ABSTRACT
Our study investigated the role of WTAP in colon cancer. We employed experiments including m6A dot blot hybridization, methylated RNA immunoprecipitation, dual-luciferase, and RNA immunoprecipitation to investigate the regulatory mechanism of WTAP. Western blot was performed to analyze the expression of WTAP, FLNA and autophagy-related proteins in cells. Our results confirmed the up-regulation of WTAP in colon cancer and its promoting effect on proliferation and inhibiting effect on apoptosis. FLNA was the downstream gene of WTAP and WTAP-regulated m6A modification led to post-transcriptional repression of FLNA. The rescue experiments showed that WTAP/FLNA could inhibit autophagy. WTAP-mediated m6A modification was confirmed to be crucial in colon cancer development, providing new insights into colon cancer therapy.
KEYWORDS:
Introduction
The number of new cases of colon cancer ranks the second highest among females and the third highest among males worldwide [Citation1], characterized by high morbidity and high mortality [Citation2]. Currently, the commonly used diagnostic and treatment methods include endoscopic inspection, surgical treatment, molecular targeted therapy, radiotherapy and chemotherapy [Citation3]. However, the early diagnosis rate and treatment rate for colon cancer are low now. Hence, there is an urgent need to promote better accesses to colon cancer treatment.
As an important mRNA dynamic modification, N6-methyladenosine (m6A), by regulating mRNA translocation, translation, and stability, takes an essential part in various biological processes [Citation4]. Methyltransferases catalyze the formation of m6A. Methyltransferase like 3 (METTL3) and methyltransferase like 4 (METTL4) form a core methyltransferase-catalyzed complex that is stabilized by WT1 associated protein (WTAP). Dynamic and reversible m6A modulation are associated with multiple physiological and pathological processes, such as stem cell differentiation, adipogenesis, and sperm development [Citation5–8]. Recent years have witnessed increasing evidence of the significant part that m6A modification takes in cancer development [Citation9–13]. For example, METTL3 promotes tumor progression in colon cancer with a m6A-IGF2BP2-dependent mechanism [Citation14]. Wang et al [Citation15]. found that METTL3-mediated m6A modification of HDGF mRNA facilitated gastric cancer development. WTAP-mediated m6A modification propels hepatocellular carcinoma (HCC) progression via the HuR-ETS1-p21/p27 axis [Citation16]. In conclusion, m6A modification has a crucial part in tumor development. However, the part that m6A modification takes in colon cancer remains an unanswered question given the limited studies. Therefore, our primary focus is figuring out the m6A modification mechanism when diving into the regulatory role of m6A modification in colon cancer.
Filamin A (FLNA), also called actin-binding protein 280 (ABP 280), is a sizable actin-binding protein that stabilizes the actin network [Citation17]. FLNA has been found identified as a biomarker in various cancers like HCC, prostate cancer and breast cancer [Citation18–20]. Recent studies have shown the downregulated FLNA expression in colorectal and gastric cancers [Citation21,Citation22]. Sun et al [Citation23]. discovered that FLNA, by regulating Matrix Metallopeptidase 9 (MMP-9) expression, hampered the migratory and invasive abilities of prostate cancer cells. Wang et al [Citation24]. also found that in bladder cancer, overexpressed FLNA showed a suppressive role in the malignant behaviors of tumor cells but a promotive role in facilitating autophagy. These findings suggest a close connection between FLNA with human cancer progression, indicating the potential of FLNA as a therapeutic target in treating cancer. However, the part of FLNA in colon cancer has not been researched enough. We found by bioinformatics analysis that FLNA expression had a close connection with m6A methylation. Hence, studying the molecular mechanism of FLNA is also our focus through which we can dive deeper into its effect on the colon cancer development.
The significance of autophagy in cancer development has been demonstrated intensively by recent studies [Citation25,Citation26]. Autophagy exerts an integral role in cellular growth and metabolic processes as a significant cellular component of degradation and recycling mechanism [Citation27]. Autophagy can either be a promoter (help cancer cells develop stress resistance to promote cell survival) or a suppressor (hinder the growth of cancer) in the cancer development during intracellular or extracellular stress, depending on the type, stage, grade, genetic background, and cellular microenvironment of the tumor [Citation25]. For example, miR-30d, by inhibiting autophagy and promoting apoptosis, is able to inhibit colon cancer cell proliferation [Citation28]. Zheng et al [Citation29]. found that miR-142-3p, by targeting tumor protein p53 inducible nuclear protein 2 (TP53INP2), impeded the autophagy and facilitated the progression of colon cancer. The findings mentioned above suggest the essential role of autophagy in cancer development. Additionally, FLNA, by interacting with sequestosome 1 (SQSTM1), has a part in the autophagic process of DNA repair [Citation30]. Whether FLNA has a regulatory effect on colon cancer remains an unanswered question. Hence, we aimed to explore the regulatory effect of FLNA on autophagy in colon cancer tumor cells.
