ABSTRACT
The skeletal muscle regeneration occurs due to the presence of tissue specific stem cells - satellite cells. These cells, localized between sarcolemma and basal lamina, are bound to muscle fibers and remain quiescent until their activation upon muscle injury. Due to pathological conditions, such as extensive injury or dystrophy, skeletal muscle regeneration is diminished. Among the therapies aiming to ameliorate skeletal muscle diseases are transplantations of the stem cells. In our previous studies we showed that Sdf-1 (stromal derived factor −1) increased migration of stem cells and their fusion with myoblasts in vitro. Importantly, we identified that Sdf-1 caused an increase in the expression of tetraspanin CD9 - adhesion protein involved in myoblasts fusion. In the current study we aimed to uncover the details of molecular mechanism of Sdf-1 action. We focused at the Sdf-1 receptors - Cxcr4 and Cxcr7, as well as signaling pathways induced by these molecules in primary myoblasts, as well as various stem cells - mesenchymal stem cells and embryonic stem cells, i.e. the cells of different migration and myogenic potential. We showed that Sdf-1 altered actin organization via FAK (focal adhesion kinase), Cdc42 (cell division control protein 42), and Rac-1 (Ras-Related C3 Botulinum Toxin Substrate 1). Moreover, we showed that Sdf-1 modified the transcription profile of genes encoding factors engaged in cells adhesion and migration. As the result, cells such as primary myoblasts or embryonic stem cells, became characterized by more effective migration when transplanted into regenerating muscle.
Introduction
The skeletal muscle regeneration occurs due to the presence of stem cells called satellite cells (SCs) that are localized between sarcolemma and basal lamina. The role of SCs in skeletal muscle repair is unquestionable (reviewed in ref. Citation1). Unfortunately, due to aging, extensive damages or various pathological states, for example muscular dystrophy, muscle reconstruction is diminished.Citation2-4 Stem cells transplantation belongs to the therapeutic approaches aiming to improve muscle regeneration (reviewed in ref. Citation5). In the initial studies, focusing on the skeletal muscle cell therapies, SCs and primary myoblasts, due to their natural function, were the first choice of cells tested (reviewed in refs. Citation6, 7). Under physiological conditions, after muscle injury, SCs become activated what leads to the cell cycle re-entry, proliferation, and finally their differentiation into myoblasts that fuse and reconstruct myotubes and then muscle fibers. In the 80s of XX century Partridge and collaborators documented that wild-type myoblasts injected to muscle of dystrophic mice (mdx mice) were able to reconstruct muscle fibers and restore the dystrophin synthesis.Citation8 As demonstrated later, the improvement in the skeletal muscle regeneration was observed after transplantation of undifferentiated, purified satellite cells population, rather than satellite cells derived myoblasts.Citation9,10 In the 90s many clinical trials based on the model described by Partridge were conducted, however, the results were not satisfactory (reviewed in refs. Citation6, 11). Transplanted cells were able to participate in the muscle regeneration and partially restore dystrophin expression but no functional long-term improvement was observed.Citation12-14
The most important obstacles in myoblast transplantation include specific immune response against transplanted cells, limited migration within the muscle, and massive apoptosis of transplanted cells (reviewed in refs. Citation7, 15, 16). The limited migration ability of transplanted myoblasts was shown in many studies.Citation17-20 Thus, many lines of evidence documented that injected myoblasts accumulate within the site of injection and only few reports showed that they could migrate up to 1 cm in depth from the monkey (Macaca mulata) muscle surface.Citation21 Importantly, co-injected growth factors such as bFGF (basic fibroblast growth factor) and IGF-1 (insulin like growth factor), improved migration of monkey (Macaca mulata) myoblasts transplanted into biceps brachii. However, myofibers formed with the participation of these cells were detectable only near the injection site. Moreover, analyzed myoblasts were not able to fuse with undamaged muscle fibers, regardless of the growth factors used.Citation22 In our own studies we showed that the Sdf-1 could improve migration of satellite cell derived myoblasts and C2C12 myoblasts in vitro in metalloproteinase (MMP) dependent manner.Citation23 We also documented that Sdf-1 treatment enhanced embryonic stem cells (ESCs) and bone marrow derived mesenchymal stem cells (BM-MSC) migration and fusion with myoblasts in vitro, what was connected with the increase in tetraspanin CD9 expression.Citation24
In the current study we investigated which molecular pathways induced by Sdf-1 lead to the increased migration. We hypothesized that stimulation of transplanted cells migration using Sdf-1 improves their ability to participate in muscle repair. To verify this hypothesis we analyzed various stem cell populations - mouse primary myoblasts derived from SCs, human mesenchymal stem cells isolated from umbilical cord connective tissue, i.e., Wharton's jelly (WJ-MSCs), and mouse embryonic stem cells (ESCs). Our choice based on the previous studies in that we documented that these stem cells are able to undergo myogenic differentiation and also to participate in the skeletal muscle regeneration.Citation24-26
Multipotent mesenchymal stem cells (MSCs) could be derived from different sources, such as bone marrow, adipose tissue, Wharton jelly (umbilical cord connective tissue), umbilical cord blood, skin, dental pulp, spleen, lung, and also skeletal muscles (reviewed in refs. Citation27, 28). Various populations of mesenchymal stem cells were able to improve skeletal muscle reconstruction.Citation29-31 Myogenic differentiation of the pluripotent stem cells, such as embryonic stem cells (ESCs), which are characterized by unlimited potential to proliferate and ability to differentiate into any given tissue, has been also documented (reviewed in refs. Citation32, 33). An efficient protocol allowing derivation of myoblasts from ESCs, based on the supplementation of culture medium with factors inducing mesoderm formation and myogenic differentiation, was proposed only recently and obtained myoblasts were tested both in vitro and in vivo.Citation34 Cells derived from ESCs when transplanted into tibialis anterior muscles of mdx mice were able to form muscle fibers and also to differentiate into Pax7-expressing cells that resembled SCs.Citation34 However, methods improving homing of these cells to the site of the injury via improvement of their migration are still not readily available.
In the current study we compared the reaction of primary myoblasts, WJ-MSC, as well as ESCs to Sdf-1 treatment, which—as we previously shown—is a potent factor improving skeletal muscle regeneration.Citation23,24 First we analyzed changes in transcription profile and the signaling pathways engaged in stem cells response to Sdf-1 treatment. Next, we concentrated on the role of Sdf-1 receptors i.e. CXCR7 and CXCR4 in stem cells migration both in vitro and in vivo. Then, we examined if Sdf-1 pretreatment of stem cells with Sdf-1 or co-injection of these cytokine could improve participation of tested cells in the skeletal muscle regeneration.
Materials and methods
All the experiments were performed with the approval of Local Ethical Commission No 1 in Warsaw – permission no 240/2012.
Cells cultures
Satellite cells – derived myoblasts (primary myoblasts)
Satellite cells were isolated from the gastrocnemius muscles of 3 months old C57Bl6N male mice carrying the lacZ transgene in the ROSA26 locus. Mice were sacrificed by cervical dislocation. Muscle fibers were isolated according to previously described protocol.Citation35 Briefly, muscles were dissected and digested with 0.2% collagenase type I (Sigma-Aldrich) in Dulbecco's modified Eagle's medium (DMEM, Life Technologies) at 37°C in 5% CO2 for 60 min. Next, single muscle fibers were transferred to DMEM containing 10% horse serum (HS, Life Technologies) and 1% penicillin/streptomycin antibiotics (AB, Life Technologies). Suspension of muscle fibers was passed through a syringe needle (21G) and cleared by filtration through 40 µm cell strainer. Obtained satellite cells were plated in 6-well culture dishes coated with Matrigel Matrix Growth Factor Reduced (BD Biosciences). Primary myoblasts were maintained in so called “growth medium,” i.e., DMEM supplemented with 20% fetal bovine serum (FBS), 10% HS, 0.5% chicken embryo extract (CEE, Life Technologies) and 1% AB.
Mesenchymal stem cells derived from Wharton jelly (WJ-MSCs)
WJ-MSCs were kindly provided by prof. Zygmunt Pojda (Department of Molecular and Translational Oncology, Maria Skłodowska-Curie Memorial Cancer Center and Institute of Oncology, Warsaw, Poland). WJ-MSCs were seeded and cultured in DMEM (Life Technologies) supplemented with 15% heat inactivated FBS (hiFBS) and 1% AB.
