 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Any cellular response leading to morphological changes is highly tuned to balance the force generated from structural reorganization, provided by actin cytoskeleton. Actin filaments serve as the backbone of intracellular force, and transduce external mechanical signal via focal adhesion complex into the cell. During migration, cells not only undergo molecular changes but also rapid mechanical modulation. Here we focus on determining, the role of spatial distribution of mechanical changes of actin filaments in epithelial, mesenchymal, fibrotic and cancer cells with non-migration, directional migration, and non-directional migration behaviors using the atomic force microscopy. We found 1) non-migratory cells only generated one type of filament elasticity, 2) cells generating spatially distributed two types of filament elasticity showed directional migration, and 3) pathologic cells that autonomously generated two types of filament elasticity without spatial distribution were actively migrating non-directionally. The demonstration of spatial regulation of filament elasticity of different cell types at the nano-scale highlights the coupling of cytoskeletal function with physical characters at the sub-cellular level, and provides new research directions for migration related disease.
Introduction
Cell migration is an essential process in all multicellular organisms. Cell migration participates in morphogenetic movements during development and contributes to tissue renewal, immune responses, wound healing, and cancer metastasis.Citation1 Different cell types exhibit different migratory patterns. Generally speaking, normal epithelial cells or endothelial cells that provide form the sturdy architectural foundation for tissues and organs are more stable and less motile. Mesenchymal cells, such as fibroblasts, on the other hand, are usually more motile due to their nature to migrate within the mesenchyme.Citation2 The contrast in the ability to migrate is featured in the migration assay, in which epithelial cells that increases migration speed after stimulation are termed undergoing epithelial-to-mesenchymal transition, EMT.Citation3 Abnormal cells such as cancer cells, are also motile, and the degree of their motility often correlates with the level of malignancy. There are also cell types that could shift between static and motile states autonomously, such as the U2OS osteosarcoma cells that could either be in the non-polarized static state or polarized and undergo directional migration.Citation4
To characterize the migration behavior of a series of cells with the same origin differing by their malignancy, a static, non-motile normal epithelial cell would have low speed, low velocity, and low persistence. A low-grade, non-directional migration (or random migration) transformed version of this cell would have a moderate speed, low velocity, and low persistence. A highly malignant cancer cell that display directional migration would have high speed, high velocity, and high persistence.Citation4 Highly migratory cells do not always migrate directionally; in the case of keloid fibroblasts, a disease characterized by abnormal fibroblasts migrating and expanding beyond the original wound boarder, are highly motile but also non-directional.Citation5 These cells would have a high speed, low velocity and low persistence.
Furthermore, depending on how the classification is defined, there are various modes of migratory behavior. Distinct modes of migration range from the movement of single cells to collective cell migration, where groups of cells move coordinately (Friedl and Wolf 2010). The movement of a single cell can further be described as either random or directional migration, depending on the persistence of its migrating track. The exact mechanism of how a cell can directionally migrate with persistence has yet to be fully understood; however, the principle is that cells containing polarity signaling machinery influence directional cell motility. These signals may come from the topography of the ECM, constituents of the matrix, distribution of soluble or substrate-bound guidance cues and/or other factors.Citation6 Morphologically speaking, in order for a cell to directionally migrate persistently, the new protrusions and subsequent new adhesions need to be forming on the same front-rear axis as the cell body. In other words, if new lamellipodia were formed away from the leading edge, the cell would move away from the original line of migration. Multiple cycles of such spontaneous formation of non-directionally persisting protrusions would lead to the random migration of the cell.Citation7
To migrate efficiently, cells must possess asymmetric reorganization of cytoskeleton in order to generate a leading and a trailing edge.Citation8 This asymmetry can also be observed mechanically; invasive cancer cells migrate with peaks in high traction forces exerted on the substrate, while less invasive cells develop traction stresses almost constant in time.Citation9 In live and polarized Dictyostelium discoideum, the front end of the cell is stiffer than the rear end.Citation10 These reports suggest that during migration, cells undergo spatial reorganization of force generation, which could be a result of change in actin filament elasticity.
Actin cytoskeleton is responsible for a cell's ability to change and adapt to their environment, migrate, divide.Citation11 The mechanical properties of the actomyosin cytoskeleton and its contractility play an important role in cell motility, especially for movement within tissue or in confined space. Our previous study on the highly migratory keloid fibroblasts have shown that migrating cells display two distinct types of filament elasticity. However, no spatial distribution of these filament elasticity were observed. Whether spatial distribution of filament elasticity is important in regulating different modes of migration remains to be investigated.
