http://dx.doi.org/10.1016/j.celrep.2014.08.063
- Paulson A, Prasad M, Thuringer A, Manzerra P. Regulation of cadherin expression in nervous system development. Cell Adh Migr 2014; 8:19-28; PMID:24526207; http://dx.doi.org/10.4161/cam.27839
- Sun Y, Aiga M, Yoshida E, Humbert PO, Bamji SX. Scribble interacts with beta-catenin to localize synaptic vesicles to synapses. Mol Biol Cell 2009; 20:3390-400; PMID:19458197; http://dx.doi.org/10.1091/mbc.E08-12-1172
- Sun Y, Bamji SX. β-Pix modulates actin-mediated recruitment of synaptic vesicles to synapses. J Neurosci 2011; 31:17123-33; PMID:22114281; http://dx.doi.org/10.1523/JNEUROSCI.2359-11.2011
- Aiga M, Levinson JN, Bamji SX. N-cadherin and neuroligins cooperate to regulate synapse formation in hippocampal cultures. J Biol Chem 2011; 286:851-8; PMID:21056983; http://dx.doi.org/10.1074/jbc.M110.176305
- Stan A, Pielarski KN, Brigadski T, Wittenmayer N, Fedorchenko O, Gohla A, Lessmann V, Dresbach T, Gottmann K. Essential cooperation of N-cadherin and neuroligin-1 in the transsynaptic control of vesicle accumulation. Proc Nat Acad Sci 2010; 107:11116-21; PMID:20534458; http://dx.doi.org/10.1073/pnas.0914233107
- Hill E, Broadbent ID, Chothia C, Pettitt J. Cadherin superfamily proteins in Caenorhabditis elegans and Drosophila melanogaster. J Mol Biol 2001; 305:1011-24; PMID:11162110; http://dx.doi.org/10.1006/jmbi.2000.4361
- Petrovic M, Hummel T. Temporal identity in axonal target layer recognition. Nature 2008; 456:800-3; PMID:18978776; http://dx.doi.org/10.1038/nature07407
- Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol Cell Neurosci 2004; 27:509-21; PMID:15555928; http://dx.doi.org/10.1016/j.mcn.2004.08.008
- Huntley GW, Benson DL. Neural (N)-cadherin at developing thalamocortical synapses provides an adhesion mechanism for the formation of somatopically organized connections. J Comp Neurol 1999; 407:453-71; PMID:10235639; http://dx.doi.org/10.1002/(SICI)1096-9861(19990517)407:4<453::AID-CNE1>3.0.CO;2-4
- Fiederling A, Ewert R, Andreyeva A, Jüngling K, Gottmann K. E-cadherin is required at GABAergic synapses in cultured cortical neurons. Neurosci Lett 2011; 501:167-72; PMID:21782891; http://dx.doi.org/10.1016/j.neulet.2011.07.009
- Dobrosotskaya IY, James GL. MAGI-1 interacts with β-catenin and is associated with cell–cell adhesion structures. Biochem Biophys Res Commun 2000; 270:903-9; PMID:10772923; http://dx.doi.org/10.1006/bbrc.2000.2471
- Nishimura W, Yao I, Iida J, Tanaka N, Hata Y. Interaction of synaptic scaffolding molecule and Beta -catenin. J Neurosci 2002; 22:757-65; PMID:11826105
- Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, Lu B, Reichardt LF. Role of beta-catenin in synaptic vesicle localization and presynaptic assembly. Neuron 2003; 40:719-31; PMID:14622577; http://dx.doi.org/10.1016/S0896-6273(03)00718-9
- Tong C, Ohyama T, Tien A-C, Rajan A, Haueter CM, Bellen HJ. Rich regulates target specificity of photoreceptor cells and N-cadherin trafficking in theDrosophilavisual system via Rab6. Neuron 2011; 71:447-59; PMID:21835342; http://dx.doi.org/10.1016/j.neuron.2011.06.040
- Ting C-Y, Yonekura S, Chung P, Hsu S-N, Robertson HM, Chiba A, Lee C-H. Drosophila N-cadherin functions in the first stage of the two-stage layer-selection process of R7 photoreceptor afferents. Development 2005; 132:953-63; PMID:15673571; http://dx.doi.org/10.1242/dev.01661
- Nern A, Nguyen L-VT, Herman T, Prakash S, Clandinin TR, Zipursky SL. An isoform-specific allele of Drosophila N-cadherin disrupts a late step of R7 targeting. Proc Natl Acad Sci USA 2005; 102:12944-9.
- Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci 2004; 24:3879-89; PMID:15084669; http://dx.doi.org/10.1523/JNEUROSCI.4710-03.2004
- Schwabe T, Neuert H, Clandinin TR. A network of cadherin-mediated interactions polarizes growth cones to determine targeting specificity. Cell 2013; 154:351-64; PMID:23870124; http://dx.doi.org/10.1016/j.cell.2013.06.011
- Pecot MY, Tadros W, Nern A, Bader M, Chen Y, Zipursky SL. Multiple interactions control synaptic layer specificity in the drosophila visual system. Neuron [Internet] 2013; 77:299-310. Available from: http://pubget.com/site/paper/23352166?institution=
- Mendez P, De Roo M, Poglia L, Klauser P, Muller D. N-cadherin mediates plasticity-induced long-term spine stabilization. J Cell Biol 2010; 189:589-600; PMID:20440002; http://dx.doi.org/10.1083/jcb.201003007
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 2000; 101:657-69; PMID:10892652; http://dx.doi.org/10.1016/S0092-8674(00)80877-6
- Saglietti L, Dequidt C, Kamieniarz K, Rousset M-C, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron 2007; 54:461-77; PMID:17481398; http://dx.doi.org/10.1016/j.neuron.2007.04.012
- Kwon H-B, Kozorovitskiy Y, Oh W-J, Peixoto RT, Akhtar N, Saulnier JL, Gu C, Sabatini BL. Neuroligin-1–dependent competition regulates cortical synaptogenesis and synapse number. Nat Neurosci 2012; 15:1667-74; PMID:23143522; http://dx.doi.org/10.1038/nn.3256
- Pagnamenta AT, Khan H, Walker S, Gerrelli D, Wing K, Bonaglia MC, Giorda R, Berney T, Mani E, Molteni M, et al. Rare familial 16q21 microdeletions under a linkage peak implicate cadherin 8 (CDH8) in susceptibility to autism and learning disability. J Med Genet 2011; 48:48-54; PMID:20972252; http://dx.doi.org/10.1136/jmg.2010.079426
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron 2011; 70:863-85; PMID:21658581; http://dx.doi.org/10.1016/j.neuron.2011.05.002
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry 2009; 66:773-84; PMID:19581569; http://dx.doi.org/10.1001/archgenpsychiatry.2009.83
- Franke B, Neale BM, Faraone SV. Genome-wide association studies in ADHD. Hum Genet 2009; 126:13-50; PMID:19384554; http://dx.doi.org/10.1007/s00439-009-0663-4
- Wang K, Zhang H, Ma D, Bucan M, Glessner JT, Abrahams BS, Salyakina D, Imielinski M, Bradfield JP, Sleiman PMA, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 2009; 459:528-33; PMID:19404256; http://dx.doi.org/10.1038/nature07999
- Gilling M, Lind-Thomsen A, Mang Y, Bak M, Møller M, Ullmann R, Kristoffersson U, Kalscheuer VM, Henriksen KF, Bugge M, et al. Biparental inheritance of chromosomal abnormalities in male twins with non-syndromic mental retardation. Eur J Med Genet 2011; 54:e383-8; PMID:21426945; http://dx.doi.org/10.1016/j.ejmg.2011.03.008
- Singh SM, Castellani C, O'Reilly R. Autism meets schizophrenia via cadherin pathway. Schizophr Res 2010; 116:293-4; PMID:19861233; http://dx.doi.org/10.1016/j.schres.2009.09.031
- St Pourcain B, Wang K, Glessner JT, Golding J, Steer C, Ring SM, Skuse DH, Grant SFA, Hakonarson H, Smith GD, et al. Association between a high-risk autism locus on 5p14 and social communication spectrum phenotypes in the general population. Am J Psychiatry 2010; 167:1364-72; PMID:20634369; http://dx.doi.org/10.1176/appi.ajp.2010.09121789
- McCarroll SA, Hyman SE. Perspective. Neuron 2013; 80:578-87; PMID:24183011; http://dx.doi.org/10.1016/j.neuron.2013.10.046
- Arias-Vásquez A, Altink ME, Rommelse NNJ, Slaats-Willemse DIE, Buschgens CJM, Fliers EA, Faraone SV, Sergeant JA, Oosterlaan J, Franke B, et al. CDH13 is associated with working memory performance in attention deficit/hyperactivity disorder. Genes Brain Behav 2011; 10:844-51; PMID:21815997; http://dx.doi.org/10.1111/j.1601-183X.2011.00724.x
- Coussen F, Normand E, Marchal C, Costet P, Choquet D, Lambert M, Mège R-M, Mulle C. Recruitment of the kainate receptor subunit glutamate receptor 6 by cadherin/catenin complexes. J Neurosci 2002; 22:6426-36; PMID:12151522
- Gorski JA, Gomez LL, Scott JD, Dell'Acqua ML. Association of an A-Kinase-anchoring protein signaling scaffold with cadherin adhesion molecules in neurons and epithelial cells. 200516:3574-90; PMID:15930126
- Heisler FF, Lee HK, Gromova KV, Pechmann Y, Schurek B, Ruschkies L, Schroeder M, Schweizer M, Kneussel M. GRIP1 interlinks N-cadherin and AMPA receptors at vesicles to promote combined cargo transport into dendrites. PNAS 2014; 111:5030-5; PMID:24639525; http://dx.doi.org/10.1073/pnas.1304301111
- Nagaoka T, Ohashi R, Inutsuka A, Sakai S, Fujisawa N. The Wnt/Planar cell polarity pathway component Vangl2 induces synapse formation through direct control of N-cadherin. Cell Rep 2014; 6:916-27; PMID:24582966; http://dx.doi.org/10.1016/j.celrep.2014.01.044
- Nuriya M, Huganir RL. Regulation of AMPA receptor trafficking by N-cadherin. J Neurochem 2006; 97:652-61; PMID:16515543; http://dx.doi.org/10.1111/j.1471-4159.2006.03740.x