Mouse embryonic stem cells (ESCs)
ESCs constitutively expressing histone H2B-GFP were provided by Dr. Kat Hadjantonakis.Citation36 Mitomycin-inactivated mouse embryonic fibroblasts (MEFs), that served as feeder layer for ES cells, were plated on 1% gelatin coated culture dishes (Sigma-Aldrich) and cultured in DMEM supplemented with 10% FBS and 1% AB. Twenty four hours later ESCs were seeded onto the inactivated MEFs and cultured in knockout DMEM (Life Technologies) supplemented with 10% serum replacement (SR, Life Technologies), 0.1 mM nonessential amino acids (Sigma-Aldrich), 2 mM L-glutamine (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 1% AB, and 500 U/ml leukemia inhibitory factor (LIF, Chemicon). Prior to transfection with siRNA, ESCs were separated from MEFs by pre-plating and cultured in cultured dishes coated with 10% Matrigel Matrix Growth Factor Reduced (BD Biosciences) in DMEM until the time of further manipulations.
Morphological analyses
The morphology of primary myoblasts, WJ-MSCs, and ESCs was analyzed using Nikon Eclipse TE200 microscope equipped with Hoffman contrast.
Cells transfection
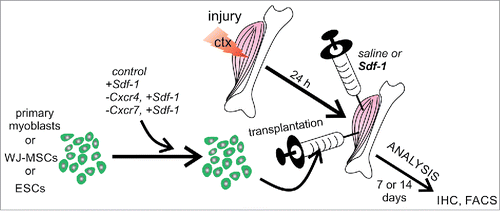
Primary myoblasts, WJ-MSCs, and ESCs were plated into culture dishes and after reaching 50-60% of confluency transfected with Silencer Select Pre-designed siRNA (Life Technologies) complementary to mRNAs encoding either Cxcr4 (ID:s64091) or Cxcr7 (ID:s64124). Appropriate negative control siRNA was used according to manufacturer's recomendation. siRNA duplexes were diluted in DMEM to 100 pmol concentration and Lipofectamine RNAiMAX (Life Technologies) was added according to manufacturer's instructions. The Sdf-1 (10 ng/µl) was added 24 h after transfection. The cells were collected 48 h post-Sdf-1 treatment and processed either for mRNA isolation, followed by qRT-PCR, immunolocalization, Western blotting, G-LISA or for transplantation into injured and regenerating gastrocniemus muscles. The efficiency of CXCR4 or CXCR7 down regulation was assessed by qRT-PCR and Western-blot.
Quantified real time PCR (qRT-PCR)
Total RNA was isolated from primary myoblasts, WJ-MSCs, and ESCs using mirVana Isolation Kit (Life Technlogies), according to the manufacturer's protocol. RNA was extracted from biological triplicates (3 independent cell cultures per each experiment). 250 ng of RNA from each sample was reverse-transcribed using the SuperScript II Reverse Transcriptase (Life Technologies) according to the manufacturer's protocol. Next, mRNA levels were examined using Quantitative real-time PCR analysis (qPCR) with TaqMan assays (Life Technologies) for the following genes: CXCR4 [Mm01996749], CXCR7 [Mm02619632], Rac-1 [Mm01331626], Cdc42 [Mm01194005], focal adhesion kinase (FAK) [Mm00552827], and actin [Mm01268569]. Hypoxanthine phosphoribosyltransferase 1 (Hprt1) [Mm01545399] was used as the reference gene. All reactions were performed in triplicates. qPCR was performed with the TaqMan Gene Expression Master Mix (Life Technologies) using LightCycler 480 (Roche Applied Sciences) according to manufacturer's instruction. The conditions of RT-qPCR were as follows: reverse transcription: 25°C for 10 min, 42°C for 60 min, 85°C for 5 min, qPCR: 50°C for 2 min, template denaturation 95°C for 10 min, 45 cycles of 95°C for 15 sec and 60°C for 60 sec. Threshold-cycle (Ct) values of the analyzed amplicons were determined with LightCycler® 480 Software (Roche Applied Science). Expression levels were calculated with 2-(ΔCT) formula using relative quantification tool in LightCycler® 480 Software. Expression levels and standard deviations for each gene was visualized as the column charts using GraphPad Software (La Jolla, CA, USA). Reference gene Hprt1 displayed high expression stability. Results were analyzed using using GraphPad Software and non-paired t-test was performed to compare treated with the control cells. The differences were considered statistically significant when p < 0.05 (marked with asterisks).
Cell proliferation assay
Primary myoblasts, WJ-MSCs, and ESCs were incubated in 0.5 µM carboxyfluorescein succinimidyl ester (CFSE, Life Technologies) in PBS at 37°C for 10 min. Cells were rinsed in PBS and cultured for 2 days in the culture medium appropriate for each cell type, under standard conditions. Next, cells were rinsed in PBS and subjected to flow cytometry analysis (BD FACSCALIBUR, BD Biosciences) using CellQuestPro software. Unlabeled cells (negative control) and cells analyzed directly after labeling with CFSE (positive control) were included into each experiment. Three independent experiments were performed. Results were analyzed using GraphPad Software and non-paired t-test was performed to compare treated with the control cells. The differences were considered statistically significant when p < 0.05 (marked with asterisks).
Migration assay
Migration of myoblasts, WJ-MSCs, and ESCs was analyzed using scratch wound healing assay.Citation37 Briefly, cells were plated in the culture dish and cultured until they reached 90% of confluency. Next, the cells were scratched from the plate using plastic tip to create the “wound.” The wound healing manifested by the ability of the cells to refill the created gap was monitored after 48h of culture. Three independent experiments were performed. Results were analyzed using using GraphPad Software and non-paired t-test was performed to compare treated with the control cells. The differences were considered statistically significant when p < 0.05 (marked on charts with asterisks).
Analysis of Rac-1 and Cdc42 activity
Primary myoblasts, ESCs and WJ-MScs were cultured as described above. Thirty min after Sdf-1 treatment cells were lysed in culture dishes, lysates collected and frozen in liquid nitrogen. Active Cdc42 and Rac-1 were analyzed using the G-LISA activation assay kit (Cytoskeleton, Inc.) according to the manufacturer's instructions. The chemiluminescence signal was detected using the μQuant (Biotek Instruments) microplate reader. Three independent experiments were performed. Results were analyzed using GraphPad Software and non-paired t-test was performed to compare treated with the control cells. The differences were considered statistically significant when p < 0.05 (marked on charts with asterisks).
Microarray analysis
ESCs were cultured and either treated with Sdf-1 or transfected with Silencer Select Pre-designed siRNA (Life Technologies) complementary to mRNAs encoding either CXCR4 (ID:s64091) or CXCR7 (ID:s64124) as described above. Total RNA was isolated using mirVana Isolation Kit (Life Technlogies). Next, its integrity was checked with 2100 Bioanalyzer (Agilent Technologies) using RNA 6000 NAno LAb Chip kit (Agilent Technologies). All RNA samples had integrity number above 8.5. 100 ng of total RNA for each sample was biotin labeled with the TargetAmp™-Nano Labeling Kit for Illumina® Expression BeadChip® (Epicentre Biotechnologies). Labeled RNA was purified with RNeasy® MinElute® Cleanup Kit (Qiagen) and hybridized onto MouseRef-8 v2.0 Expression BeadChip (Illumina Inc.) according to manufacturer's instructions. Arrays were scanned with an HiScan®SQ System (Illumina Inc.). Raw data were imported to GenomeStudio (Illumina) and the average signal intensities were analyzed in Partek Genomic Suite (Partek, Inc.) v. 6.6 after quantile normalization and Log2 transformation. Qualitative analysis was performed, e.g. Principal Component Analysis, in order to identify outliers and artifacts on the microarray. After quality check the 2-way ANOVA (Analysis of Variance) model by using Method of MomentsCitation38 was performed on the data and lists of significantly and differentially expressed genes between biological variants (with the cutoff values: p-value < 0.05, −1.3>Fold Change>1.3) were created. Fisher's Least Significant Difference (LSD) was used as the contrast methodCitation39 to compare: ES-Cxcr4 (ESCs transfected with siRNA complementary to CXCX4 mRNA) vs ES-Sdf-1 (ESCs treated with Sdf-1) and ES-Cxcr7 (ESCs transfected with siRNA complementary to CXCX7 mRNA) vs ES-Sdf-1. Unsupervised hierarchical clustering was performed on the selected lists to in order to find genes and samples with similar profiles. Gene networks were created by interposing the results onto the database of Ingenuity containing information about gene functions with the use if Ingenuity Pathway Analysis tool.
