ABSTRACT
The early endosome protein Rab5 was recently shown to promote cell migration by enhancing focal adhesion disassembly through mechanisms that remain elusive. Focal adhesion disassembly is associated to proteolysis of talin, in a process that requires calpain2. Since calpain2 has been found at vesicles and endosomal compartments, we hypothesized that Rab5 stimulates calpain2 activity, leading to enhanced focal adhesion disassembly in migrating cells. We observed that calpain2 co-localizes with EEA1-positive early endosomes and co-immunoprecipitates with EEA1 and Rab5 in A549 lung carcinoma cells undergoing spreading, whereas Rab5 knock-down decreased the accumulation of calpain2 at early endosomal-enriched fractions. In addition, Rab5 silencing decreased calpain2 activity, as shown by cleavage of the fluorogenic substrate tBOC-LM-CMAC and the endogenous substrate talin. Accordingly, Rab5 promoted focal adhesion disassembly in a calpain2-dependent manner, as expression of GFP-Rab5 accelerated focal adhesion disassembly in nocodazole-synchronized cells, whereas pharmacological inhibition of calpain2 with N-acetyl-Leu-Leu-Met prevented both focal adhesion disassembly and cell migration induced by Rab5. In summary, these data uncover Rab5 as a novel regulator of calpain2 activity and focal adhesion proteolysis leading to cell migration.
Introduction
Cell migration is a complex and highly regulated process involving a series of signaling pathways that promote cytoskeletal rearrangements and remodeling of integrin-based adhesions, referred to as focal adhesions (FAs), in order to release the cell body from the anchors that link the cytoskeleton to the extracellular matrix.Citation1 FA turnover has been extensively studied and represents an enabling characteristic for cell migration. However, while several studies have addressed the mechanisms underlying the assembly of FAs,Citation2 little is known about the molecular players involved in FA disassembly. Particularly, it has been shown that FA disassembly is not simply the reversal of the assembly process, and that disruption of tension is produced by proteolytic cleavage of FA proteins by the intracellular calcium-dependent protease calpain2.Citation3 Calpain2 cleaves talin, a FA component required for the stability of integrin-actin connections, and this proteolytic event is required for the disassembly of FAs, because expression of a calpain2-resistant talin mutant impairs the rates of FA disassembly.Citation4 In addition, calpain2 displays extended proteolytic activity towards other FA proteins, such as paxillin, cortactin, and the focal adhesion kinase (FAK), although the role of these cleavage events in FA disassembly is still a matter of debate.Citation5-Citation7 Calpain2 is activated at different levels, including the requirement of millimolar-range concentrations of intracellular Ca2+ and membrane association, which make accessible the active site cleft.Citation8,9 Under basal non-stimulated conditions, calpain2 is generally found to be inactive throughout the cytosol, whereas stimulation, such as Ca2+ entry promotes re-localization of calpain2 to the cell periphery and plasma membrane by mechanisms that remain unclear.Citation10-Citation12 Recent studies suggest an association of calpain2 with microtubules and FA proteins,Citation13,Citation14 which suggests that microtubule-dependent trafficking of calpain2 is relevant to its localization to FAs. Intriguingly, calpain2 has been detected in vesicles and clathrin-coated pits,Citation15-Citation17 whereas calpain2 activity was also detected in multivesicular endosomes following serum stimulation.Citation18 The role of vesicular/endosomal calpain2 in FA disassembly and its control by endosomal regulators remains unknown.
We have previously shown that early endosomes containing the small GTPase Rab5 become re-distributed to the cell periphery and FAs during cell migration, and that these events involve the activation of Rab5.Citation19 Indeed, Rab5 activity is required for FA disassembly, tumor cell migration and invasion,Citation19 although the mechanisms underlying the role of Rab5 in promoting FA disassembly remain unclear. Here, we show that Rab5 increases the recruitment of calpain2 to endosome compartments, leading to augmented calpain2 activation, talin cleavage and FA disassembly in a calpain2-dependent manner.