Therefore, based on the above findings, we aim to explore the potential role and possible mechanism of WTAP in colon cancer with a view of finding a promising strategy for the treatment of colon cancer.
Materials and methods
Bioinformatics
The expression data of colon cancer protein (Normal: 100, Tumor: 95) were downloaded from Clinical Proteomic Tumor Analysis Consortium (CPTAC). The mRNA expression data of colon cancer (Normal: 41, Tumor: 480) were downloaded from The Cancer Genome Atlas (TCGA). The t-test was employed on the protein for the difference between normal and tumor groups (P < 0.05) to obtain differentially expressed proteins (DEproteins). The mRNA was examined by the edgeR package for the difference between normal and tumor groups (|logFC|>2, FDR<0.05) to obtain differentially expressed mRNAs. The target m6A enzyme of the study was determined by literature citation [Citation31]. Potential proteins of the target m6A enzyme were predicted using the m6A2 Target database (http://m6a2target.canceromics.org/#/home), and target proteins were identified by the analysis of Pearson correlation.
Cell culture and transfection
Human colon cancer cells HCT116 (BNCC337692), SW620 (BNCC337664), LoVo (BNCC338601), and human normal colon epithelial cells HCoEpiC (BNCC340030) were acquired from BeNa Culture Collection (China). RPMI-1640 medium with 10% fetal bovine serum (FBS, Gibco, USA) was employed for the maintaining of all the cells above. Culture conditions were all maintained in a 37°C incubator containing 5% CO2.
The purchased si-WTAP, si-FLNA, and corresponding negative controls (si-NC, Thermo Fisher Scientific, USA) were transfected into colon cancer cells as per the instructions of the Lipofectamine 2000 kit (Thermo Fisher Scientific, USA).
Cell viability assay
The CCK-8 kit (Beyotime, China) was utilized for the detection of cell viability [Citation16]. 2 × 103 cells/well transfected colorectal cancer cells were seeded in 96-well plates (Corning, USA) for pre-culture. 10 μL of CCK-8 detection reagent was added after 0 h, 24 h, 48 h, 72 h, and 96 h of cell culture. Absorbance was detected at a wavelength of 450 nm with a microplate reader, and three experimental biological replicates were performed for each set of experiments. The formula was: cell survival rate = [(experimental well – blank well)/(control well – blank well)] × 100%.
Colony formation assay
To investigate the proliferative ability of colorectal cancer cells after transfection, colony formation experiments were done. The transfection groups were si-NC, si-WTAP, si-NC+si-NC, si-WTAP+si-NC, si-NC+si-FLNA, and si-WTAP+si-FLNA. Colon cancer cells were plated with 6-well plates (2 × 103 cells/well) and cultured in a culture medium for two weeks. Then, colonies were fixed using 4% paraformaldehyde (PFA) as previously described [Citation32] and darkly stained by 1% crystal violet.
Dual-luciferase assay
The 3’ UTR of FLNA was inserted into the pGL3 vector (Promega, USA). The putative m6A site base (A) was mutated to that in the 3’ UTR (C) by a site-directed mutagenesis kit (Thermo Fisher Scientific, USA). The WT and MUT plasmids, si-NC and si-WTAP were subcloned into colon cancer cells. Next, the luciferase activity was measured using the Dual-Luciferase Assay Kit (Promega, USA) [Citation16].
Western blot
The specific steps of Western blot were performed as described previously [Citation33]. First, the radioimmunoprecipitation assay buffer (RIPA) was used to obtain proteins, followed by protein concentration assay using bicinchoninic acid (BCA) assay kit (Beyotime, China). Next, proteins were then separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). The separated proteins were transferred to polyvinylidene (PVDF, Bio-Rad, USA) membrane. Then, 5% nonfat milk powder (Beyotime, China) was used to enclose the membrane at room temperature for 1 h. After blocking, the primary antibodies were introduced to incubate overnight at 4°C. After incubation, TBST (Beyotime, China) was used to wash the membrane, which was then incubated at room temperature with secondary antibody for 4 h. Finally, ECL hypersensitive chemiluminescence kit was used for protein color development, and pictures were taken with the visualizer. The primary antibodies were sourced from Abcam (UK): rabbit anti-human WTAP (ab195380), FLNA (ab76289), LC3I/II (LC3B, ab51520), β-actin (ab8227), p62 (ab109012). The secondary antibody was goat anti-rabbit IgG (ab6721), which was purchased from Abcam (UK).
qRT-PCR
Total RNA isolation was completed withTRIzol (Thermo Fisher Scientific, USA). For qPCR, cDNA was synthesized using the First Strand cDNA Synthesis Kit (Takara, Japan). Then, mRNA expression was analyzed using SYBR GREEN (Bio-Rad, USA). Finally, 7500 FAST instrument (Thermo Fisher Scientific, USA) was employed for computer detection. All primer sequences are in .
Table 1. Primer set for qRT-PCR.