Muscle injury and cells transplantation
To induce regeneration of skeletal muscles, 3-month old male BALB/c mice were anesthetized and their gastrocniemus muscles were injected with 50 µl of cardiotoxin (CTX) from Naja mossambica (10 µM in PBS, Sigma-Aldrich). After the procedure mice were kept under standard conditions with free access to food and water. Twenty four hours later control cells, Sdf-1 treated cells, or cells in that expression of Cxcr4 or Cxcr7 was silenced were injected into injured muscles. The number of transplanted cells varied, i.e. 0.5 million of myoblasts, 0.2 million of WJ-MSCs or 1 million of ESCs, suspended in 50 μl of PBS, was transplanted. Moreover, regenerating gastrocnemius muscle was injected with Sdf-1 (100 ng per 20 µl of 0.9% NaCl ) or 20 µl of 0.9% NaCl (saline treated muscles served controls). Sdf-1 and NaCl was injected at the opposite ends of the muscles than the transplanted cells. After 7 or 14 d after injury, i.e., days of regeneration, muscles were dissected and analyzed (immunocytochemistry and histochemistry). Localization of transplanted cells within the muscle was based on the expression of appropriate markers. Satellite cells, from which primary myoblasts were derived, were isolated from 3-month old C57Bl6N male mice carrying the lacZ transgene in the ROSA26 locus. WJ-MSCs were localized on the basis of human nuclear antigen. ESCs were localized on the basis of the expression of H2B-GFP.
Immunocytochemistry
Selected antigens were immunolocalized in in vitro cultured cells, isolated at day 7 of regeneration muscle fibers, as well as in muscle sections (cross and longitudinal). Cells or isolated muscle fibers were fixed with 3% PFA for 10 min, washed with PBS and stored in 4°C. Muscles were dissected 7 or 14 d after injury and cells transplantation. They were frozen in isopentane cooled with liquid nitrogen, transferred into −80°C, and cut into 7 μm-thick sections with cryomicrotome (Microm HM505N) and stored in 4°C. Cryosections were hydrated in PBS, fixed in 3% PFA, and washed with PBS. To obtain longitudinal sections dissected muscles were fixed with Bouin's solution, dehydrated and embed into paraffin blocks. Paraffin blocks were cut for 9 μm-thick slices and placed on covered with 0.5% gelatin in water glass slides and then dried in 40°C. Paraffin sections were stored in 4°C and rehydrated before immunolocalization.
Next, cells or muscle sections were permeabilized with 0.1% Triton X-100/PBS (Sigma-Aldrich), and incubated with 0.25% glycine (Sigma-Aldrich). Non-specific binding of antibodies was blocked with 3% bovine serum albumin (BSA, Sigma-Aldrich) supplemented with 2% donkey serum (Sigma-Aldrich) in PBS, at room temperature, for 1h. Next, samples were incubated for 2 h with primary antibodies diluted 1:100 in 3% BSA in PBS, overnight, washed with PBS, and incubated at room temperature with secondary antibodies diluted 1: 200 in 1.5 % BSA in PBS. After washing with PBS, cell nuclei were visualized by incubation with DraQ5 (Biostatus Limited) diluted 1:1000 in PBS for 10 min. Specimens were mounted with Fluorescent Mounting Medium (Dako Cytomation). After the procedure was completed samples were analyzed using confocal microscope Axiovert 100M (Zeiss) and LSM 510 software. The following primary antibodies were used: chicken polyclonal anti-β-galactosidase (Abcam), mouse monoclonal anti-human nuclear antigen (Abcam), mouse monoclonal anti-Green Fluorescent Protein (GFP) (Abcam), rabbit polyclonal anti-Myod1 (Abcam), mouse monoclonal anti-Cdc42 (SantaCruz Biotechnology), rabbit polyclonal anti-Rac-1 (SantaCruz Biotechnology), rabbit polyclonal anti-FAK (SantaCruz Biotechnology), and rabbit polyclonal anti-laminin (Sigma-Aldrich). The following secondary antibodies were used: anti-mouse igG Alexa Fluor 488, anti-rabbit igG Alexa Fluor 566, anti-rabbit igG Alexa Fluor 488, and anti-chicken igG Alexa Fluor 488. All secondary antibodies were purchased from Life Technologies. Actin cytoskeleton was localized using falloidin conjugated with TRITC (Sigma). Three independent experiments were performed for each analysis.
Western blotting
Proteins were isolated from cells or gastrocnemius muscles using cOmplete Lysis-M EDTA-free kit (Roche Applied Science). Twenty-five µg of total protein lysate were denatured by boiling in Laemmli buffer, separated using SDS-Page electrophoresis, and transferred to PVDF membranes (Roche Applied Science). The membranes were blocked with 5% Blotto (BioRad)/TBS for 1h and incubated with primary antibodies diluted 1:2000 in 5% Blotto (BioRad)/TBS, at 4°C, overnight, followed by secondary antibodies diluted 1:20000, at room temperature, for 2 h. Next, protein bands were visualized with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and exposed to chemiluminescence positive film (Amersham Hyperfilm ECL, GE Healthcare). The obtained results were analyzed with GelDoc2000 using Quantity One software (BioRad). The density of examined bands was compared to density of tubulin bands. The following primary antibodies were used: rabbit polyclonal anti-Cxcr4 (Abcam), rabbit polyclonal anti-Cxcr7 (Abcam), rabbit polyclonal anti-pFAK (Cell Signaling), rabbit polyclonal anti-FAK (SantaCruz Biotechnology), mouse monoclonal anti-Cdc42 (SantaCruz Biotechnology), rabbit polyclonal anti-Rac-1 (SantaCruz Biotechnology), mouse monoclonal anti-actin (Abcam), and mouse monoclonal anti-tubulin (Sigma-Aldrich). Secondary antibodies used were: peroxidase-conjugate rabbit anti-mouse igg (Sigma-Aldrich) and peroxidase-conjugate goat anti-rabbit igg (Sigma-Aldrich). Three independent experiments were performed.
Flow cytometry analysis
Gastrocnemius muscles that received ESCs constitutively expressing histone H2B-GFP were isolated at day 7 and 14 of regeneration. Next, they were digested with 0.15% pronase (Sigma Aldrich) in Ham's F12 medium (Life Technologies) buffered with 10 mM HEPES (Life TEchnolofies), containing 10% fetal calf serum (FCS), at 37°C, for 1.5 h. Obtained cell suspension was filtered through 40 µm cell strainer. Then, cells were fixed in a 3% PFA in PBS, washed with PBS, and analyzed with FACSCalibur (Becton-Dickinson) equipped with a 488-nm argon laser to detect GFP signal. The cells were also incubated with rabbit polyclonal anti-Myf5 antibody (Abcam) diluted in 3% BSA in PBS 1:100, at room temperature, for 1 h, followed by secondary antibody anti-rabbit igG Alexa Fluor 566. Three data parameters were acquired and stored: FSC, SSC and fluorescence 1 – FL1 (fluorescein isothiocyanate, FITC). CellQuest application, version 1.2, was used for the analysis. Three independent experiments were performed. Results were analyzed GraphPad Software and non-paired t-test was performed to compare treated with the control cells. The differences were considered statistically significant when p < 0.05 (marked on charts with asterisks).