To summarize, the ability of a cell to migrate directionally or non-directionally (randomly), is tightly regulated by the intricacies of actin cytoskeleton, microtubules, signaling mechanism, and cellular forces. Although previous studies have explored extensively into the signaling and dynamic organization of cytoskeleton during cell migration, few have investigated the spatial pattern of force distribution and the mechanical properties of actin filaments during different modes of migratory behavior. The project combined the advances in nanotechnology with the foundation in cell biology, screened various cell types from epithelial to mesenchymal, normal, diseased, to transformed, and tried to answer one question. “Could we determine the migratory behavior of a cell from the spatial distribution of elasticity of its actin filaments?”
Materials and methods
Cell culture
U2OS, NIH-3T3, NRK52E, NRK49F, M10, MCF-7, and MDA-MB-231 were cultured according to ATCC's recommendations. U2OS cells were stimulated according to Khyrul et al.Citation12 to facilitate polarization.
Primary culture of keloid fibroblasts
The experiment was approved by Institutional Review Boards in National Cheng Kung University Hospital, according to the Helsinki Declaration (NCKUH-10105017/BR-100-102). The isolation of fibroblasts from tissues was performed as previously described.Citation13 The epidermis and subcutaneous tissue were excised from the sample tissue, which was then cut into roughly 1 mm3 fragments using a blade, and placed as explants in 25 cm2 tissue culture flasks with 5 ml culture medium consisting of Dulbecco's modified Eagle's medium (DMEM), L-glutamine, streptomycin (100 mg/mL), penicillin (100 U/mL) and fetal calf serum (10%). The cell culture was maintained at 37°C in humidified incubator with 5% CO2. Growth medium was changed every 3–4 d. Some of the harvest fibroblasts were frozen in 10% DMSO containing serum at −80°C until use. Fibroblasts used in these experiments were limited to passages 3-8. The control group was from anatomical region- and age-matched healthy subjects.
Atomic force microscopy live cell imaging and indentation
The method is adopted from the work of Harn et al.Citation5 A NanoWizard II (JPK Instruments, Berlin, Germany) AFM mounted on an inverted epi-fluorescent microscope was used for all experiments. Silicon nitride cantilevers (CSC-38B, MikroMasch, Sofia, Bulgaria) with a nominal spring constant of 0.03 N/m [calibrated using the thermal noise method Citation14] and cone half angle < 15° were used for both imaging and indenting. All AFM images and measurements were obtained in contact mode in liquid phase. For AFM analysis of living cells, a temperature-controlled sample holder Bio-Cell filled with DMEM containing 10% FBS as culture media was used.
To scan and imagine a living cell, we positioned the scanning range of AFM cantilever tip to ensure full coverage of the cell under the light microscopy. The cell was then scanned with tip velocity controlled at 100 μm/sec and force applied between 0.5 ∼1 nN. The whole scanning time was kept under 15 min to minimize the changes in cytoskeleton throughout the process. The image file was processed using JPK data analysis software (JPK Instruments, Berlin, Germany), where the deflection, height, 3-dimensional, and cross-section profile images were extracted.
After obtaining the scanned image, the indentation points were selected according to the scanned fluorescent image, where prominent actin filament could be identified (). We indented the cell at 1 nN around the periphery at 3˜5 μm apart. During elasticity measurements, force–distance curves were recorded continuously at 1 Hz (1 s per μm during approach–retract force curve). The determination of the elastic modulus from the force–distance curves was performed using the Sneddon's variation of the Hertz model Citation15,16 through JPK data analysis software (JPK Instruments, Berlin, Germany). The force on the cantilever F(h) is given by:
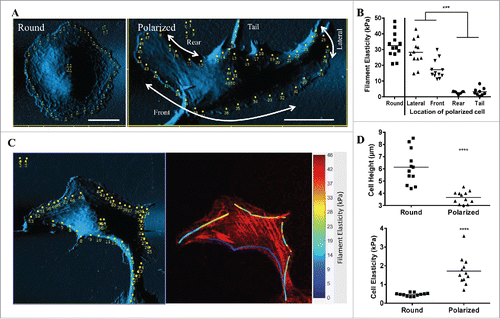
Figure 1. Migrating osteosarcoma cells display a polarized distribution of different filament elasticity. (A) Contact-mode AFM deflection images and indentation points (yellow dots) of a living round and polarized U2OS cell. Designated locations of a polarized U2OS cell are indicated by the arrows and texts. (B) Dot plot showing filament elasticity with respect to different locations of the cell in round and polarized state. Each dot indicates an average of filament elasticity from at least 4-10 indentation points. (C) Left panel: AFM scanned images of a polarized U2OS cell. The yellow dots indicate the indentation points measured. Red images: Right panel: Overlay image of filament elasticity displayed as color heatmap and confocal image of polarized U2OS cells stained with phalloidin for filamentous actin. (D) Dot plots showing cell height and elasticity in round and polarized state. Data from at least 8 cells were collected in each group.