Results
Calpain2 is recruited to early endosomes in a Rab5-dependent manner
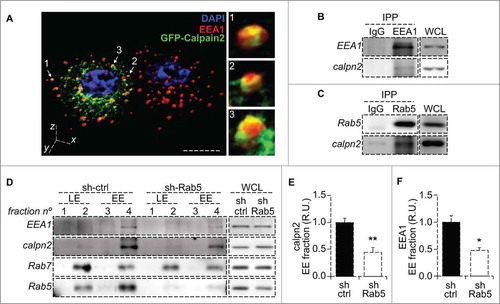
Different studies have shown an endosome/vesicular pool of calpain2 in resting cells.Citation15-Citation18 However, sorting of this protease into the different endosomal compartments and the consequences of such compartmentalization has not been described. Hence, we first evaluated the localization of calpain2 in A549 lung carcinoma cells undergoing spreading onto fibronectin matrix. Since endogenous calpain2 detected by immunofluorescence showed a diffuse and punctate pattern throughout the cytoplasm (data not shown, in disagreement with previous studiesCitation20), we expressed a GFP-tagged form of calpain2, previously shown to be functional,Citation4,Citation21 and evaluated its sub-cellular localization by confocal fluorescence microscopy. GFP-calpain2 showed a perinuclear distribution and partially co-localized with EEA1-positive early endosomes (). By z-stack 3D reconstruction we observed some calpain2 decorated early endosomes (, magnification panels), which suggested that endosomes could be a platform for mobilizing a membrane-bound fraction of calpain2, as previously suggested.Citation14,Citation15 To confirm this possibility, we immunoprecipitated early endosome markers and evaluated the putative association with calpain2. Calpain2 was found to co-immunoprecipitate with both early endosomal antigen 1 (EEA1) and Rab5, but it was not detected in IgG control precipitates (, ). These data suggest that calpain2 associates in a complex with early endosome markers, which might be accounting for its detection at early endosomes. Since Rab5 is a master regulator of early endosome dynamics and it was previously shown to promote FA disassemblyCitation19,Citation22 (an event that is known to require calpain2,Citation4), we next sought to evaluate the requirement of Rab5 in calpain2 recruitment to endosomal pools. To this end, endogenous Rab5 was knocked-down by shRNA, as previously describedCitation23 and calpain2 levels were assessed in endosome-enriched fractions, obtained by subcellular fractionation in discontinuous sucrose gradients, as previously described.Citation23 In control cells (treated with a scramble shRNA), calpain2 was readily detected at early endosomal, EEA1-enriched fractions, but not at late endosomal, Rab7-enriched fractions (). Intriguingly, upon Rab5 knock-down, calpain2 levels were decreased at the EEA1-enriched fractions, with no apparent changes at the Rab7-enriched fractions (). Importantly, no changes in total calpain2 levels were observed upon Rab5 knock-down (Supplementary Fig. 1), whereas a significant decrease was noted for both calpain2 and EEA1 in early endosome fractions, as measured by scanning densitometric analysis with their respective total protein levels (, ). Taken together, these data suggest that Rab5 is required for calpain2 localization at early endosomal pools, which might be relevant for its activation and function.
Figure 1. Calpain2 is recruited to endosomes in a Rab5 dependent manner. (A) Intracellular localization of calpain2 and the early endosomal network was assessed by three-dimensional reconstruction of confocal z-stacks in A549 lung cancer cells expressing GFP-Calpain2 (green) and stained for endogenous EEA1 with a polyclonal antibody (red). Calpain2-positive endosomes are indicated by arrows and magnifications are shown in the right panels. Bar represents 10 µm. (B, C) Immunoprecipitation of early endosome proteins EEA1 (B) and Rab5 (C). Immunoprecipitations were performed with rabbit polyclonal antibodies and endogenous calpain2 was detected by Western blotting. Whole cell lysates (WCL) are shown as reference. (D) Endosome fractionation and immunoblot detection of calpain2 and the endosomal markers Rab5, Rab7 and EEA1. A549 cells were treated with either control shRNA (sh-ctrl) or a Rab5-specific shRNA (sh-Rab5), as previously described,Citation19 whole cell lysates (WCL) were obtained and endosomes were enriched in discontinuous sucrose gradients, as described in the materials and methods. Fraction 2 contained the interphase between 8 and 30% sucrose (low density, Rab7-positive late endosomes, LE); fraction 4 contained the interphase between 30 and 35% sucrose (high density, EEA1-positive early endosomes, EE). Whole cell lysates (20 µg) are shown as reference. Images are representative from three independent experiments. Relative amount of calpain2 (E) and EEA1 (F) at the early endosome-enriched fraction was assessed by scanning densitometry of immunoblots and normalized to total calpain2 and EEA1 in the whole cell lysates, respectively. Data represent the average of three independent experiments (mean ± s.e.m.; *p < 0.05, **p < 0.01.
Rab5 is required for calpain2 activation
To evaluate the dependence of Rab5 on calpain2-mediated proteolysis, we used two different approaches, the first based on a synthetic fluorogenic substrate and the second based on the endogenous substrate talin.Citation4,Citation24 The fluorogenic substrate tBOC-LM-CMAC is a cell-permeable molecule that fluoresces upon cleavage of L-leucyl-L-methionine linker, which attaches a tBOC quencher.Citation24-Citation26 A549 cells were incubated with the substrate for different periods of time and then samples were fixed and mounted, to measuring the fluorescence intensity by microscopy. Accordingly, shRNA-control cells showed a time-dependent increase in fluorescence intensity. However, shRNA-Rab5 cells showed a delay in the kinetics of fluorescence appearance, which was particularly evident at 15 minutes, where shRNA-Rab5 cells reached near half the intensity observed in shRNA-control cells (, ). By linear correlation of scatter graphs, slopes were determined and showed a significant reduction in cumulative fluorescence production (), which suggests that calpain2 activity is decreased in Rab5-depleted cells. These observations were further confirmed in live cells by time-lapse video microscopy, as tBOC-LM-CMAC was readily cleaved in both shRNA-control and shRNA-Rab5 cells (), although shRNA-Rab5 cells showed delayed kinetics, as observed by the cumulative fluorescence signal () and the velocity of cleavage (). Importantly, these effects on calpain activity were recapitulated by using another shRNA sequence targeting endogenous Rab5 (Supplementary Fig. 1). Together, these observations confirm that Rab5 is important for calpain2 activity.