Results
Sdf-1 induces stem cells migration but not proliferation
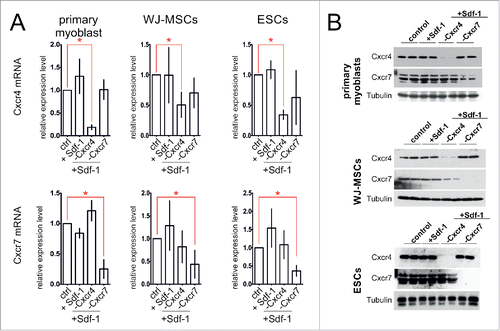
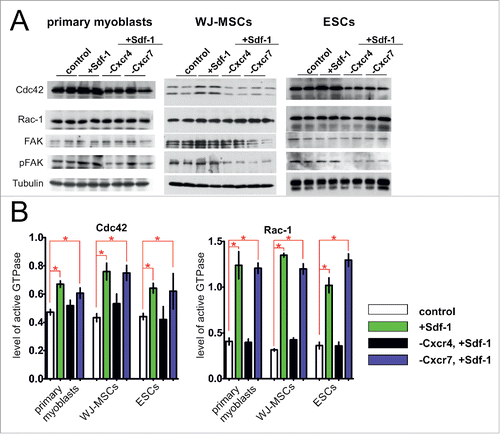
We analyzed primary myoblasts, WJ-MSCs, or ESCs which were treated with Sdf-1 alone or transfected with siRNA complementary to mRNAs encoding either Cxcr4 or Cxcr7 and treated with Sdf-1, along with control, i.e. untreated cells. The Cxcr4 or Cxcr7 silencing assessed at mRNA level was proved to be efficient. siRNA complementary to Cxcr4 mRNA decreased the level of this transcript to 28% +/− 8% in primary myoblasts, 49% +/− 16% in WJ-MSCs, and to 34% +/− 8% in ESCs, as compared to control, i.e., cells of each type that where neither treated with Sdf-1 nor transfected with siRNAs (). siRNA complementary to mRNA encoding Cxcr7 decreased the level of Cxcr7 transcripts to 34% +/− 11% in primary myoblasts, 38% +/− 18% in WJ-MSCs and 41 % +/− 13% in ESCs, as compared to control (). Sdf-1 treatment did not significantly change the level of Cxcr4 or Cxcr7 mRNA (). The changes in CXCR4 and CXCR7 level in treated cells were also pronounced at protein level ().
Figure 1. The Cxcr4 and Cxcr7 level in in vitro cultured primary myoblasts, WJ-MSCs, and ESCs 48h after transfection with siRNA and Sdf-1 treatment. (A) The level of mRNA encoding Cxcr4 and Cxcr7. Obtained data is presented as mean ± standard deviation. Student's non-paired t-test was used for statistical analyses. Asterisk marks significant differences (P < 0.05). (B) Western blotting of Cxcr4, Cxcr7, and tubulin.
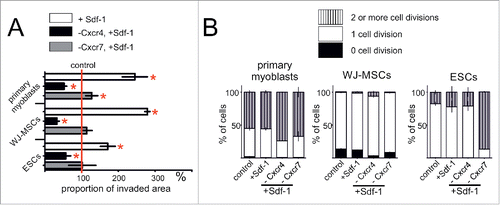
Scratch migration assay revealed that in the response to Sdf-1 gradient primary myoblasts, WJ-MSC and ESCs migrate more effectively (). Migration of all types of examined cells depended on Cxcr4 receptor - silencing of its expression decreased this process (). Cxcr7 silencing did not significantly impact at the cell migration in performed assay. Next, we tested whether Sdf-1 controls the ability of primary myoblasts, WJ-MSCs, and ESCs to proliferate. CFSE test allowed us to estimate the proportion of cells that did not divide, divided once or more than twice. It proved that Sdf-1 treatment did not change myoblasts, WJ-MSCs, as well as ESCs proliferation rate. Neither Cxcr4 nor Cxcr7 expression silencing affected primary myoblasts and WJ-MSCs divisions (). Interestingly, Cxcr7 silencing significantly increased ESCs proliferation ().
Figure 2. Sdf-1 impact at in vitro cultured primary myoblasts, WJ-MSCs, and ESCs migration and proliferation. (A) Proportion of invaded area calculated from the results of scratch wound assay. Analysis was performed at day second after the scratch wound formation. (B) Results of CFSE test documenting the proliferation rate of studied cells. Analysis was performed after 2 d of culture subsequently CFSE staining. Obtained data is presented as mean ± standard deviation. Student's non-paired t-test was used for statistical analyses. Asterisk marks significant differences (P < 0.05).
The signaling pathways in actin organization in stem cells after Sdf-1 treatment
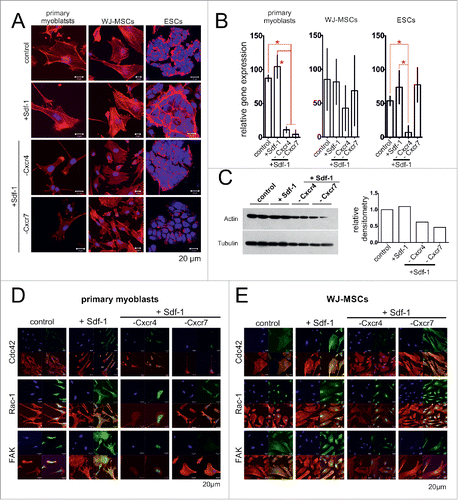
Next, we analyzed the changes in actin cytoskeleton organization and which signaling pathways were involved in the activation of the analyzed cells migration in the response to Sdf-1. We chose to analyze Cdc42 (cell division control protein 42), Rac-1 (Ras-Related C3 Botulinum Toxin Substrate 1), and focal adhesion kinase (FAK), i.e. the factors known to participate in the processes associated with cell migration such as actin polymerization and focal contacts formation.Citation40-42 After Sdf-1 stimulation the morphology of the cells and organization of actin cytoskeleton changed, i.e., all analyzed cells formed numerous stress fibers and filopodia (). In Sdf-1 treated cells the actin filaments were more abundant (). This effect was reversed by Cxcr4 silencing, what correlated with the decrease in actin expression at mRNA and protein levels (). The SCs and WJ-MSCs in that Cxcr4 expression was silenced were characterized by changes in Cdc42, Rac-1 and FAK localization ().
Figure 3. Sdf-1 impact at actin, FAK, Rac-1 and Cdc42 in in vitro cultured primary myoblasts, WJ-MSCs, and ESCs. (A) Immunolocalization of actin (red - actin, blue - chromatin). (B) The level of mRNA encoding actin. Obtained data is presented as mean ± standard deviation. Student's non-paired t-test was used for statistical analyses. Asterisk marks significant differences (P < 0.05). (C) Western blotting of actin in ESCs. (C) Localization of FAK, Rac-1 and Cdc42 in primary myoblasts (blue – chromatin, red – immunolocalization of actin, green – immunolocalization of studied proteins). (D) Localization of FAK, Rac-1 and Cdc42 in WJ-MSCs (blue – chromatin, red – immunolocalization of actin, green – immunolocalization of studied proteins).
However, Sdf-1 did not impact at the levels of Cdc42, Rac-1, and FAK proteins in primary myoblasts, WJ-MSCs, or ESCs (). Silencing of Cxcr4 or Cxcr7 expression slightly decreased the level of Cdc42 protein (). Importantly, Sdf-1 caused the significant changes in the activity of studied proteins (). The activity of Cdc42 and Rac-1 GTPases was higher in Sdf-1 treated myoblasts, WJ-MSCs, and also ESCs, as compared to the untreated cells (). Silencing of Cxcr4 but not Cxcr7 abolished the impact of Sdf-1 at the activity of Cdc42 and Rac-1 GTPases. As far as active, phosphorylated form of FAK (pFAK), is concerned it was detectable in control myoblasts, WJ-MSCs, and ESCs (). Sdf-1 caused increase in the level of pFAK in myoblasts and WJ-MSCs, but not in ESCs. However, in all studied cell types the effect of Sdf-1 on FAK phosphorylation was lost when expression of both its receptors, i.e. Cxcr4 and Cxcr7, was silenced ().
Figure 4. Sdf-1 impact at FAK, Rac-1 and Cdc42 level and activation in vitro cultured primary myoblasts, WJ-MSCs, and ESCs. (A) Western blotting of Cxcr4, Cxcr7, FAK, phosphorylated FAK (pFAK), Rac-1, Cdc42, and tubulin. (B) The activity of Rac-1 and Cdc42 in primary myoblasts, WJ-MSCs and ESCs. Obtained data is presented as mean ± standard deviation. Student's non-paired t-test was used for statistical analyses. Asterisk marks significant differences (P < 0.05).