Confocal images and three-dimensional reconstruction
The cells were fixed by 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. Fixed cells were blocked with SuperBlock blocking buffer (Thermo Scientific, MA, USA) for 1 h and then incubated with designated primary antibody (BD, CA, USA) for overnight at 4°C and incubated with a anti-mouse or rabbit IgG conjugated with Alexa 488 (Molecular Probe, Oregon, USA), phalloidin-TRITC (Sigma-Aldrige, MO, USA) and Hoechst 33258 (Sigma, MO, USA). The serial sections of immunofluorescence images were taken under confocal microscopy (Olympus, FV-1000, Tokyo, Japan) at thickness < 0.3 µm per section. The serial images were reconstructed using Avizo standard software (VSG, MA, USA) to obtain the final 3D images.
Fibroblast wound healing assay
3T3 cells were seeding in 6 cm dish at the density of 3 × 106 and kept until confluent. A wound was created by using a pipet tip to scratch off a line of cells across the center of the dish. The dish was then incubated for 8 h or 24 h until the cells migrate into the gap.
Inhibitor treatment
CytoD or ML-7 (Sigma-Aldrige, MO, USA) was added to the culture medium according to the desired dose 8 hours after the cells were seeded. The cells were measured after 12 hours of treatment. The effect of CytoD or ML-7 on actin filaments were confirmed with immunofluorescence imaging of the filamentous actin.
Real-time cell recording and tracking
Real-time cell recording and tracking were performed according to Huang et al.Citation4 Briefly, the cells were cultured in 3 cm dish and placed on an inverted microscope under a temperature and CO2-controlled environment. Cell images were taken every 30 min for 12 hours. These captured images were complied, and the migratory pattern were analyzed using Leica MDW software (Leica, Wetzlar, Germany).
Statistics
All the numbers presented are displayed as mean ± SE. At least 16 filaments in at least 10 cells were measured for each condition. Unpaired t test was performed using Prism 6 (GraphPad Software, La Jolla, CA, USA), and values of P < 0.05 were considered statistically significant. *P< 0.05, **P<0 .01, ***P<0 .001.
Results
Cell polarization and migration are characterized by generating distinct and spatial filament elasticity
Migration is characterized by rapid actin cytoskeletal reorganization, focal adhesion turnover, and traction force generation.Citation18 To study changes in the mechanical properties of actin filaments and cells during cell migration, we had previously set up a bio-AFM system with which we could scan and indent a living cell and filaments in a stable temperature-controlled environment.Citation5 We adopted U2OS osteosarcoma cell into our system due to its autonomous polarizing nature in directional migration.Citation12 In the round, non-polarized state, the AFM images showed linear filamentous structures on cell periphery with a prominent nucleus protrusion at the center (, Round). In the polarized cell, the AFM image showed the cell displaying a long and short axis, with linear filaments at the lateral edges of the cell. In this study, the locations of a polarized U2OS cell are categorized into front, lateral, rear and tail (, Polarized). The thicknesses of the two lateral edges were similar, while the front (leading) edge of the cell is relatively flatter in comparison to the rear edge.
The changes in the mechanical properties of the filaments and the cell during polarization could be observed on the dot plot (). Round cell displayed only one filament elasticity throughout cell periphery at 32.6 ± 2.2 kPa, while polarized cell showed at least 4 distinct filament elasticity on different parts of the cell. The lateral edge of the cell showed the highest elasticity (29.4 ± 3.0 kPa), followed by front (23.0 ± 5.04 kPa), tail (3.90 ± 1.81 kPa) and rear (2.71 ± 0.29 kPa). The respective spatial distribution of filament elasticity and filamentous actin is demonstrated in ). Cell polarization also affected nucleus elasticity, as it increased from 0.47 ± 0.03 kPa in round state to 1.73 ± 0.32 kPa when polarized. Cell height was inversely correlated with nucleus elasticity, as it was higher in round (6.16 ± 0.51 µm) than polarized state (3.69 ± 0.19 µm) (). These results showed the re-distribution of actin filaments and the change in their elasticity could play an important role in cell polarization.