Figure 2. Rab5 is required for calpain2 activation. (A) Calpain2 activity in fixed cells. A549 cells treated with control shRNA (sh-ctrl) or a Rab5-specific shRNA (sh-Rab5) were grown on glass coverslips, serum-starved for 30 minutes and incubated with the calpain2 substrate tBOC-LM-CMAC for 0, 5, 10 or 15 min, and then fixed and mounted. Mean fluorescence intensity was measured in background-subtracted images. (B, C) Data obtained in (A) were analyzed by linear correlation (B), and the slopes corresponding to the velocity of substrate cleavage were calculated (C). Data represent the average of three independent experiments (mean ± s.e.m.; *p < 0.05). (D) Calpain2 activity in live cells. Fluorescence intensity of the cleaved substrate tBOC-LM-CMAC was evaluated in time-lapse experiments, by incubating A549 cells grown on glass-bottom dishes with the substrate and recorded directly in an epifluorescence microscope. Representative images obtained at 0 and 15 min time-frames are shown. (E, F) Data described in (D) were quantified and plotted as the background-subtracted intensities of fluorescence (E) (***p < 0.001; two-way ANOVA), while the slopes obtained at the initial time points corresponded to the velocity of substrate cleavage (F). Data represent the average of three independent experiments (mean ± s.e.m.; *p < 0.05). (G, H) Intracellular Ca2+ response. Intracellular Ca2+ levels were evaluated in time-lapse experiments, by incubating A549 cells grown in glass-bottom dishes with the cytosolic Ca2+ probe Fluo-4AM (5 μM). Ca2+ signal was recorded directly under an epifluorescence microscope. 30 seconds of baseline were recorded and then cells were stimulated with FBS 10% for 2 minutes. Relative fluorescence (Fluorescence difference between stimulated cells and baseline (F-F0) to baseline value (F0)) was calculated as a function of time. The time to reach the peak of Ca2+ signal (i.e. the time to reach the maximal response ((F-F0)/F0)) and the time of activation (i.e. the lapsed time for Ca2+ signal to increase from 10% to 90% of the maximal responseCitation28) were determined from three independent experiments (n.s. non significant).
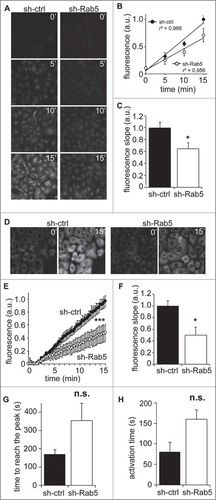
Calpain2 activation and membrane association depend on Ca2+ levels,Citation8,Citation10,Citation12,Citation27 raising the possibility that the effects of Rab5 knock-down are associated with altered Ca2+ fluctuations. To evaluate this possibility, we measured intracellular Ca2+ levels upon serum stimulation, using the fluorescent sensor Fluo-4. We found that Rab5-silenced cells tend to delay both, the serum-induced time to reach the maximal responses of Ca2+ signal (signal peak of Ca2+, ) and the serum-induced activation time (lapsed time for Ca2+ signal to increase from 10% to 90% of the maximal response,Citation28 ) when compared to control cells. Alternatively, we used as stimulus histamine (100 mM), a reported promoter of endoplasmic reticulum Ca2+ release and of extracellular Ca2+ entry.Citation29,Citation30 In these experiments, we did not observe any significant changes in the intracellular Ca2+ signal responses induced by histamine in sh-Rab5 silenced cells compared to control cells (Supplementary Fig. 1).
Rab5-dependent FA disassembly and cell migration require calpain2
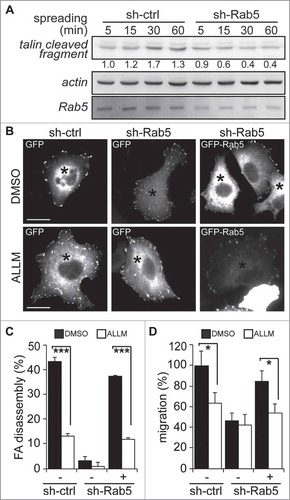
While recent studies have shown that Rab5 is required for FA disassembly and cell migration,Citation19,Citation22,Citation23 early studies showed that calpain2 mediates the proteolytic cleavage of FA proteins, leading to FA disassembly.Citation3-Citation5,Citation7,Citation17 Since Rab5 expression promotes calpain2 activity (), we sought to evaluate the role of Rab5 in the cleavage of talin, an endogenous substrate of calpain2.Citation17 To this end, A549 cells were allowed to spread onto fibronectin-coated plates, which is known to increase talin cleavage, leading to the accumulation of the processed N-terminal fragment of 47 kDa.Citation4 As anticipated, cell spreading increased the levels of talin cleaved fragment in control cells, whereas Rab5 knock-down decreased the accumulation of this fragment (). This result reinforces the idea of a requirement of Rab5 in calpain2 activity, in accordance with our fluorogenic substrate cleavage results (). Since cleavage of talin by calpain2 is relevant for FA disassembly,Citation4,Citation17 we next evaluated the requirement of calpain2 in Rab5-driven FA disassembly, by using Rab5-depleted cells (shRNA-Rab5) transfected with either GFP alone or GFP-Rab5. Calpain2 activity was inhibited with the selective inhibitor N-acetyl-Leu-Leu-Met (ALLM, 50 µM) and the rates of FA disassembly were measured in nocodazole-synchronized cells.Citation19,Citation31,Citation32 A549 cells were seeded onto coverslips, starved, and incubated with 10 µM nocodazole for 4 hours to disrupt the microtubule network and synchronize mature FAs, which upon removal of nocodazole disassembled in a synchronized manner.Citation31,Citation32 FAs, visualized by vinculin staining, were almost completely disassembled at 15 minutes after nocodazole removal in shRNA-control cells, but not shRNA-Rab5 cells, which is in agreement with previous studiesCitation19 (). Importantly, expression of GFP-tagged Rab5 recovered the disassembly capacity and this was found to be dependent on calpain2 activity, since ALLM abrogated these effects (, ). Since FA disassembly is relevant to cell migration and Rab5 was previously shown to be required for tumor cell migration, we evaluated the effects of inhibiting calpain2 in cells induced to migrate by the expression of GFP-Rab5. Rab5 driven cell migration was found to require calpain2 activity, because ALLM abolished Rab5-induced transmigration in Boyden chambers (). Taken together, these results indicate that Rab5-dependent FA disassembly and cell migration require calpain2.