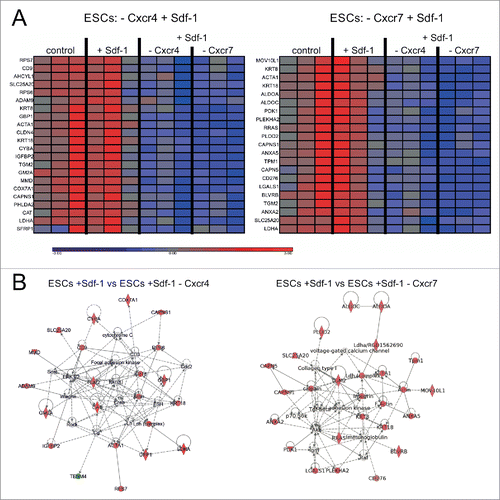
Changes in transcription profile in ESCs after Sdf-1 treatment
To analyze the changes in the transcriptome provoked by Sdf-1 we decided to use ESCs because in these cells the changes in morphology and cytoskeleton organization was the best pronounced after Sdf-1 treatment. mRNA isolated from control ESCs, as well as cells that were Sdf-1 treated, Sdf-1 treated and transfected with siRNA complementary to mRNAs encoding either Cxcr4 or Cxcr7, was analyzed using microarray technique (). Analysis of variance (ANOVA) allowed to create lists of genes significantly changed in ESCs that were Sdf-1 treated and transfected with appropriate siRNA, as compared to cells treated only with Sdf-1 (with the cutoff values: p-value< 0.05, −1.3>Fold Change>1.3). This analysis revealed that Sdf-1, acting via Cxcr4 receptor, regulates the expression of 90 transcripts, while acting via Cxcr7 receptor affects the expression of 113 transcripts (Fig. S1). Using Ingenuity Pathway Analysis we showed that Sdf-1 impacts at the expression of many genes encoding proteins engaged in cells adhesion and migration (), including transcripts encoding proteins engaged directly or indirectly in actin and adhesion proteins expression. Sdf-1 acting via Cxcr4, but not Cxcr7, regulates the expression of mRNA encoding adhesion proteins such as tetraspanin CD9 and ADAM9 (a disintegrin and metalloproteinase 9). Sdf-1 acting via Cxcr4 influences the expression of transcripts encoding cytoskeleton proteins present in skeletal muscle fibers, such as actin or α actin (ACTA1). Activation of this signaling pathway also induced the expression of calpain small subunit 1 (CAPNS1) that belongs to the family of calcium-dependent, non-lysosomal cysteine proteases. Sdf-1 acting via Cxcr7 impacted the levels of mRNAs encoding F-actin and ACTA1. It also reduced the expression of mRNAs encoding calpains, such as calpain 5 (CAPN 5) and CAPNS1. Summarizing, transcriptome analysis confirmed important role of Sdf-1 in the activation of migration and allowed us to pinpoint and distinguish the targets of pathways activated by Sdf-1 binding to Cxcr4 or Cxcr7.
Figure 5. Sdf-1 impact at global gene expression in in vitro cultured ESCs. (A) Transcription profile of genes in ESCs. Blue color indicates low and red color indicates high expression levels of mRNA transcripts. (B) Gene networks created by interposing the results onto database of Ingenuity containing information about the gene function with the use of Ingenuity Pathway Analysis tool.
Regeneration of injured skeletal muscles treated with Sdf-1 and stem cells
Next, we decided to analyze if Sdf-1 pretreatment of transplanted cells or Sdf-1 co-injection with transplanted cells could improve participation of stem cells in muscle regeneration. Again we also focused at the role of Cxcr4 and Cxcr7 in the migration of tested cells in regenerating muscle. Control, i.e., untreated primary myoblasts, WJ-MSCs, or ESCs, as wells as cells treated with Sdf-1 alone or treated with Sdf-1 and transfected with siRNA complementary to mRNAs encoding either Cxcr4 or Cxcr7 were transplanted to control muscles injected with 0.9% NaCl (saline) or muscles injected with Sdf-1 in 0.9% NaCl (). One muscle of each muscle pair was injected with saline, while another, i.e. contralateral, with Sdf-1. Importantly, Sdf-1 was injected at the opposite end of the muscle in the relation to the site of cells transplantation (approximately 1 cm distance). Sdf-1 was co-injected with transplanted cells.
First, we focused at the morphology of all groups of treated muscles, followed the localization of transplanted cells, as well as, the efficiency of their migration and muscle homing (). Primary myoblast were identified as β-galactosidase positive cells, human WJ-MSCs on the basis of human nuclear antigen immunolocalization, and ESCs on the basis of histone H2B-GFP fluorescence. The efficiency of the participation of transplanted cells in the muscle regeneration was assessed on the basis of the number of cells able to home injured muscle and/or to form new muscle fibers. Transplanted primary myoblasts formed new muscle fibers with the highest efficiency, as compared to other cells analyzed (). Control primary myoblasts transplanted to muscles injected with saline participated in the formation of 4.7% +/− 3% muscle fibers. In Sdf-1 treated muscles this proportion reached 8.2% +/− 3.5%. Finally, Sdf-1-treated myoblasts injected to Sdf-1-injected muscles participated in the formation of 12.1% +/− 5.5% fibers (). Silencing of Cxcr4 expression significantly decreased the number of myofibers formed with the participation of transplanted myoblasts (). Silencing of Cxcr7 expression did not significantly change the number of myofibers formed with the participation of transplanted myoblasts (). Thus, co-injection of myoblasts and Sdf-1 improved participation of myoblast in formation of new myofibers.
Figure 7. The localization of primary myoblasts and ESCs after transplantation to injured gastrocnemius muscle. (A) The localization of transplanted primary myoblasts expressing β-galactosidase in cross section of muscle at day 7 of regeneration (green - β-galactosidase, blue – chromatin, red – immunolocalization of laminin). (B) The proportion of muscle fibers formed with the participation of transplanted myoblasts in cross sections of muscle at day 7 of regeneration (n = 5). (C) The localization of transplanted ESCs expressing Green Fluoresent protein (GFP) in longitudinal section of muscle at day 7 of regeneration (green - GFP, blue – chromatin, red – immunolocalization of laminin). (D) The localization of mononucleated cells at muscle fiber isolated from skeletal muscle engrafted with ESCs expressing GFP analyzed at day 7 of regeneration (green – GFP, red – immunolocalization of GFP using anti-GFP antibody, blue – immunolocalization of Myod1, yellow - chromatin). (E) Proportion of ESCs expressing GFP in the population of mononucleated cells isolated from the muscle at day 7 and 14 of regeneration (n = 3). (F) Proportion of ESCs expressing Myf5 in the population of GFP expressing ESCs (n = 3). FACS analysis of results is presented as mean ± standard deviation. Student's non-paired t-test was used for statistical analyses. Asterisk marks significant differences (P < 0.05).