Generating distinct and spatial filament elasticity is essential for cell polarization and migration
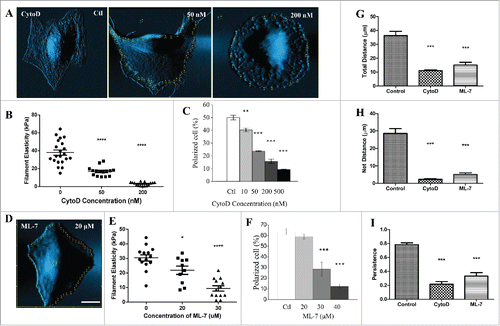
Next we would like to know whether generating distinct spatial filament elasticity is essential for cell polarization and migration. We treated U2OS cells with actin polymerization inhibitor Cytochalasin D (CytoD) and observed that cell morphology, filament elasticity, and the ability to polarize over time were all affected. As indicated by AFM-deflection images, the linear filamentous structures on cell surface were reduced when cells were treated with CytoD (). 50 and 200 nM of CytoD reduced the filament elasticity of round U2OS cells from 37.9 ± 2.9 kPa to 16.9 ± 1.3 kPa and 3.05 ± 0.52 kPa, respectively (). These numbers are important because they set the upper limit of filament elasticity under each dose of CytoD treatment. We further specifically measured filaments of the polarized U2OS cells after treated with CytoD, and found that these cells displayed distinct filament elasticity despite the change in morphology and reduced polarization. Treating the cells with ML-7, the inhibitor of myosin light chain kinase, resulted in similar morphological changes (), reduction in filament elasticity (), and altered polarized percentage (). Lastly, we showed the total distance, net distance, and persistence profiles of the CytoD or ML-7 treated cells were all decreased significantly (). In parallel, these results showed the ability of actin filaments to re-distribute and change their elasticity is essential to the polarization of the cell.
Figure 2. Inhibition of actin polymerization or myosin light chain kinase reduce filament elasticity, cell polarization and directional migration in U2OS cells. (A) Contact-mode AFM deflection images and indentation points (yellow dots) of living U2OS cell treated with different doses of CytoD. (B) Dot plot showing filament elasticity of round U2OS cells treated with different doses of CytoD. (C) The changes in percentage of polarized cells with respect to different doses of CytoD. (D) Contact-mode AFM deflection images and indentation points (yellow dots) of living U2OS cell treated with 20 μM of ML-7. (E) Dot plot showing filament elasticity of round U2OS cells treated with different doses of ML-7. (F) The changes in percentage of polarized cells with respect to different doses of ML-7. Each dot indicates an average of filament elasticity from at least 4–10 indentation points. Cells treated with 200 nM of CytoD, 30 µm of ML-7 were compared with control for total distance (G), net distance (H), and persistence (I). Persistence is defined as net distance over total distance. Cell activity was recorded in 5-min cycles for 4 h. Bar: 20 μm. Data from at least 10 cells were collected in each group. The percentage of polarization were calculated from 3 sets of independent experiments where at least 50 cells were counted in each set.
Spatial distribution of filament elasticity reflects the migrating modes of the cells
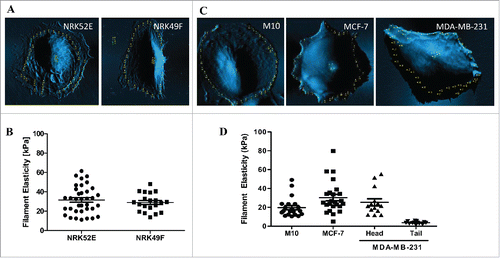
To further understand the relationship between spatial distribution of distinct filament elasticity and the migratory behavior of the cell, we scanned a series of cell lines, from epithelial cells, fibroblasts, to different levels of malignant cancer cells. NRK52E and NRK49F, kidney epithelial cells and fibroblasts derived from rat, respectively, were non-migratory and only showed one type of filament elasticity ( and ). M10, normal breast epithelial cells were also non-migratory and displayed one type of filament elasticity. MCF-7, the less malignant breast cancer cells, showed a wider range of filament elasticity (from 8kPa ∼81kPa), but no spatial distribution of distinct filament elasticity were observed. Lastly, in the highly malignant breast cancer cells MDA-MB-231, the cells displayed distinct filament elasticity with the stiff ones located at the head end (∼22kPa) while the tail end showed soft filament elasticity (∼5kPa) (). These findings indicate that spatial distribution of distinct filament elasticity reflects the migratory modes of the cell.