RNA m6A dot blot assay and RNA m6A quantification
Total RNA was isolated from colon cancer cells with TRIzol (Thermo Fisher Scientific, USA), and then RNA mass was tested using NanoDrop3000. M6A content was measured by the EpiQuik m6A RNA Methylation Quantification Kit (Epigentek, USA).
The poly (A) RNA (600, 300, and 150 ng) dots were subjected to RNA m6A dot hybridization analysis on nylon membranes (GE Healthcare, USA) and then probed with m6A antibody (Millipore, USA) overnight at 4°C. Then, the membranes were combined with secondary antibody (ab97051, Abcam, UK). Color rendering was performed with ECL chemiluminescence kit. The exposure was then completed using an imaging system (Bio-Rad, USA) [Citation31]. Finally, they were dyed with methylene blue and photographed.
MeRIP-qRT-PCR
Total RNA was extracted from colon cancer cells with TRIzol. DNA-free fragment RNA was probed with the anti-m6A antibody (Abcam, UK) bound to magnetic Dynabeads to concentrate mRNA with m6A. Then, the beads were processed with proteinase K, and RNA was isolated for qRT-PCR validation [Citation31]. Primers are shown in .
Immunofluorescence
This study performed immunofluorescence experiments using the method described previously [Citation34]. Cell slides were made first, then fixed with 4% paraformaldehyde, and then permeated with 0.5% Triton X-100 (prepared with PBS) at room temperature for 20 min. Then goat serum was introduced and kept at room temperature for 30 min. Next, rabbit anti-human LC3 antibody (ab51520, Abcam, UK) was added and incubated overnight at 4°C. On the second day, fluorescent secondary antibody (ab150077, Abcam, UK) was added and incubated at 37°C for 1 h away from light. Then, DAPI was added and incubated for 5 min away from light. Finally, the slides were sealed with the sealing solution containing anti-fluorescence quench agent, and the images were observed and collected under a fluorescence microscope.
Flow cytometry
The cells were collected and cell suspension was digested by trypsin (Beyotime, China). The single-cell suspension was then collected in a 15 mL centrifuge tube for centrifugation (2000 rpm/5 min). The cells were washed with PBS, and resuspended with propidium iodide (PI; 40%, sigma, USA) and RNase A. Next, the treated cells were incubated for 30 min at 37°C. Finally, apoptotic cells in 5 mL centrifuge tubes were analyzed using a BD FACS Celesta flow cytometer (BD Biosciences) using Annexin V-FITC/PI Apoptosis Detection Kit (Yeasen Bio, China) staining. FITC had a maximum excitation wavelength of 488 nm and a maximum emission wavelength of 525 nm, while PI-DNA complex had a maximum excitation wavelength of 535 nm and a maximum emission wavelength of 615 nm.
Statistical analysis
All the assays were run three times independently and the data in this study were expressed as mean ± standard deviation. Analysis of variance test was used to test the significance of differences among multiple groups. The Student’s t-test was used to test the significance of differences between two groups. In this paper, P < 0.05 was defined as significant difference.
Results
WTAP displays a high expression in colon cancer tissues and cells
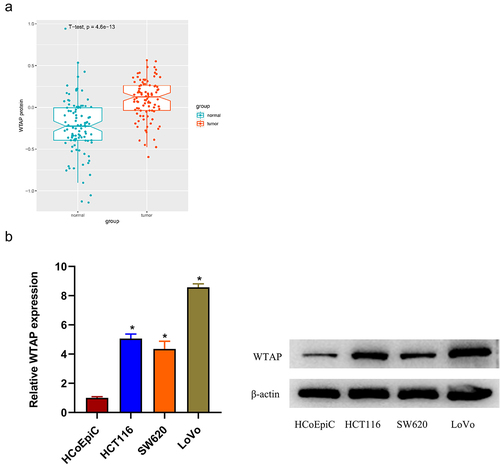
The colon cancer protein expression data were downloaded from CPTAC, DEproteins were obtained by differential analysis using t-test, and the research object m6A enzyme was determined by literature citation. T-test analysis demonstrated a high expression of WTAP protein in colon cancer tissues (). Based on previous finding [Citation35], we assumed that WTAP exerted a promotive role in colon cancer progression. To validate it, qRT-PCR and western blot was firstly employed to detect the expression status of WTAP in colon cancer cell lines and human normal colon epithelial cells. The result illustrated a remarkably upregulated expression of WTAP in colon cancer cell lines compared with that in normal colon epithelial cells, with relatively higher expression in HCT116 and LoVo cell lines. These two cell lines were selected for subsequent functional experiments (). The results all pointed that WTAP was markedly upregulated in colon cancer tissues and cells.
Figure 1. WTAP is highly expressed in colon cancer tissues and cells. a: The expression of WTAP protein in colon cancer tissues; b: The expression of WTAP in colon cancer cell lines (HCT116, SW620, and LoVo) and human normal colon epithelial cells (HCoEpiC) was detected by qRT-PCR and western blot; * indicates P < 0.05.