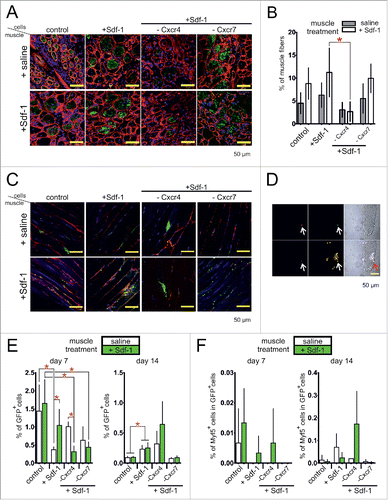
The WJ-MSCs transplanted into injured muscles, control or Sdf-1 injected, only very rarely were found within regenerating tissue. Only few of them were able to participate in formation of new muscle fibers (data not shown). On the other hand, ESCs were able to home regenerating tissue and were easily detectable between muscle fibers. However, these cells also only very rarely were found to participate in the formation of muscle fibers. In control, saline-injected muscles most of the control, untreated ESCs formed aggregates surrounded with muscle fiber basal lamina. Only single cells were localized along basal lamina. The localization of ESCs changed when muscles were injected with Sdf-1. Under such conditions ESCs were able to migrate and localize along muscle fiber basal lamina. In this case they very rarely formed aggregates and were mostly visible as a single cells (). Similar behavior characterized Sdf-1 treated ESCs transplanted either into control or Sdf-1 treated muscles. Silencing of Cxcr4 but not Cxcx7 expression led to the decrease of ESCs migration. As a result transplanted cells were localized mostly in aggregates. FACS analysis allowed us to verify the proportion of ESCs present within the muscle at days 7 and 14 of regeneration. Generally, in the population of mononucleated cells isolated from the regenerating muscle we were able to detect between 0.32% and 1.65% of ESCs at day 7 and only 0.08% – 0.64% of ESCs at day 14 (). ESCs were identified on the basis of histone H2B-GFP fluorescence. At day 7 of regeneration the proportion of ESCs detectable within the muscles injected with saline was 0.37%–1.44% and it reached 0.32%–1.65% in the muscles treated with Sdf-1. At day 14 of regeneration the proportion of ESCs was very low (less than 0.64%) and it was comparable between control and Sdf-1 treated muscles. Silencing of Cxcr4 or Cxcr7 expression did not decreased the proportion of ESCs present in regenerating muscles (). Regardless of their localization, ESCs cells very rarely expressed myogenic transcription factors, such as Myod1, as demonstrated by immunolocalization (). At day 7 of regeneration up to 0.01% of GFP positive cells i.e., ESCs isolated from the muscles expressed Myf5, as shown by FACS analysis (). At day 14 of regeneration the proportion of Myf5 expressing ESCs reached 0.17%. Sdf-1 did not change this proportion. Summarizing, Sdf-1 improved the ability of ESCs to migrate in injured muscle. However, these cells only very rarely initiated myogenic differentiation when transplanted into injured muscle.
Discussion
Our study shows that Sdf-1, acting via Cxcr4, increased primary myoblast, WJ-MSC, and ESCs ability to migrate in vitro. Except increasing the expression of CD9Citation24 Sdf-1 also impacts at the expression and activation of other proteins engaged in cell adhesion and migration. Sdf-1 treatment also resulted in the activation of FAK, i.e. non-receptor tyrosine kinase present in focal contacts composed of proteins anchoring integrins with actin cytoskeleton (reviewed in ref. Citation40). Sdf-1 dependent FAK activation could be achieved by stimulating both Cxcr4 and Cxcr7 receptors. Active FAK kinase is a key component of many signal transduction pathways (reviewed in ref. Citation43). However, from our point of view, the role of FAK in the activation of cell motility is the most important one (reviewed in ref. Citation44). Among such functions of FAK could be its positive impact at the MMP-2 and -9 (matrix metalloproteinases-2 and -9) expression and activity influencing extracellular matrix degradation during cells migration.Citation45,46 Next, FAK signaling controls the formation and turnover of focal contactsCitation46 and also activates Rho GTPases leading to actin stress fiber formation.Citation47 In our study, Sdf-1 treatment of cells led also to the activation of GTPases: Rac-1 and Cdc42 belonging to Rho GTPases family. Importantly, activation of these proteins depended only at Cxcr4. It was shown previously that Rac-1 mediates actin polymerization in lamellipodia at the front of migrating cells and Cdc42 induced actin polymerization in filopodia and invadopodia.Citation48
Our study also reveals the differences in cell signaling mediated by Sdf-1 - Cxcr4 and Cxcr7 pathways. Cxcr4 interacts with Sdf-1 but Cxcr7 except Sdf-1 also binds chemokine I-TAC (CXCL11) (reviewed in ref. Citation49). By silencing each of these receptors we were able to distinguish which one is involved in the regulation of certain genes. Thus, in ESCs expression of CD9 is regulated via Sdf-1 activating Cxcr4, but not Cxcr7, what was in agreement with our previous results documenting Sdf-1 dependent expression of CD9 in C2C12 myoblasts, bone marrow derived MSC, and ESCs.Citation24 Here we showed that also expression of ADAM9 is induced in ESCs by Sdf-1 in Cxcr4 dependent manner. The role of ADAM-9 in the cell migration was previously documented for keratinocytes and fibroblasts.Citation50,51 Analysis of keratinocytes showed that ADAM-9 regulates cells migration by interaction with integrin β1 and regulation of MMPs synthesis.Citation50 Thus, activation of FAK, Rac-1, and Cdc42, as well as induction of CD9 and ADAM-9 expression, underlay the ability of studied cells to migrate.
We show that Sdf-1, acting via Cxcr4, increased myoblast ability to migrate in vitro and participate in the formation of new muscle fibers in vivo when transplanted intramuscularly. The effect of Sdf-1 treatment was manifested better when myoblasts and Sdf-1 were co-injected than when myoblast were pre-treated with Sdf-1. On the other hand, it was shown that the pre-incubation of myoblasts with bFGF or Concanavalin A was shown to increase the efficiency of myoblasts transplantation.Citation52-54 The effect we observed was also similar to that documented for other cell types, such as mesoangioblasts, which pre-treatment with Sdf-1 or tumor necrosis factor α (TNF-α) enhanced their delivery and led to a complete reconstitution of skeletal muscles in mice that serve as a mouse model of severe muscular dystrophy.Citation55 In our hands the pre-treatment of ESCs with Sdf-1 or co-injection of ESCs and Sdf-1 into skeletal muscles increased their ability to migrate within the regenerating tissue. Previously, we showed that ESCs pretreatment improved the ability of ESCs to migrate and fuse with myoblasts in vitro.Citation24 Since, Sdf-1 increased the expression of CD9 in ESCs we postulated that it might facilitate the fusion.Citation24 Currently, we also documented that Sdf-1 promoted migration of ESCs within injured muscle and stimulated these cells to align in the manner characteristic for fusing myoblasts. Unfortunately, it did not affect the ESCs ability to initiate myogenic differentiation and fusion with myoblasts in vivo. However, it was previously shown that in order to induce myogenic differentiation of ESCs one has to either overexpress myogenic factors such as MyoD, Pax3 or Pax7 or treat the them with precisely designed cocktail of factors (reviewed in ref. Citation33). ESCs that were not subjected to such treatments fail to effectively differentiate and fuse with myoblasts most probably due to the fact that they do not initiate the expression of M-cadherin or vascular cell adhesion molecule (V-CAM1) that are also crucial for fusion.Citation25
MSCs isolated from Wharton jelly (WJ-MSCs), as well as adherent fraction of human umbilical cord blood cells, i.e., the cells that constitute the subpopulation enriched in MSCs, were shown by us to be able to follow myogenic program both in vitro and in vivo.Citation26,57 In our hands WJ-MSCs were able to colonize injured skeletal muscle and, with frequency of 5.3%, participate in the formation of new muscle fibers. Pre-treatment of WJ-MSCs with Sdf-1 did not impact their ability to form new muscle fibers but significantly increased muscle mass. Interestingly, in vitro these cells manifested myogenic potential and formed hybrid myotubes with C2C12 myoblasts.Citation26 Currently we documented that Sdf-1 treatment induced migration of WJ-MSC in vitro. However, this stimulation was not sufficient to improve their participation in the muscle reconstruction. Our result is in bright contrast to other study which showed that MSCs isolated from rat or human bone marrow participated in the formation of as many as 60–70% of new muscle fibers and restored expression of dystrophin in mdx mice muscles.Citation57 Thus, MSCs isolated form varied sources could differ in their myogenic potential.
Summarizing, Sdf-1 improved the ability of primary myoblasts and ESCs to migrate within the injured muscle. Moreover, injected intramuscularly Sdf-1 stimulated the transplanted primary myoblasts to participate in the formation of new muscle fibers. Mechanisms controlling cells migration activated by Sdf-1 rely at Cxcr4-dependent signaling pathways leading to the activation of proteins engaged in the focal contacts formation and actin polymerization, such as FAK, Rac-1, and Cdc42, as well as the expression of CD9 and ADAM-9. Sdf-1 - Cxcr7 interactions change the expression and activation of proteins engaged in cell migration, however, these changes does not result in the alternation of cell motility. Sdf-1 certainly improves migration of transplanted cells, however, fails to efficiently induce their myogenic differentiation. However, one has to remember that, as we shown previously, it greatly impacts at the homing of endogenous stem cells and by doing that improves muscle regeneration.Citation23,57
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_materials.zip
Download Zip (1.3 MB)Acknowledgments
Authors would like to acknowledge constant encouragement and support of Katarzyna Bocian, Karolina Archacka, Iwona Grabowska, Eugeniusz Krzysztof Machaj, Zygmunt Pojda, Jerzy Moraczewski, Maria Jolanta Rędowicz, and Malgorzata Zimowska.