Figure 3. Spatial distribution of filament elasticity reflects migrating modes of cells. (A) AFM-scanned image of NRK52E and NRK49F cells. (B) Dot plot showing filament elasticity of NRK52E and NRK49F cells. (C) AFM-scanned image of M10, MCF-7, and MDA-MB-231 cells. (D) Dot plot showing filament elasticity of M10, MCF-7, and MDA-MB-231 cells. Data from at least 10 cells were collected in each group.
Keloid fibroblasts show non-spatially distributed filament elasticity and non-directional migration
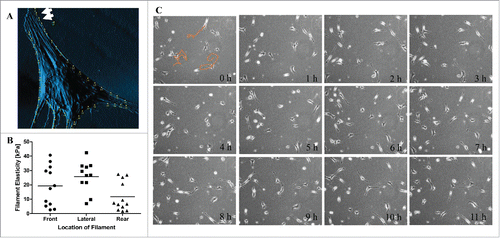
To further explore the changes in distribution of filament elasticity in different modes of migration, we used keloid fibroblast as a model. Our previous findings have shown that the ability to generate distinct filament elasticity autonomously could be an important feature contributing to the migrating nature of these pathologic cells.Citation5 In several of the scanned images of keloid fibroblasts, we noticed lamellipodia forming at the narrow end of the cell (, white arrows). This is quite a contrast to our previous observation where lamellipodia were usually formed at the wider “front” end of the fibroblast. This hints that the cell was forming a new leading edge at what may had been an old rear end. In order to understand the spatial distribution of filament elasticity in keloid fibroblasts, we analyzed the filament elasticity of keloid fibroblasts according to its respective location on the cell (). Despite autonomously generating 2 distinct types of filament elasticity, both elasticities of these filaments were distributed at all locations of the cell (front, lateral and rear). We also compiled time-lapse images of keloid fibroblasts migration over 12 h, and most of the cells migrated non-directionally (, orange lines). This migrating pattern indicates that new lamellipodia were spontaneously forming at all locations of the cell, hence the mixing pattern of filament elasticity. Collectively speaking, keloid fibroblasts show non-spatially distributed filament elasticity and non-directional migration.
Figure 4. Keloid fibroblasts show non-spatially distributed filament elasticity and non-directional migration. (A) An AFM-scanned image of keloid fibroblast shows lamellipodia forming at the narrow end of the cell, marked by the white arrows. (B) Dot graph displaying the different filament elasticities measured from different location of the keloid fibroblasts. (C) Compiled time-lapse image of keloid fibroblasts migration over 12 h. Orange lines indicate the migrating track of the designated cells.
Spatial distribution of filament elasticity determine the migratory mode of the cell
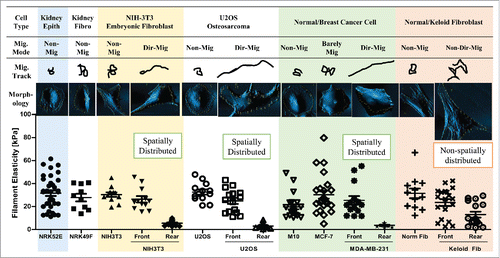
To examine how filament elasticity affect the migratory mode of the cell, we compared the filament elasticity and recorded time-lapse video to observe the migratory behavior of various types of cells from epithelial, fibroblast, cancer and pathologic origins (). Both kidney epithelial cells NRK52E and fibroblasts NRK49F were not migratory and displayed only one stiff type of filament elasticity at around 30 kPa (, Kidney Epith, Kidney Fibro). Embryonic fibroblast NIH-3T3 cells were also non-migratory; however, when stimulated to undergo directional migration during wound closure assay, the cells showed spatial distribution of filament elasticity with stiff filaments in the front and soft ones in the rear end (, NIH-3T3). Osteosarcoma U2OS cells showed one stiff filament type when non-polarized, and like the stimulated 3T3 fibroblasts, displayed a stiff front edge and a soft rear end when polarized and directionally migrated (, U2OS). Normal breast epithelial cell M10 were non-migratory and only displayed one filament elasticity. The low-malignant breast cancer cell line MCF-7 were barely migratory, thus showing a few filaments with soft elasticity (between 8∼11 kPa). The highly malignant breast cancer cell line MDA-MB-231 display prominent directional migration behavior, and also showed distinct and spatially distributed filament elasticity at the front and rear end of the cell (, Normal/Breast Cancer Cell). Lastly, primary normal fibroblasts from human skin did not migrate much and showed only one stiff filament type. However, in primary keloid fibroblasts, filaments of both elasticity were non-spatially distributed in all locations of the cell, while showing an active yet non-directionally migrating pattern. (, Normal/Keloid Fibroblast)
Figure 5. Spatial distribution of filament elasticity determine the migratory mode of the cell. Summary of migratory mode, migratory track, morphology, and spatial distribution of filament elasticity of kidney epithelial cell (Kidney Epith), kidney fibroblast (Kidney Fibro), NIH-3T3 embryonic fibroblast, U2OS osteosarcoma cell, normal and breast cancer cell, and normal fibroblast (Norm Fib) and keloid fibroblast (Keloid Fib). Migratory track images are illustrations of migratory distance and pattern recorded over 12 hours using time-lapse cell tracing software. The morphology images are AFM-scanned images. Spatial distribution of filament elasticity of each cell type is displayed by the dot plot. Mig. Mode: Migratory mode. Non-Mig: non migratory. Dir-Mig: Directional migration. Non-Dir Mig: Non-directional migration. Data from at least 10 cells were collected in each group.