Silencing WTAP significantly inhibits the proliferation of colon cancer cells and promotes cell apoptosis
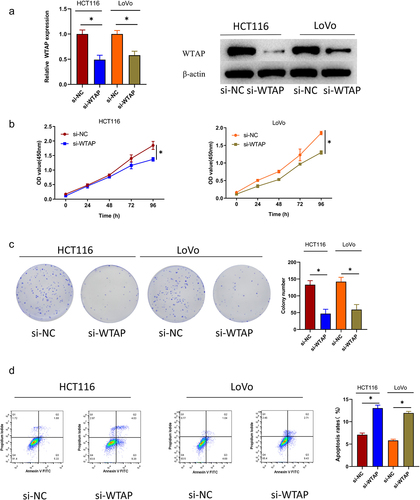
To dive deeper into how WTAP exactly works in colon cancer progression, we transfected si-NC and si-WTAP into HCT116 and LoVo cells, respectively, and examined the transfection efficiency of si-WTAP by qRT-PCR and western blot. The results demonstrated that the expression level of WTAP in the si-WTAP group was considerably lowered compared with that in the control group (). The CCK-8 assay suggested that silencing WTAP attenuated the cell viability of colon cancer compared to that in the control group (). The colony formation assay illustrated that silencing WTAP inhibited the cell proliferation of colon cancer cells (). The flow cytometry assay showed that silencing WTAP could promote apoptosis of colon cancer cells (). In summary, WTAP acted as a pro-oncogene in colon cancer progression. Silencing WTAP could inhibit colon cancer cell proliferation and promote apoptosis.
Figure 2. Silencing WTAP significantly inhibits the proliferation of colon cancer cells and promotes apoptosis. a: Expression of WTAP after transfected was assessed by qRT-PCR and western blot; b: Cell viability after transfected was measured by CCK-8 assay; c: Cell proliferation after transfected was tested by colony formation assay; d: Cell apoptosis after transfected was tested by flow cytometry; * indicates P < 0.05.
FLNA is a potential target of WTAP
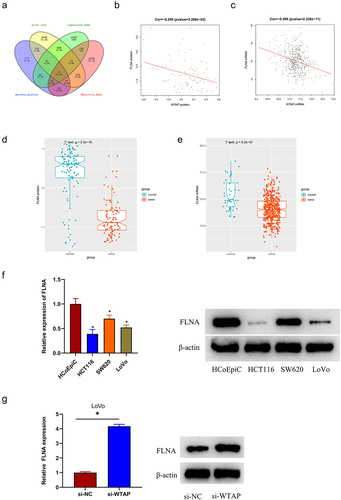
To figure out the mechanism underlying the regulatory effect of WTAP in colon cancer progression, the m6A2 Target database was utilized to predict target proteins and took an intersection with down-regulated DEproteins to obtain 9 differentially potential target proteins (). Pearson correlation analysis revealed that WTAP protein had a negative correlation with FLNA protein (), and WTAP mRNA was also negatively correlated with FLNA mRNA (). Next, FLNA was analyzed by t-test to determine that its protein and mRNA levels were lowly expressed in tumor tissues (). The qRT-PCR and western blot was employed for the analysis on FLNA expression in colon cancer cell lines and human normal colon epithelial cells. The results demonstrated that the FLNA expression was remarkably down-regulated in colon cancer cell lines with a contrast to that in normal epithelial cells (). Finally, the relationship between WTAP and FLNA expression in LoVo cells was assessed. This assessment illustrated that FLNA expression was considerably up-regulated in the cells of the si-WTAP group with a contrast to that in the control group, and it was further confirmed that WTAP was negatively correlated with FLNA expression (). These findings suggested that FLNA was a potential target of WTAP, and FLNA was lowly expressed in colon cancer tissues and cells. LoVo cell line was used in subsequent assays due to the highest expression of WTAP in colon cancer cell line.
Figure 3. FLNA is a potential target of WTAP. a: Venn plot of the target protein predicted by m6A2 Target database and down-regulated DEproteins; b: Correlation analysis of WTAP protein and FLNA protein; c: Correlation analysis of WTAP mRNA and FLNA mRNA; d: Expression of FLNA protein in colon cancer tissues; e: Expression of FLNA mRNA in colon cancer tissues; f: qRT-PCR and western blot was used to detect the expression of FLNA in colon cancer cell lines (HCT116, SW620, and LoVo) and human normal colonic epithelial cells (HcoEpiC); g: qRT-PCR and western blot were used to detect the expression of FLNA in si-WTAP-treated LoVo cells; * indicates P < 0.05.