Figure 3. Rab5-mediated focal adhesion disassembly and cell migration require calpain2. (A) A549 cells treated with either shRNA-control (sh-ctrl) or shRNA-Rab5 (sh-Rab5) were allowed to attach and spread on fibronectin-coated plates (2 µg/mL) and whole cell lysates were analyzed by Western blotting of talin cleaved fragment (47 kDa), Rab5 and actin. Representative images are shown and numbers below the panel correspond to the quantification of talin signals by scanning densitometry. Data were normalized to actin (loading control) and shown as the mean of three independent experiments. (B) A549 cells treated with shRNA-control or shRNA-Rab5 were transfected with either GFP (−) or GFP-Rab5 (+), grown on glass coverslips and serum starved overnight. Focal adhesions were synchronized by treatment with 10 µM nocodazole, and focal adhesion disassembly was assessed upon 15 min of nocodazole washout in the presence of either control vehicle (DMSO) or the calpain inhibitor ALLM (50 μM). Focal adhesions were visualized by vinculin staining and representative images obtained after nocodazole washout are shown. Asterisks indicate GFP-positive cells (whole composite images are provided as Supplementary Figure 2). Bar represents 10µm. (C) Focal adhesion disassembly was quantified as the difference between the total number of synchronized focal adhesions per cell (time point 0) and residual focal adhesion number after nocodazole washout. Representative images (B) and data (C) were obtain from three independent experiments (mean ± s.e.m.; ***p<0.001). Note that at least 100 cells were analyzed per condition. (D) A549 cells treated with shRNA-control (sh-ctrl) or shRNA-Rab5 (sh-Rab5) were transfected with either GFP (−) or GFP-Rab5 (+) and allowed to migrate for 120 min in Transwell chambers coated with 2 μg/mL fibronectin in the presence of either control vehicle (DMSO) or the calpain inhibitor ALLM (50 μM). Cells that migrated were visualized by crystal violet staining. Data represent the average from three independent experiments (mean ± s.e.m; *p < 0.05).
Discussion
This study reinforces the existence of a functional relationship between the endosomal compartment and cell migration,Citation19,Citation22, Citation23,Citation33 by showing that the endosomal protein Rab5 promotes FA disassembly through the activation of the cysteine protease calpain2. By using A549 lung carcinoma cells, which is a convenient model for the analysis of FA dynamics and migration in tumor cells,Citation34-Citation36 we found that a pool of calpain2 resides in early endosomes, as shown by confocal microscopy, immunoprecipitation and fractionation approaches. These data are in agreement with previous studies showing that calpain2 associates with endomembranous compartments, such as clathrin-coated vesicles and multivesicular endosomes.Citation15,Citation18 Our microscopy data allowed us to reconstruct early endosomes and visualize the distribution of GFP-calpain2, which was generally accumulated at the perinuclear region, however a fraction of this protease showed a punctate pattern towards the cell periphery and co-localizing with early endosomes. The latter suggests that these organelles may act as a platform to mobilize active pools of calpain2 to FAs committed to undergo disassembly. Three-dimensional reconstruction of confocal images showed the presence of GFP-calpain2 decorated early endosomes, mainly located near the cell periphery. Alternatively, immunoprecipitation and fractionation data showed that Rab5 is required for calpain2 localization at early endosome pools, which suggests a participation of Rab5 in selective targeting of calpain2 to endosomes. Interestingly, the decreased amount of EEA1 observed at early endosome fractions obtained from Rab5 knocked-down cells is in accordance with the accepted model where GTP-bound Rab5 recruits EEA1 to endosomal membranes.Citation37-Citation40 These observations are interesting because EEA1 is an effector protein of Rab5 and this raises the possibility that the recruitment of calpain2 to the early endosomes due to Rab5 might be an EEA1-dependent process, although additional studies are required to sustain this hypothesis. Our biochemical measurements showed that Rab5 knock-down was followed by a decreased rate of calpain activity (i.e. substrate cleavage), which correlated with the decreased amount of calpain2 recruited to early endosome fractions. Nevertheless, decreased calpain2 activity could also be due to altered Ca2+ fluctuations. Our data suggest that Rab5 does not regulates the total intracellular Ca2+ response, although they showed a tendency that might involve Rab5 in the time response of serum-induced Ca2+ signal. These trends might be explained by the possible contribution of Rab5/endosome system-dependent regulation on the plasma membrane residence of different populations of ion channels, and thus, modulating the dynamics of calpain2 activity and FAs dynamics. Accordingly, store-operated Ca2+ entry (SOCE) and Ca2+ channels have been reported as calpains and FAs dynamics regulators.Citation41-Citation43 Then, we cannot exclude the possibility that Rab5-dependent effects on calpain2 activation have a partial contribution based on the availability of these channels at the cell surface. Moreover, monovalent non-selective cationic channels (e.g. TRPM4) and K+ channels have been involved on the modulation of FAs dynamics.Citation44-Citation46 As such, changes on the availability and local activities of these channels might cause changes on the local membrane potential, subsequent Ca2+ signaling and subsequent FAs dynamics alterations.