Microtubules may play an important role in determining the migratory mode of cells
Reports have described that the crosstalk of microtubules and actin play an important role in cell polarity and directional migration.Citation19 To explore the possible factor affecting cell polarity during cell migration in our model, we examined the organization of microtubules during non-migrating, directional migration, and non-directional migration. The microtubules in round, non-polarized U2OS cells were mostly distributed in the central region of the cytosol with a prominent MTOC located proximal to the nucleus. On the other hand, in its polarized state, the microtubules were mainly distributed in the rear end of the cell (SI ). We then performed the same experiments on non-polarized and polarized 3T3-fibroblasts. The 3D-reconstructed images show that in the non-polarized fibroblast, the distribution of microtubules are symmetrical and evenly distributed throughout the cytosol. On the other hand, in its polarized state, more microtubules are distributed in the rear end (SI ). Generally speaking, the location of microtubules and actin filaments show complementary pattern. We further treated the fibroblasts with a microtubule polymerization inhibitor, nocodazol, and showed that the fibroblast lost its mesenchymal-like spindle shape feature and became rounded (SI ). Furthermore, the distribution of microtubules changed from located at the spindle endings of the cell to an even distribution around the outer cytosol. Treating cells with microtubule inhibitor not only affected the polymerization and distribution of microtubules, but also the organization of actin filaments and cell shape and polarity as a whole. Lastly, we examined the expression of microtubules in the non-directionally migrating keloid fibroblasts. The results show that microtubules were downregulated in comparing to normal fibroblasts (SI ). These results implicate the complementary relationships of microtubules and actin cytoskeletons, and signify the important role of microtubule in generating the polarity and migratory modes of the cell.
Discussion
This study shows that 1) non-migratory cells only generated one type of filament elasticity, 2) cells generating spatially distributed two types of filament elasticity showed directional migration, and 3) pathologic cells that autonomously generated two types of filament elasticity without spatial distribution were actively migrating non-directionally. We postulate that migrating cells (U2OS, breast cancer cells, or keloid fibroblasts) showed a softer population of filaments because the focal adhesions of those filaments were disassembling, which also led to the depolymerization of the associated actin filaments, and allowed the cells to migrate forward. This is also the reason why the softer population of filaments was not identified in the population of non-migratory cells we examined (NRK52E, NRK49F, non-stimulated NIH3T3, non-polarized U2OS cells, primary skin normal fibroblasts, ). In our previous work we used PF573228, an inhibitor of FAK phosphorylation at tyrosine 397, to treat keloid fibroblast and show that all of the filament elasticity were lowered, and the cells could not migrate effectively.Citation5
One of the most significant findings of this study is how spatial distribution of distinct filament elasticity leads to different modes of migration. As illustrated in , although keloid fibroblasts also generated 2 types of filament elasticity autonomously, these filaments were not spatially distributed as in the cases of migrating polarized U2OS cells or migrating malignant breast cancer MDA-MB-231 cells. Since the nature of keloid pathogenesis is characterized by keloid fibroblasts migrating beyond the original boarder of the wound, we speculate that these active keloid fibroblasts are constantly generating new lamellipodia in order to “probe” and “reach out” from any given location of the cell to the new surroundings, and hence this cytoskeleton dynamics were reflected in the non-spatial distribution of filament elasticity. On the other hand, both polarized U2OS cells and MDA-MB-231 cells showed high persistency in their migrating behavior, which correspond to the high spatially distributed filament elasticity. These results all point to not only the mechanical but also spatial regulation of actin filaments are finely tuned in regulating the behavior of the cell.