WTAP regulates FLNA expression by m6A
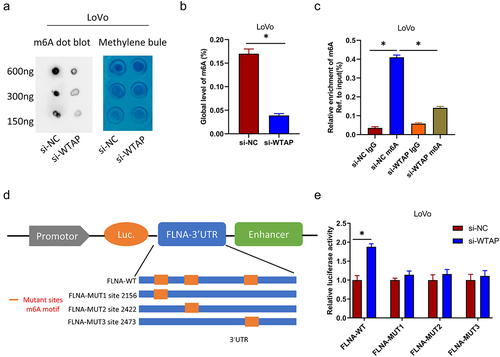
The analysis of m6A2 Target data suggested that WTAP may regulate FLNA expression depending on m6A. To determine whether WTAP can mediate m6A modification of FLNA, we initially examined total levels of m6A in the negative control and stable WTAP knockdown groups by two different methods (m6A dot blotting and RNA methylation quantification). The results demonstrated that in LoVo cells, the m6A level was considerably reduced in the WTAP knockdown group compared with that in the control group (). The enrichment of m6A in FLNA examined by MeRIP-qPCR, and the results revealed that the m6A-specific antibody efficiently enriched FLNA in LoVo cells compared with the IgG control. In contrast, the enriched FLNA in LoVo cells was considerably reduced in the WTAP knockdown group (). On the basis of these findings, we considered that WTAP had a regulatory effect on FLNA m6A levels. The m6A RNA sequence results illustrated that the m6A modification was located at the 3′ UTR of FLNA, sequence-based RNA adenosine methylation site predictor (SRAMP) (http://www.cuilab.cn/sramp), and three m6A sites with the highest confidence were predicted at the 3’ UTR of FLNA (Table S1, Figure S1). To further demonstrate the targeting of FLNA by WTAP, the FLNA 3’ UTR with these three m6A sites was cloned into the pGL3 vector (). Dual-luciferase demonstrated that the luciferase activity of FLNA-WT in LoVo cells was enormously elevated in response to WTAP silencing, whereas silencing WTAP could not affect luciferase activity of FLNA-Mut (). All those findings suggested that WTAP suppressed FLNA expression by modulating the m6A modification of FLNA at the 3’ UTR.
Figure 4. WTAP regulates FLNA expression by m6A. a: m6A dot blot analysis of m6A levels of isolated poly (a) + RNAs in total RNA from WTAP-knockdown LoVo cells. Corresponding RNA was loaded equally at 2-fold serial dilutions of 600 ng, 300 ng, and 150 ng, and methylene blue staining was used as loading control; b: RNA methylation quantitative analysis was used to detect the total content of m6A; c: MeRIP analysis and qRT-PCR were used to assess the m6A modification of FLNA in WTAP-silenced LoVo cells. The enrichment of m6A in each group was calculated by m6A IP/input and IgG IP/input. d: Wild-type or m6A site mutant FLNA was cloned in pGL3; e: Dual-luciferase reporter assay showed targeting of WTAP to the 3’ UTR of FLNA; * indicates P < 0.05.
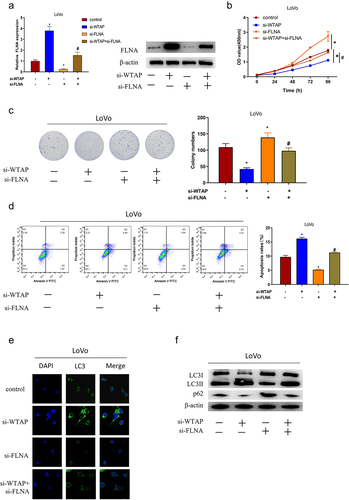
FLNA, a tumor suppressor, reverses the effect of WTAP in colon cancer
We then looked deeper into whether WTAP had a promotive role in colon cancer by regulating FLNA expression. Silencing WTAP greatly increased the expression of FLNA, and silencing FLNA considerably attenuated FLNA expression compared with the control group, while silencing FLNA and WTAP restored the expression of FLNA in LoVo cells (). CCK-8 result demonstrated that cell viability was enormously hampered by silencing WTAP in LoVo cells with a contrast to that in the control group, while silencing FLNA could reverse the effect of WTAP gene silencing on LoVo cell viability (). Similarly, the result of the colony formation assay showed that silencing WTAP significantly reduced the proliferation ability of LoVo cells, while silencing FLNA could reverse the suppressive effect of WTAP gene silencing on LoVo cell proliferation (). Flow cytometry result suggested that cell apoptosis was significantly promoted by silencing WTAP in LoVo cells compared with that in the control group, while silencing FLNA could reverse the effect of WTAP gene silencing on LoVo cell apoptosis (). Since it was previously found that FLNA could promote tumor cell autophagy in bladder cancer and hindered the growth of tumor cells [Citation24], we speculated that in colon cancer, the WTAP/FLNA axis could have an effect on colon cancer development by regulating the autophagic process. Subsequently, the accumulation of autophagy marker (LC3) was examined by immunofluorescence, which illustrated that silencing WTAP led to a substantial increase in the accumulation of LC3 in LoVo cells with a contrast to that in the control group; silencing FLNA decreased the LC3 accumulation in LoVo cells. And concurrent silencing of FLNA and WTAP restored the LC3 accumulation in LoVo cells (). Additionally, we examined autophagy-related protein expression in LoVo cells. It was showed that silencing WTAP considerably elevated the LC3II protein expression and attenuated the p62 protein expression in LoVo cells with a contrast to that in the control group. Silencing FLNA significantly reduced the expression of LC3II protein and significantly increased the expression of p62 protein in LoVo cells, and silencing FLNA was able to reverse the effect that silencing WTAP took on autophagy in LoVo cells (). In conclusion, WTAP promoted colon cancer cell proliferation, inhibited apoptosis and autophagy through FLNA.