Finally, pharmacological inhibition assays showed that calpain2 is required for Rab5-dependent FA disassembly and cell migration, establishing a causal relationship between calpain2 proteolytic function towards FA components and early endosomes through Rab5. Together, our results show for the first time a specific association between the protease responsible for adhesion detachment calpain2, and the early endosomal network, proposing a novel level of regulation of the focal adhesion proteolytic degradation at the endosomal level.
Materials and methods
Materials
Antibodies raised against Rab5 (sc-46692), Rab7 (sc-376362), EEA1 (sc-33585), HSP90 (sc-7947) and calpain2 (sc-373966) were from Santa Cruz Biotechnology (Santa Cruz, CA). Other antibodies included anti-vinculin (number V4505, Sigma-Aldrich), anti-actin (A5060, Sigma-Aldrich) and anti-talin (MAB1676, Merck-Millipore). Goat anti-rabbit and goat anti-mouse antibodies coupled to horseradish peroxidase (HRP) were from KPL. Alexa-Fluor-488 and Alexa-Fluor-568-conjugated secondary antibodies were from Invitrogen (Carlsbad, CA). Tissue culture medium, antibiotics, and fetal bovine serum (FBS) were from Corning Mediatech. The calpain2 substrate tBOC-LM-CMAC was from Thermofisher (Cat NºA6520) and the calpain2 inhibitor II (ALLM, 50 uM) was from Santa Cruz Biotechnology (Cat Nº: sc-201268).
Plasmids
The pEGFP-C1 plasmid encoding wild-type Rab5 was previously describedCitation19 and the pEGFP-C2 plasmid encoding calpain2 was kindly provided by Dr. Anna Huttenlocher (University of Wisconsin, Madison, USA).
Cell culture
A549 lung carcinoma cells were cultured in DMEM-high glucose, supplemented with 10% FBS and antibiotics. Down-regulation of endogenous Rab5A by different shRNA constructs was previously described,Citation19 (shRab5 B5 was the mainly sequence used in this work). A549 cells expressing a nonspecific shRNA sequence (plasmid 1864; Addgene) were used as experimental control.
Immunofluorescence
Cells were grown for 24 h on glass coverslips and following each treatment, samples were fixed with 4% formaldehyde in PBS for 15 minutes, then permeabilized with 0.1% triton X100 in PBS for 2 minutes, washed with PBS and blocked with 5% BSA/PBS. Samples were then incubated with primary antibodies for 1 h at 37ºC, washed 3 times (PBS, 5 minutes) and incubated with secondary antibodies for 1 h, washed 3 times and mounted. Samples were visualized by fluorescence microscopy, using either a Nikon Eclipse Ti epifluorecence microscope for focal adhesion analysis or a Nikon C2 plus confocal microscope for early endosome analysis.
Focal adhesion synchronization and disassembly
For focal adhesion synchronization experiments, cells were grown on glass coverslips, starved overnight in medium containing 1% serum and incubated with 10 µM nocodazole in serum-free medium for 4 h to depolymerize microtubules, as previously described.Citation32 Nocodazole was washed-out with serum-free medium and cells were incubated at 37ºC for the indicated periods of time. Then, cells were fixed and prepared for immunofluorescence staining. The analysis of focal adhesion disassembly was performed by manually counting the punctate and elongated structures, positive for vinculin. The number of focal adhesions was normalized with respect to the number of cells analyzed. Data were normalized with respect to the number of focal adhesions in control cells.
Co-localization analysis
Co-localization data was evaluated in z-stacks of images obtained by confocal microscopy (Nikon Eclipse Ti C2plus). Analysis was performed with the Nikon Imaging software. Stacks were used to reconstruct three-dimensional endosome network. Vesicles/endosomes which were positive for calpain2 were zoomed and modelled separately.