More importantly, since the actin stress fibers are linked to the two major modules controlling mechanotransduction and response, the focal adhesions and the nucleus, the elasticity of the actin filaments could provide very insightful information regarding the status quo, of the state of the cell behavior. A screening of various cell types with different migratory behavior as performed in this study demonstrates that (). Epithelial cells are the building blocks of our organs, and thus the cells are less motile, in which the filament elasticity are uniform. Mesenchymal cells are migratory, and therefore a high and low of distinct filament elasticity could be observed in the same cell, reflecting the net imbalance force in order for the cell to move. Furthermore, depending on the spatial distribution of these filaments, the migratory behavior – direction or non-directional – could be delineated.
The role of microtubule in cell polarization and directional migration is another intriguing finding in this study. We showed that upon treatment of nocodazole, cells failed to polarize, and microtubules were downregulated in the non-directional migrating keloid fibroblasts. Ganguly et al. have used tubulin mutations or low concentrations of drugs that suppress microtubule dynamics, and showed that migrating cells still extended lamillipodia but were inhibited in their ability to retract their tails. These cells could still move at near normal speeds, but showed more random, non-directional migration.Citation20 The continual targeting of microtubules to focal adhesion therefore is essential for the directional migration of the cell. Evidences suggest that microtubules may track along actin filaments in stress fibers to achieve this targeting process.Citation21 Stehbens et al. also observed that in migrating cells, dynamic microtubules repeatedly target the mature focal adhesions located in the rear end, leading to the disassembly of the focal adhesions.Citation22 The intricate interaction of microtubules-focal adhesions-actin cytoskeleton could provide another perspective in understanding the migratory behavior of pathologic cells such as cancer and keloid fibroblast.
This is the first study to demonstrate the spatial generation of filament of distinct elasticity in the same cell, and providing a significant implication to it. Previous studies on cell migration were limited to how the cytoskeleton are organized and how the cell establish the polarity in order to migrate persistently, yet few have explored the regional changes in mechanical properties of the actin cytoskeleton. This study expands the implications of biological measurements to facilitate our understanding of cell migration, from normal to pathologic cells.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Figures_1-2.zip
Download Zip (891.8 KB)Acknowledgments
We would like to thank Dr. Chao-Yuan Yeh for constructing the elasticity heatmap, Dr. Yu-Wei Chiou for assisting with AFM operation, Ms. Tzi-Ling Chen for administrative help and Ms. Wei-Ying Lin for invaluable discussions.
Funding
This study was supported by grants from Ministry of Science and Technology, Taiwan, (MoST-101-2320-B-006-011-MY3) to M-JT.
References
- Bravo-Cordero JJ, Hodgson L, Condeelis J. Directed cell invasion and migration during metastasis. Curr Opin Cell Biol 2012; 24:277-83; PMID:22209238; http://dx.doi.org/10.1016/j.ceb.2011.12.004
- Vicente-Manzanares M, Webb DJ, Horwitz AR. Cell migration at a glance. J Cell Sci 2005; 118:4917-9; PMID:16254237; http://dx.doi.org/10.1242/jcs.02662
- Wu TH, Li CH, Tang MJ, Liang JI, Chen CH, Yeh ML. Migration speed and directionality switch of normal epithelial cells after TGF-beta1-induced EMT (tEMT) on micro-structured polydimethylsiloxane (PDMS) substrates with variations in stiffness and topographic patterning. Cell Communication Adhesion 2013; 20:115-26; PMID:24053415; http://dx.doi.org/10.3109/15419061.2013.833194