Funding
This research was supported by Foundation for Polish Science (Parent Bridge Program co-financed by the European Union within European Regional Development Fund, grant number: POMOST/2011–4/3) and European Union project number: POKL.04.03.00-00-060/12.
References
- Relaix F, Zammit PS. Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development 2012; 139:2845-56; PMID:22833472; https://doi.org/10.1242/dev.069088
- Gibson MC, Schultz E. Age-related differences in absolute numbers of skeletal muscle satellite cells. Muscle Nerve 1983; 6:574-80; PMID:6646160; https://doi.org/10.1002/mus.880060807
- Barani AE, Durieux AC, Sabido O, Freyssenet D. Age-related changes in the mitotic and metabolic characteristics of muscle-derived cells. J Appl Physiol 2003; 95:2089-98; PMID:14555672; https://doi.org/10.1152/japplphysiol.00437.2003
- Kharraz Y, Guerra J, Pessina P, Serrano AL, Munoz-Canoves P. Understanding the Process of Fibrosis in Duchenne Muscular Dystrophy. BioMed Res Int 2014; 2014:965631; PMID:24877152; https://doi.org/10.1155/2014/965631
- Sirabella D, De Angelis L, Berghella L. Sources for skeletal muscle repair: from satellite cells to reprogramming. J Cachexia Sarcopenia Muscle 2013; 4:125-36; PMID:23314905; https://doi.org/10.1007/s13539-012-0098-y
- Bajek A, Porowinska D, Kloskowski T, Brzoska E, Ciemerych MA, Drewa T. Cell therapy in Duchenne Muscular Dystrophy treatment. Crit Rev Eukaryot Gene Expr 2015; 25:1-11; PMID:25955813; https://doi.org/10.1615/CritRevEukaryotGeneExpr.2015011074
- Briggs D, Morgan JE. Recent progress in satellite cell/myoblast engraftment – relevance for therapy. FEBS J 2013; 280:4281-93; PMID:23560812; https://doi.org/10.1111/febs.12273
- Partridge TA, Morgan JE, Coulton GR, Hoffman EP, Kunkel LM. Conversion of mdx myofibres from dystrophin-negative to -positive by injection of normal myoblasts. Nature 1989; 337:176-9; PMID:2643055; https://doi.org/10.1038/337176a0
- Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell 2008; 134:37-47; PMID:18614009; https://doi.org/10.1016/j.cell.2008.05.049
- Xu X, Wilschut KJ, Kouklis G, Tian H, Hesse R, Garland C, Sbitany H, Hansen S, Seth R, Knott PD, et al. Human satellite cell transplantation and regeneration from diverse skeletal muscles. Stem Cell Rep 2015; 5:419-34; PMID:26352798; https://doi.org/10.1016/j.stemcr.2015.07.016
- Costamagna D, Berardi E, Ceccarelli G, Sampaolesi M. Adult stem cells and skeletal muscle regeneration. Curr Gene Ther 2015; 15:348-63; PMID:26122100; https://doi.org/10.2174/1566523215666150630121024
- Gussoni E, Pavlath GK, Lanctot AM, Sharma KR, Miller RG, Steinman L, Blau HM. Normal dystrophin transcripts detected in Duchenne muscular dystrophy patients after myoblast transplantation. Nature 1992; 356:435-8; PMID:1557125; https://doi.org/10.1038/356435a0
- Huard J, Roy R, Bouchard JP, Malouin F, Richards CL, Tremblay JP. Human myoblast transplantation between immunohistocompatible donors and recipients produces immune reactions. Transplant Proc 1992; 24:3049-51; PMID:1466052
- Huard J, Bouchard JP, Roy R, Malouin F, Dansereau G, Labrecque C, Albert N, Richards CL, Lemieux B, Tremblay JP. Human myoblast transplantation: preliminary results of 4 cases. Muscle Nerve 1992; 15:550-60; PMID:1584246; https://doi.org/10.1002/mus.880150504
- Palmieri B, Tremblay JP, Daniele L. Past, present and future of myoblast transplantation in the treatment of Duchenne muscular dystrophy. Pediat Transplant 2010; 14:813-9; PMID:20963914; https://doi.org/10.1111/j.1399-3046.2010.01377.x
- Partridge T. The current status of myoblast transfer. Neurol Sci 2000; 21:S939-42; PMID:11382193; https://doi.org/10.1007/s100720070007
- El Fahime E, Torrente Y, Caron NJ, Bresolin MD, Tremblay JP. in vivo migration of transplanted myoblasts requires matrix metalloproteinase activity. Exp Cell Res 2000; 258:279-87; PMID:10896779; https://doi.org/10.1006/excr.2000.4962
- Quenneville SP, Chapdelaine P, Skuk D, Paradis M, Goulet M, Rousseau J, Xiao X, Garcia L, Tremblay JP. Autologous transplantation of muscle precursor cells modified with a lentivirus for muscular dystrophy: human cells and primate models. Mol Ther 2007; 15:431-8; PMID:17235323; https://doi.org/10.1038/sj.mt.6300047
- Skuk D, Goulet M, Tremblay JP. Use of repeating dispensers to increase the efficiency of the intramuscular myogenic cell injection procedure. Cell Transplant 2006; 15:659-63; PMID:17176617; https://doi.org/10.3727/000000006783981648
- Skuk D, Goulet M, Roy B, Tremblay JP. Efficacy of myoblast transplantation in nonhuman primates following simple intramuscular cell injections: toward defining strategies applicable to humans. Exp Neurol 2002; 175:112-26; PMID:12009764; https://doi.org/10.1006/exnr.2002.7899
- Skuk D, Goulet M, Tremblay JP. Transplanted myoblasts can migrate several millimeters to fuse with damaged myofibers in nonhuman primate skeletal muscle. J Neuropathol Exp Neurol 2011; 70:770-8; PMID:21865885; https://doi.org/10.1097/NEN.0b013e31822a6baa
- Lafreniere JF, Caron MC, Skuk D, Goulet M, Cheikh AR, Tremblay JP. Growth factor coinjection improves the migration potential of monkey myogenic precursors without affecting cell transplantation success. Cell Transplant 2009; 18:719-30; PMID:19523340; https://doi.org/10.3727/096368909X470900
- Brzoska E, Kowalewska M, Markowska-Zagrajek A, Kowalski K, Archacka K, Zimowska M, Grabowska I, Czerwinska AM, Czarnecka-Gora M, Streminska W, et al. Sdf-1 (CXCL12) improves skeletal muscle regeneration via the mobilisation of Cxcr4 and CD34 expressing cells. Biol Cell 2012; 104:722-37; PMID:22978573; https://doi.org/10.1111/boc.201200022
- Brzoska E, Kowalski K, Markowska-Zagrajek A, Kowalewska M, Archacki R, Plaskota I, Streminska W, Janczyk-Ilach K, Ciemerych MA. Sdf-1 (CXCL12) induces CD9 expression in stem cells engaged in muscle regeneration. Stem Cell Res Ther 2015; 6:46; PMID:25890097; https://doi.org/10.1186/s13287-015-0041-1
- Archacka K, Denkis A, Brzoska E, Swierczek B, Tarczyluk M, Janczyk-Ilach K, Ciemerych MA, Moraczewski J. Competence of in vitro cultured mouse embryonic stem cells for myogenic differentiation and fusion with myoblasts. Stem Cells Dev 2014; 23:2455-68; PMID:24940624; https://doi.org/10.1089/scd.2013.0582
- Grabowska I, Brzoska E, Gawrysiak A, Streminska W, Moraczewski J, Polanski Z, Hoser G, Kawiak J, Machaj EK, Pojda Z, et al. Restricted myogenic potential of mesenchymal stromal cells isolated from umbilical cord. Cell Transplant 2012; 21:1711-26; PMID:22525423; https://doi.org/10.3727/096368912X640493
- Farini A, Sitzia C, Erratico S, Meregalli M, Torrente Y. Clinical applications of mesenchymal stem cells in chronic diseases. Stem Cells Int 2014; 2014:306573; PMID:24876848; https://doi.org/10.1155/2014/306573