Endosome enrichment in sucrose gradients
Fractionation in discontinuous sucrose gradients was performed as previously described.Citation47 Briefly, cellular homogenates from 4 × 10-cm plates (7 × 106 cells per plate) were obtained by scrapping the cell layer with 500 µL of ice cold homogenization buffer containing 8% sucrose, 2 mM imidazole, pH 7.4 and protease inhibitors, and passed 8 times through a 22 1/2G needle in a 1 ml syringe, followed by centrifugation at 13,000xg during 5 min. Post-nuclear supernatants were transferred to 5 ml ultracentrifugation tubes and sucrose concentration was adjusted to 40%, followed by addition of 1.5 ml layers of 35%, 30% and 8% sucrose solutions, respectively. Samples were centrifuged at 125,000 x g for 66 min in a Sorvall WX plus ultracentrifuge, using a TH-660 rotor. Ten fractions were obtained, while the interphases between 8%-30% (fraction 2) and 30%-35% layers (fraction 4) corresponded to late and early endosome-enriched fractions, respectively.
Time-lapse microscopy
Time-lapse microscopy and live-cell imaging was performed using the Nikon Eclipse Ti inverted epifluorescence microscope with a heated and humidified sealed chamber connected to a 5% CO2 flux-meter. Cells were grown for 24 h on glass-bottom tissue culture plates (MatTek, Mattek Corporation), serum starved for 1 h and then pulsed with the fluorogenic substrate tBOC-LM-CMAC, followed by immediate recording. Pictures were captured for 15 min with intervals of 1 min. For calcium spike analysis, cells were serum-starved 30 minutes, followed by incubation with 5 µM of Fluo-4 in serum-free medium during 30 minutes, carefully washed and then placed in imaging chambers with Ca2+ free PBS. Samples were imaged during 30 seconds to establish a baseline, and then a PBS solution containing 10% FBS was added directly and imaged for additional 2 min. Alternatively, Fluo-4-loaded cells were placed in imaging chambers with Ca2+-containing media, imaged during 30 seconds to establish a baseline, and then a Ca2+-containig media suplemmented with histamine (100 nM) was added directly and imaged for additional 1.5 min. Fluctuating Fluo-4 intensities were measured in indicated regions of interest with the NIS software.
Immunoprecipitation
Immunoprecipitations were performed as previously described.Citation19 Briefly, cells were homogenized in 1% NP-40 buffer containing protease inhibitors, samples were centrifuged at 13,000 x g for 1 min at 4ºC and post-nuclear supernatants were precipitated for 30 min with polyclonal antibodies immobilized on protein A/G beads. Precipitated beads were solubilized in Laemmli buffer, boiled, separated by SDS-PAGE and analyzed by Western blotting.
Transwell migration assay
Assays were performed in fibronectin coated Boyden chambers (Transwell Costar, 6.5-mm diameter, 8 um pore size). Cells (50,000 cells per condition) were re-suspended in 100 µL of serum-free medium, plated on top of Transwell inserts and allowed to migrate towards lower chambers containing medium supplemented with 10% serum. After 2 h, inserts were removed, washed and cells that migrated to the bottom side of the inserts were stained with 0.1% Crystal Violet in 2% methanol and counted in an inverted microscope.
Statistical analysis
Where pertinent, results were compared using either unpaired t test with Welch's correction or two-way ANOVA, by using the GraphPad Prism 5 software (San Diego, CA). Values averaged from at least three independent experiments were compared. A p value < 0.05 was considered significant.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary Files
Download Zip (15.5 MB)Acknowledgments
We acknowledge Dr. Anna Huttenlocher for providing the pEGFP-C2 plasmid encoding for calpain2.
Funding
This work was supported by National Fund for Scientific and Technological Development (FONDECYT) #1140907 (to VAT), CONICYT-FONDAP #15130011 (to VAT), FONDECYT #1160518 (to O. C.), and FONDECYT #3170660 (to P. S.).