Figure 5. FLNA is involved in WTAP-mediated colon cancer progression as a tumor suppressor. a: qRT-PCR and western blot were used to detect the expression of FLNA in LoVo cells of each group; b: CCK-8 was used to detect cell viability in each group; c: Colony formation assay was used to detect cell proliferation in each group; d: Flow cytometry was used to detect cell apoptosis in each group; e: Immunofluorescence was used to detect LC3 aggregation in each group; f: Western blot was used to detect the expression of autophagy-related proteins (LC3 I, LC3 II, p62) in each group; the specific experimental groups were: control group (si-NC+si-NC), si-WTAP group (si-WTAP+si-NC), si-FLNA group (si-NC+si-FLNA), and si-WTAP+si-FLNA group; * VS si-NC+si-NC group, # VS si-WTAP+si-NC group. */ # indicates P < 0.05.
Discussion
Many studies have mentioned the involvement of m6A in multiple human diseases, including cancer [Citation9]. However, what kind of the role of m6A in colon cancer remains unanswered. Our research is centered on the underlying mechanism of WTAP and its mediated m6A modification in colon cancer progression. We found an upregulation of WTAP in colon cancer tissues and cells. Functionally, WTAP promoted colon cancer cell proliferation. Mechanistically, FLNA was verified to be a downstream target gene of WTAP, regulated by WTAP via m6A modification at the 3’ UTR. Final studies revealed that WTAP promoted colon cancer cell proliferation. inhibited apoptosis and autophagy by regulating FLNA expression through m6A modification.
In fact, WTAP has been mentioned in many studies to be an oncogene in various cancers, including cholangiocarcinoma, osteosarcoma, and liver cancer [Citation16,Citation31,Citation36]. The upregulation of WTAP was also found in colon cancer tissues and cells. Yu et al [Citation37]. found a high expression level of WTAP in ovarian cancer cells and the silencing WTAP could hinder the growth of cancer cells, indicating the potential of WTAP to be a prognostic marker for high-grade serous ovarian cancer. Our results verified the above findings. We found that the WTAP expression was higher in HCT116 and LoVo, but low in SW620 cells. The reason for this result may be that the differences in gene expression profiles and phenotypes of cell lines led to different expression levels of WTAP in different cell lines [Citation38]. Meanwhile, we discovered that silencing WTAP was able to substantially impede the proliferation of colon cancer cells. In addition, the close association between WTAP and a functional m6A methylation complex has been mentioned in recent studies [Citation39]. And the critical role of WTAP as an m6A regulator has also been covered in some studies. For instance, Chen et al [Citation31]. found that WTAP inhibited the expression of HMBOX1 in an m6A-dependent manner, thus promoting tumorigenesis in osteosarcoma. Our findings argued that in colon cancer, WTAP was up-regulated and played a key role in m6A modification. Our findings also elucidated that FLNA was a downstream target gene of WTAP-mediated m6A modification, and WTAP inhibited FLNA expression through m6A modification.
FLNA is an actin filament cross-linking protein that has a part in cell-cell contact and adherens junctions during blood, brain and heart organs [Citation40,Citation41]. FLNA plays both cancer-promoting and cancer-inhibiting roles in tumors which is considered an oncoprotein in metastatic melanoma, lung cancer, and HCC [Citation42–44]. However, many studies have also found that FLNA can inhibit tumor progression. For example, Xu et al [Citation45]. reported that FLNA showed a decreased expression in breast cancer tissues, which had a negative correlation with lymph node metastasis. Their findings suggest that silencing FLNA facilitates migration and invasion of breast cancer cells. Meanwhile, there are also related studies in colorectal cancer research. However, the research in colorectal cancer only focused on the inhibiting role of FLNA on the migration and invasion abilities of colorectal cancer cells, and the contents related to apoptosis and autophagy of colorectal cancer cells were not involved. In our results, FLNA also displayed a downregulated expression in colon cancer cells, and silencing FLNA could facilitate colon cancer cell proliferation. It was also found that WTAP promoted colon cancer cell proliferation, inhibited apoptosis and autophagy through FLNA. These findings suggest that the role FLNA plays in human malignancies remains controversial, possibly due to the heterogeneity of FLNA in different cancer species [Citation46].