- Huang YW, Chang SJ, I-Chen Harn H, Huang HT, Lin HH, Shen MR, Tang MJ, Chiu WT. Mechanosensitive store-operated calcium entry regulates the formation of cell polarity. J Cell Physiol 2015; 230:2086-97; PMID:25639747; http://dx.doi.org/10.1002/jcp.24936
- Harn HI, Wang YK, Hsu CK, Ho YT, Huang YW, Chiu WT, Lin HH, Cheng CM, Tang MJ. Mechanical coupling of cytoskeletal elasticity and force generation is crucial for understanding the migrating nature of keloid fibroblasts. Exp Dermatol 2015; 24(8):579-84; PMID:25877039
- Huttenlocher A, Horwitz AR. Integrins in cell migration. Cold Spring Harbor Perspectives Biol 2011; 3:a005074; http://dx.doi.org/10.1101/cshperspect.a005074
- Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol 2009; 10:538-49; PMID:19603038; http://dx.doi.org/10.1038/nrm2729
- Ridley A, Heald R. Cell structure and dynamics. Curr Opin Cell Biol 2011; 23:1-3; PMID:21190823; http://dx.doi.org/10.1016/j.ceb.2010.12.003
- Peschetola V, Laurent VM, Duperray A, Michel R, Ambrosi D, Preziosi L, Verdier C. Time-dependent traction force microscopy for cancer cells as a measure of invasiveness. Cytoskeleton 2013; 70:201-14; PMID:23444002; http://dx.doi.org/10.1002/cm.21100
- Haupt BJ, Osbourn M, Spanhoff R, de Keijzer S, Muller-Taubenberger A, Snaar-Jagalska E, Schmidt T. Asymmetric elastic properties of Dictyostelium discoideum in relation to chemotaxis. Langmuir 2007; 23:9352-7; PMID:17661497; http://dx.doi.org/10.1021/la700693f
- Lee SH, Dominguez R. Regulation of actin cytoskeleton dynamics in cells. Mol Cells 2010; 29:311-25; PMID:20446344; http://dx.doi.org/10.1007/s10059-010-0053-8
- Khyrul WA, LaLonde DP, Brown MC, Levinson H, Turner CE. The integrin-linked kinase regulates cell morphology and motility in a rho-associated kinase-dependent manner. J Biol Chem 2004; 279:54131-9; PMID:15485819; http://dx.doi.org/10.1074/jbc.M410051200
- Zhang Q, Oh CK, Messadi DV, Duong HS, Kelly AP, Soo C, Wang L, Le AD. Hypoxia-induced HIF-1 α accumulation is augmented in a co-culture of keloid fibroblasts and human mast cells: involvement of ERK1/2 and PI-3K/Akt. Exp Cell Res 2006; 312:145-55; PMID:16289155; http://dx.doi.org/10.1016/j.yexcr.2005.10.006
- Butt HJ, Jaschke M. Calculation of Thermal Noise in Atomic-Force Microscopy. Nanotechnology 1995; 6:1-7; http://dx.doi.org/10.1088/0957-4484/6/1/001
- Laurent VM, Kasas S, Yersin A, Schaffer TE, Catsicas S, Dietler G, Verkhovsky AB, Meister JJ. Gradient of rigidity in the lamellipodia of migrating cells revealed by atomic force microscopy. Biophysical J 2005; 89:667-75; http://dx.doi.org/10.1529/biophysj.104.052316
- Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA, Zhdanov RI. Atomic force microscopy probing of cell elasticity. Micron 2007; 38:824-33; PMID:17709250; http://dx.doi.org/10.1016/j.micron.2007.06.011
- Martens JC, Radmacher M. Softening of the actin cytoskeleton by inhibition of myosin II. Pflugers Archiv 2008; 456:95-100; PMID:18231808; http://dx.doi.org/10.1007/s00424-007-0419-8
- Kuo JC. Mechanotransduction at focal adhesions: integrating cytoskeletal mechanics in migrating cells. J Cell Mol Med 2013; 17:704-12; PMID:23551528; http://dx.doi.org/10.1111/jcmm.12054
- Akhshi TK, Wernike D, Piekny A. Microtubules and actin crosstalk in cell migration and division. Cytoskeleton 2014; 71:1-23; PMID:24127246; http://dx.doi.org/10.1002/cm.21150
- Ganguly A, Yang H, Sharma R, Patel KD, Cabral F. The role of microtubules and their dynamics in cell migration. J Biol Chem 2012; 287:43359-69; PMID:23135278; http://dx.doi.org/10.1074/jbc.M112.423905
- Bowen JR, Hwang D, Bai X, Roy D, Spiliotis ET. Septin GTPases spatially guide microtubule organization and plus end dynamics in polarizing epithelia. J Cell Biol 2011; 194:187-97; PMID:21788367; http://dx.doi.org/10.1083/jcb.201102076
- Stehbens S, Wittmann T. Targeting and transport: how microtubules control focal adhesion dynamics. J Cell Biol 2012; 198:481-9; PMID:22908306; http://dx.doi.org/10.1083/jcb.201206050