References
- Webb DJ, Parsons JT, Horwitz AF. Adhesion assembly, disassembly and turnover in migrating cells – over and over and over again. Nat Cell Biol. 2002;4:E97-100. doi:10.1038/ncb0402-e97. PMID:11944043
- Broussard JA, Webb DJ, Kaverina I. Asymmetric focal adhesion disassembly in motile cells. Curr Opin Cell Biol. 2008;20:85-90. doi:10.1016/j.ceb.2007.10.009. PMID:18083360
- Bhatt A, Kaverina I, Otey C, Huttenlocher A. Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J Cell Sci. 2002;115:3415-25. PMID:12154072
- Franco SJ, Rodgers MA, Perrin BJ, Han J, Bennin DA, Critchley DR, Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol. 2004;6:977-83. doi:10.1038/ncb1175. PMID:15448700
- Cortesio CL, Boateng LR, Piazza TM, Bennin DA, Huttenlocher A. Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J Biol Chem. 2011;286:9998-10006. doi:10.1074/jbc.M110.187294. PMID:21270128
- Perrin BJ, Amann KJ, Huttenlocher A. Proteolysis of cortactin by calpain regulates membrane protrusion during cell migration. Mol Biol Cell. 2006;17:239-50. doi:10.1091/mbc.E05-06-0488. PMID:16280362
- Chan KT, Bennin DA, Huttenlocher A. Regulation of adhesion dynamics by calpain-mediated proteolysis of focal adhesion kinase (FAK). J Biol Chem. 2010;285:11418-26. doi:10.1074/jbc.M109.090746. PMID:20150423
- Hanna RA, Campbell RL, Davies PL. Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature. 2008;456:409–12. doi:10.1038/nature07451. PMID:19020623
- Moldoveanu T, Gehring K, Green DR. Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains. Nature. 2008;456:404-8. doi:10.1038/nature07353. PMID:19020622
- Reverter D, Strobl S, Fernandez-Catalan C, Sorimachi H, Suzuki K, Bode W. Structural basis for possible calcium-induced activation mechanisms of calpains. Biol Chem. 2001;382:753-66. doi:10.1515/bchm.2001.382.5.753. PMID:11517928
- Storr SJ, Carragher NO, Frame MC, Parr T, Martin SG. The calpain system and cancer. Nat Rev Cancer. 2011;11:364-74. doi:10.1038/nrc3050. PMID:21508973
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731-801. doi:10.1152/physrev.00029.2002. PMID:12843408
- Hoshino D, Nagano M, Saitoh A, Koshikawa N, Suzuki T, Seiki M. The phosphoinositide-binding protein ZF21 regulates ECM degradation by invadopodia. PLoS One. 2013;8:e50825. doi:10.1371/journal.pone.0050825. PMID:23382803
- Nagano M, Hoshino D, Sakamoto T, Akizawa T, Koshikawa N, Seiki M. ZF21 is a new regulator of focal adhesion disassembly and a potential member of the spreading initiation center. Cell Adh Migr. 2011;5:23-8. doi:10.4161/cam.5.1.13492. PMID:20890123
- Sato K, Saito Y, Kawashima S. Identification and characterization of membrane-bound calpains in clathrin-coated vesicles from bovine brain. Eur J Biochem 1995;230:25-31. doi:10.1111/j.1432-1033.1995.tb20529.x. PMID:7601107
- Su W, Kowalczyk AP. The VE-cadherin cytoplasmic domain undergoes proteolytic processing during endocytosis. Mol Biol Cell. 2017;28:76-84. doi:10.1091/mbc.E16-09-0658. PMID:27798242
- Franco S, Perrin B, Huttenlocher A. Isoform specific function of calpain 2 in regulating membrane protrusion. Exp Cell Res. 2004;299:179-87. doi:10.1016/j.yexcr.2004.05.021. PMID:15302585
- Rintanen N, Karjalainen M, Alanko J, Paavolainen L, Maki A, Nissinen L, et al. Calpains promote alpha2beta1 integrin turnover in nonrecycling integrin pathway. Mol Biol Cell. 2012;23:448-63. doi:10.1091/mbc.E11-06-0548. PMID:22160595
- Mendoza P, Ortiz R, Diaz J, Quest AF, Leyton L, Stupack D, Torres VA. Rab5 activation promotes focal adhesion disassembly, migration and invasiveness in tumor cells. J Cell Sci. 2013;126:3835-47. doi:10.1242/jcs.119727. PMID:23813952
- Leloup L, Shao H, Bae YH, Deasy B, Stolz D, Roy P, Wells A. m-Calpain activation is regulated by its membrane localization and by its binding to phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2010;285:33549-66. doi:10.1074/jbc.M110.123604. PMID:20729206
- Nuzzi PA, Senetar MA, Huttenlocher A. Asymmetric localization of calpain 2 during neutrophil chemotaxis. Mol Biol Cell. 2007;18:795-805. doi:10.1091/mbc.E06-09-0876. PMID:17192410
- Palamidessi A, Frittoli E, Ducano N, Offenhauser N, Sigismund S, Kajiho H, Parazzoli D, Oldani A, Gobbi M, Serini G, et al. The GTPase-activating protein RN-tre controls focal adhesion turnover and cell migration. Curr Biol. 2013;23:2355-64. doi:10.1016/j.cub.2013.09.060. PMID:24239119
- Torres VA, Mielgo A, Barbero S, Hsiao R, Wilkins JA, Stupack DG. Rab5 mediates caspase-8-promoted cell motility and metastasis. Mol Biol Cell. 2010;21:369-76. doi:10.1091/mbc.E09-09-0769. PMID:19923319
- Rosser BG, Powers SP, Gores GJ. Calpain activity increases in hepatocytes following addition of ATP. Demonstration by a novel fluorescent approach. J Biol Chem 1993;268:23593-600. PMID:8226886