Autophagy has an environment-dependent role in cancer, and interventions that activate and suppress autophagy can be taken as approaches for cancer treatment [Citation47]. Current methods to inhibit autophagy in the clinical practice have focused on the inhibition of lysosome using chloroquine (CQ) or related hydroxychloroquine (HCQ). And inhibitors that inhibit other autophagy regulators such as VPS3412-14, ULK115 and 16 have been discovered to hinder tumor cell growth or facilitate apoptosis in vitro and preclinical mouse models [Citation47,Citation48]. Apoptosis and autophagy often occur in the same cells and autophagy develops prior to apoptosis. The involvement of multiple signaling pathways and regulators contributes to a complex relationship between the apoptosis and autophagy. And whether autophagy can induce or inhibit apoptosis is bound up with cell type and the duration of the stimulus [Citation49]. Recently, many articles have mentioned that autophagy has regulatory part in the progression and chemosensitivity of various malignancies such as lung cancer, colon cancer, and liver cancer [Citation50–52]. Wei et al [Citation53]. found that miR-126 inhibited the viability of colon cancer cells by inhibiting mTOR-induced apoptosis and autophagy. Lu et al [Citation54]. found that CD24 affected sorafenib resistance by activating autophagy in HCC. FLNA silencing was found to inhibit autophagy in colon cancer cells, which is consistent with Wang et al [Citation24]. Further investigations revealed that WTAP inhibited autophagy in colon cancer cells by inhibiting FLNA through m6A.
In summary, our study has discovered a high-level expression of WTAP in colon cancer tumor tissues and cells, and its potential downstream target is FLNA, down-regulated in tumor tissues and cells. WTAP remarkably facilitates the proliferation and inhibit apoptosis of colon cancer, and hinders autophagy through m6A-dependent regulation of FLNA mRNA stability. Admittedly, there are limitations in our study, we only investigated the regulation of FLNA by WTAP at the cellular level, but did not validate it at the animal and clinical levels. Additionally, FLNA downstream regulatory mechanisms are not intensely mined, which is a top priority for our future research. In a word, we identified WTAP/FLNA as a promising therapeutic target for colon cancer.
Authors’ contributions
L H contributed to conceptualization and data curation. S Z conducted the literature search. XG G contributed to methodology and formal analysis. JF S and XJ X contributed to investigation and validation. WW H contributed to writing. WF Y contributed to visualization. All authors have reviewed and approved the final manuscript.
Declaration of conflicting interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
Not applicable.
Supplemental Material
Download Zip (55.5 KB)Acknowledgments
Not applicable.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19336918.2023.2180196
Additional information
Funding
References
- Shahi F, Gorji M, Payandeh M, et al. Post-Marketing Surveillance of a generic Oxaliplatin (Alvoxal(Ⓡ)) in Iranian Patients with Cancer. Curr Ther Res Clin Exp. 2022;96(100657):100657.
- Qiao H, Liu L, Chen J, et al. The functions of N6-methyladenosine (m6A) RNA modifications in colorectal cancer. Med Oncol. 2022;39(235). DOI:10.1007/s12032-022-01827-4
- Lordick F, Hacker U, Hoffmeister A, et al. [What is confirmed in the treatment of colon cancer?]. Inn Med. 2022;63(1250–1256):1250–1256.
- Paris J, Morgan M, Campos J, et al. Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25(1):137–148 e136.
- Wang CX, Cui G-S, Liu X, et al. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16(e2004880):e2004880.
- Dorn LE, Lasman L, Chen J, et al. The N 6 -Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation. 2019;139(533–545):533–545.
- Li Z, Qian P, Shao W, et al. Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res. 2018;28(9):904–917. DOI:10.1038/s41422-018-0072-0
- Lin Z, Tong M. m(6)A mRNA modification regulates mammalian spermatogenesis. Biochim Biophys Acta Gene Regul Mech. 2019;1862:403–411.
- Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18(103). DOI:10.1186/s12943-019-1033-z
- Vu LP, Cheng Y, Kharas MG. The Biology of m(6)A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer Discov. 2019;9(25–33):25–33.
- Li Y, Xiao J, Bai J, et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol Cancer. 2019;18(137). DOI:10.1186/s12943-019-1066-3
- Lang F, Singh RK, Pei Y, et al. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 2019;15(e1007796):e1007796.
- Panneerdoss S, Eedunuri VK, Yadav P, et al. Cross-talk among writers, readers, and erasers of m 6 A regulates cancer growth and progression. Sci Adv. 2018;4(eaar8263). DOI:10.1126/sciadv.aar8263
- Li T, Hu P-S, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6) A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(112). DOI:10.1186/s12943-019-1038-7
- Wang Q, Chen C, Ding Q, et al. METTL3-mediated m 6 A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(1193–1205):1193–1205.
- Chen Y, Peng C, Chen J, et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol Cancer. 2019;18(127). DOI:10.1186/s12943-019-1053-8
- Stossel TP, Condeelis J, Cooley L, et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol. 2001;2(138–145):138–145.
- Ai J, Huang H, Lv X, et al. FLNA and PGK1 are two potential markers for progression in hepatocellular carcinoma. Cell Physiol Biochem. 2011;27(207–216):207–216.