- Moroni L, Fornasari PM. Human mesenchymal stem cells: a bank perspective on the isolation, characterization and potential of alternative sources for the regeneration of musculoskeletal tissues. J Cell Physiol 2013; 228:680-7; PMID:22949310; https://doi.org/10.1002/jcp.24223
- Meyer S, Yarom R. Muscle regeneration and transplantation enhanced by bone marrow cells. Br J Exp Pathol 1983; 64:15-24; PMID:6340712
- Shi D, Reinecke H, Murry CE, Torok-Storb B. Myogenic fusion of human bone marrow stromal cells, but not hematopoietic cells. Blood 2004; 104:290-4; PMID:15010375; https://doi.org/10.1182/blood-2003-03-0688
- Leroux L, Descamps B, Tojais NF, Seguy B, Oses P, Moreau C, Daret D, Ivanovic Z, Boiron JM, Lamaziere JM, et al. Hypoxia preconditioned mesenchymal stem cells improve vascular and skeletal muscle fiber regeneration after ischemia through a Wnt4-dependent pathway. Mol Ther 2010; 18:1545-52; PMID:20551912; https://doi.org/10.1038/mt.2010.108
- Grabowska I, Archacka K, Czerwinska AM, Krupa M, Ciemerych MA. Mouse and human pluripotent stem cells and the means of their myogenic differentiation. Results Probl Cell Differ 2012; 55:321-56; PMID:22918815; https://doi.org/10.1007/978-3-642-30406-4_18
- Swierczek B, Ciemerych MA, Archacka K. From pluripotency to myogenesis: a multistep process in the dish. J Muscle Res Cell Motil 2015; PMID:26715014
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat Biotechnol 2015; 33:962-9; PMID:26237517; https://doi.org/10.1038/nbt.3297
- Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. in vitro Cell Dev Biol Anim 1995; 31:773-9; PMID:8564066
- Hadjantonakis AK, Macmaster S, Nagy A. Embryonic stem cells and mice expressing different GFP variants for multiple non-invasive reporter usage within a single animal. BMC Biotechnol 2002; 2:11; PMID:12079497; https://doi.org/10.1186/1472-6750-2-11
- Goetsch KP, Niesler CU. Optimization of the scratch assay for in vitro skeletal muscle wound healing analysis. Anal Biochem 2011; 411:158-60; PMID:21146491; https://doi.org/10.1016/j.ab.2010.12.012
- Eisenhart C. The assumptions underlying the analysis of variance. Biometrics 1947; 3:1-21; PMID:20240414; https://doi.org/10.2307/3001534
- Heiberger RM, Holland B. Statistical analysis and data display: an intermediate course with examples in R. Springer-Verlag New York; 2015.
- Graham ZA, Gallagher PM, Cardozo CP. Focal adhesion kinase and its role in skeletal muscle. J Muscle Res Cell Motil 2015; 36:305-15; PMID:26142360
- Lee JG, Heur M. Interleukin-1beta-induced Wnt5a enhances human corneal endothelial cell migration through regulation of Cdc42 and RhoA. Mol Cell Biol 2014; 34:3535-45; PMID:25022753; https://doi.org/10.1128/MCB.01572-13
- Sadok A, Marshall CJ. Rho GTPases: masters of cell migration. Small GTPases 2014; 5:e29710; PMID:24978113; https://doi.org/10.4161/sgtp.29710
- Guan JL. Role of focal adhesion kinase in integrin signaling. Int J Biochem Cell Biol 1997; 29:1085-96; PMID:9416004; https://doi.org/10.1016/S1357-2725(97)00051-4
- Schlaepfer DD, Mitra SK. Multiple connections link FAK to cell motility and invasion. Curr Opin Genet Dev 2004; 14:92-101; PMID:15108811; https://doi.org/10.1016/j.gde.2003.12.002
- Zhang Y, Thant AA, Hiraiwa Y, Naito Y, Sein TT, Sohara Y, Matsuda S, Hamaguchi M. A role for focal adhesion kinase in hyluronan-dependent MMP-2 secretion in a human small-cell lung carcinoma cell line, QG90. Biochem Biophys Res Commun 2002; 290:1123-7; PMID:11798192; https://doi.org/10.1006/bbrc.2001.6321
- Hsia DA, Mitra SK, Hauck CR, Streblow DN, Nelson JA, Ilic D, Huang S, Li E, Nemerow GR, Leng J, et al. Differential regulation of cell motility and invasion by FAK. J Cell Biol 2003; 160:753-67; PMID:12615911; https://doi.org/10.1083/jcb.200212114
- Schaller MD. Biochemical signals and biological responses elicited by the focal adhesion kinase. Biochim Biophys Acta 2001; 1540:1-21; PMID:11476890; https://doi.org/10.1016/S0167-4889(01)00123-9
- Ridley AJ. Rho GTPase signalling in cell migration. Curr Opin Cell Biol 2015; 36:103-12; PMID:26363959; https://doi.org/10.1016/j.ceb.2015.08.005
- Sanchez-Martin L, Sanchez-Mateos P, Cabanas C. CXCR7 impact on CXCL12 biology and disease. Trends Mol Med 2013; 19:12-22; PMID:23153575; https://doi.org/10.1016/j.molmed.2012.10.004
- Zigrino P, Steiger J, Fox JW, Loffek S, Schild A, Nischt R, Mauch C. Role of ADAM-9 disintegrin-cysteine-rich domains in human keratinocyte migration. J Biol Chem 2007; 282:30785-93; PMID:17704059; https://doi.org/10.1074/jbc.M701658200
- Nath D, Slocombe PM, Webster A, Stephens PE, Docherty AJ, Murphy G. Meltrin gamma(ADAM-9) mediates cellular adhesion through alpha(6)beta(1 )integrin, leading to a marked induction of fibroblast cell motility. J Cell Sci 2000; 113(Pt 12):2319-28; PMID:10825303
- Kinoshita I, Vilquin JT, Tremblay JP. Pretreatment of myoblast cultures with basic fibroblast growth factor increases the efficacy of their transplantation in mdx mice. Muscle Nerve 1995; 18:834-41; PMID:7630343; https://doi.org/10.1002/mus.880180806
- Kinoshita I, Vilquin JT, Roy R, Tremblay JP. Successive injections in mdx mice of myoblasts grown with bFGF. Neuromuscul Disord 1996; 6:187-93; PMID:8784807; https://doi.org/10.1016/0960-8966(96)00004-1
- Ito H, Hallauer PL, Hastings KE, Tremblay JP. Prior culture with concanavalin A increases intramuscular migration of transplanted myoblast. Muscle Nerve 1998; 21:291-7; PMID:9486857; https://doi.org/10.1002/(SICI)1097-4598(199803)21:3%3c291::AID-MUS2%3e3.0.CO;2-5
- Galvez BG, Sampaolesi M, Brunelli S, Covarello D, Gavina M, Rossi B, Constantin G, Torrente Y, Cossu G. Complete repair of dystrophic skeletal muscle by mesoangioblasts with enhanced migration ability. J Cell Biol 2006; 174:231-43; PMID:16831885; https://doi.org/10.1083/jcb.200512085
- Grabowska I, Streminska W, Janczyk-Ilach K, Machaj EK, Pojda Z, Hoser G, Kawiak J, Moraczewski J, Ciemerych MA, Brzoska E. Myogenic potential of mesenchymal stem cells - the case of adhesive fraction of human umbilical cord blood cells. Curr Stem Cell Res Ther 2013; 8:82-90; PMID:23270632; https://doi.org/10.2174/1574888X11308010010
- Dezawa M, Ishikawa H, Itokazu Y, Yoshihara T, Hoshino M, Takeda S, Ide C, Nabeshima Y. Bone marrow stromal cells generate muscle cells and repair muscle degeneration. Science 2005; 309:314-7; PMID:16002622; https://doi.org/10.1126/science.1110364
- Kowalski K, Archacki R, Archacka K, Stremińska W, Paciorek A, Gołąbek M, Ciemerych MA, Brzoska E. Stromal derived factor-1 and granulocyte-colony stimulating factor treatment improves regeneration of Pax7−/− mice skeletal muscles. J Cachexia Sarcopenia Muscle 2015; PMID:27239402