- Niapour M, Berger S. Flow cytometric measurement of calpain activity in living cells. Cytometry A. 2007;71:475-85. doi:10.1002/cyto.a.20399. PMID:17458879
- Farr C, Berger S. Measuring calpain activity in fixed and living cells by flow cytometry. J Vis Exp. 2010. doi:10.3791/2050. PMID:20644512
- Friedrich P. The intriguing Ca2+ requirement of calpain activation. Biochem Biophys Res Commun. 2004;323:1131-3. doi:10.1016/j.bbrc.2004.08.194. PMID:15451413
- Mackay L, Mikolajewicz N, Komarova SV, Khadra A. Systematic Characterization of Dynamic Parameters of Intracellular Calcium Signals. Front Physiol. 2016;7:525. doi:10.3389/fphys.2016.00525. PMID:27891096
- Lee KC, Chang HT, Chou KJ, Tang KY, Wang JL, Lo YK, Huang JK, Chen WC, Su W, Law YP, et al. Mechanism underlying histamine-induced intracellular Ca2+ movement in PC3 human prostate cancer cells. Pharmacol Res. 2001;44:547-52. doi:10.1006/phrs.2001.0891. PMID:11735364
- Zhou MH, Zheng H, Si H, Jin Y, Peng JM, He L, Zhou Y, Muñoz-Garay C, Zawieja DC, Kuo L, et al. Stromal interaction molecule 1 (STIM1) and Orai1 mediate histamine-evoked calcium entry and nuclear factor of activated T-cells (NFAT) signaling in human umbilical vein endothelial cells. J Biol Chem. 2014;289:29446-56. doi:10.1074/jbc.M114.578492. PMID:25190815
- Kaverina I, Krylyshkina O, Small JV. Microtubule targeting of substrate contacts promotes their relaxation and dissociation. J Cell Biol 1999;146:1033–44. doi:10.1083/jcb.146.5.1033. PMID:10477757
- Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581-90. doi:10.1038/ncb1262. PMID:15895076
- Chao WT, Kunz J. Focal adhesion disassembly requires clathrin-dependent endocytosis of integrins. FEBS Lett. 2009;583:1337-43. doi:10.1016/j.febslet.2009.03.037. PMID:19306879
- Liu G, Meng X, Jin Y, Bai J, Zhao Y, Cui X, Chen F, Fu S. Inhibitory role of focal adhesion kinase on anoikis in the lung cancer cell A549. Cell Biol Int. 2008;32:663-70. doi:10.1016/j.cellbi.2008.01.292. PMID:18343694
- Kogan TV, Jadoun J, Mittelman L, Hirschberg K, Osherov N. Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J Infect Dis. 2004;189:1965-73. doi:10.1086/420850. PMID:15143461
- Beinke C, Van Beuningen D, Cordes N. Ionizing radiation modules of the expression and tyrosine phosphorylation of the focal adhesion-associated proteins focal adhesion kinase (FAK) and its substrates p130cas and paxillin in A549 human lung carcinoma cells in vitro. Int J Radiat Biol. 2003;79:721-31. doi:10.1080/09553000310001610231. PMID:14703945
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494-8. doi:10.1038/28879. PMID:9697774
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999;397:621-5. doi:10.1038/17618. PMID:10050856
- Christoforidis S, Zerial M. Purification of EEA1 from bovine brain cytosol using Rab5 affinity chromatography and activity assays. Methods Enzymol. 2001;329:120-32. doi:10.1016/S0076-6879(01)29073-9. PMID:11210528
- Murray DH, Jahnel M, Lauer J, Avellaneda MJ, Brouilly N, Cezanne A, Morales-Navarrete H, Perini ED, Ferguson C, Lupas AN, et al. An endosomal tether undergoes an entropic collapse to bring vesicles together. Nature. 2016;537:107-11. doi:10.1038/nature19326. PMID:27556945
- Jacquemet G, Baghirov H, Georgiadou M, Sihto H, Peuhu E, Cettour-Janet P, He T, Perälä M, Kronqvist P, Joensuu H, et al. L-type calcium channels regulate filopodia stability and cancer cell invasion downstream of integrin signalling. Nat Commun. 2016;7:13297. doi:10.1038/ncomms13297. PMID:27910855
- Fiorio Pla A, Gkika D. Emerging role of TRP channels in cell migration: from tumor vascularization to metastasis. Front Physiol. 2013;4:311. doi:10.3389/fphys.2013.00311. PMID:24204345
- Schwab A, Stock C. Ion channels and transporters in tumour cell migration and invasion. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130102. doi:10.1098/rstb.2013.0102. PMID:24493750
- Caceres M, Ortiz L, Recabarren T, Romero A, Colombo A, Leiva-Salcedo E, Varela D, Rivas J, Silva I, Morales D, et al. TRPM4 Is a Novel Component of the Adhesome Required for Focal Adhesion Disassembly, Migration and Contractility. PLoS One. 2015;10:e0130540. doi:10.1371/journal.pone.0130540. PMID:26110647
- Pardo LA, Stuhmer W. The roles of K(+) channels in cancer. Nat Rev Cancer. 2014;14:39-48. doi:10.1038/nrc3635. PMID:24336491
- Huang X, Jan LY. Targeting potassium channels in cancer. J Cell Biol. 2014;206:151-62. doi:10.1083/jcb.201404136. PMID:25049269
- Torres VA, Mielgo A, Barila D, Anderson DH, Stupack D. Caspase 8 promotes peripheral localization and activation of Rab5. J Biol Chem. 2008;283:36280-9. doi:10.1074/jbc.M805878200. PMID:18974049