- Lin JF, Xu J, Tian H-Y, et al. Identification of candidate prostate cancer biomarkers in prostate needle biopsy specimens using proteomic analysis. Int J Cancer. 2007;121(2596–2605):2596–2605.
- Tian HM, LIU X-H, HAN W, et al. Differential expression of filamin A and its clinical significance in breast cancer. Oncol Lett. 2013;6(681–686):681–686.
- Wang K, Zhu TN, Zhao RJ. Filamin A regulates EGFR/ERK/Akt signaling and affects colorectal cancer cell growth and migration. Mol Med Rep. 2019;20(3671–3678). DOI:10.3892/mmr.2019.10622
- Sun GG, Sheng SH, Jing SW, et al. An antiproliferative gene FLNA regulates migration and invasion of gastric carcinoma cell in vitro and its clinical significance. Tumour Biol. 2014;35(2641–2648). DOI:10.1007/s13277-013-1347-1
- Sun GG, Lu YF, Zhang J, et al. Filamin A regulates MMP-9 expression and suppresses prostate cancer cell migration and invasion. Tumour Biol. 2014;35(3819–3826). DOI:10.1007/s13277-013-1504-6
- Wang Z, Li C, Jiang M, et al. Filamin A (FLNA) regulates autophagy of bladder carcinoma cell and affects its proliferation, invasion and metastasis. Int Urol Nephrol. 2018;50(263–273):263–273.
- Song Y, Zhang P, Sun Y, et al. AMPK activation-dependent autophagy compromises oleanolic acid-induced cytotoxicity in human bladder cancer cells. Oncotarget. 2017;8(40):67942–67954. DOI:10.18632/oncotarget.18980
- Zeng Q, Liu J, Cao P, et al. Inhibition of REDD1 Sensitizes Bladder Urothelial Carcinoma to Paclitaxel by Inhibiting Autophagy. Clin Cancer Res. 2018;24(445–459):445–459.
- Yin H, Yang X, Gu W, et al. HMGB1-mediated autophagy attenuates gemcitabine-induced apoptosis in bladder cancer cells involving JNK and ERK activation. Oncotarget. 2017;8(71642–71656):71642–71656.
- Zhang R, Xu J, Zhao J, et al. Mir-30d suppresses cell proliferation of colon cancer cells by inhibiting cell autophagy and promoting cell apoptosis. Tumour Biol. 2017;39(1010428317703984):101042831770398.
- Zheng J, Cheng C, Xu J, et al. miR-142-3p Regulates Tumor Cell Autophagy and Promotes Colon Cancer Progression by Targeting TP53INP2. Chemotherapy. 2021;(2):57–66. DOI:10.1159/000520750.
- Hewitt G, Carroll B, Sarallah R, et al. SQSTM1/p62 mediates crosstalk between autophagy and the UPS in DNA repair. Autophagy. 2016;12(10):1917–1930. DOI:10.1080/15548627.2016.1210368
- Chen S, Li Y, Zhi S, et al. WTAP promotes osteosarcoma tumorigenesis by repressing HMBOX1 expression in an m(6) A-dependentmanner. Cell Death Dis. 2020;11(659). DOI:10.1038/s41419-020-02847-6
- Ren T, Zheng B, Huang Y, et al. Osteosarcoma cell intrinsic PD-L2 signals promote invasion and metastasis via the RhoA-ROCK-LIMK2 and autophagy pathways. Cell Death Dis. 2019;10(4):261. DOI:10.1038/s41419-019-1497-1
- Chen W, Zhai L, Liu H, et al. Downregulation of lncRNA ZFAS1 inhibits the hallmarks of thyroid carcinoma via the regulation of miR3023p on cyclin D1. Mol Med Rep. 2021;23(1). DOI:10.3892/mmr.2020.11640.
- Wang M, Han D, Yuan Z, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9(1149). DOI:10.1038/s41419-018-1187-4
- Liu X, Liu L, Dong Z, et al. Expression patterns and prognostic value of m(6) A-related genes in colorectal cancer. Am J Transl Res. 2019;11(7):3972–3991.
- Jo HJ, Shim H-E, Han M-E, et al. WTAP regulates migration and invasion of cholangiocarcinoma cells. J Gastroenterol. 2013;48(1271–1282):1271–1282.
- Yu HL, Ma X-D, Tong J-F, et al. WTAP is a prognostic marker of high-grade serous ovarian cancer and regulates the progression of ovarian cancer cells. Onco Targets Ther. 2019;12(6191–6201):6191–6201.
- Venturi G, Gomes Ferreira I, Pucci M, et al. Impact of sialyltransferase ST6GAL1 overexpression on different colon cancer cell types. Glycobiology. 2019;29(684–695):684–695.
- Sorci M, Ianniello Z, Cruciani S, et al. METTL3 regulates WTAP protein homeostasis. Cell Death Dis. 2018;9(796). DOI:10.1038/s41419-018-0843-z
- Song M, He Q, Berk B-A, et al. An adventitious interaction of filamin A with RhoGDI2(Tyr153Glu). Biochem Biophys Res Commun. 2016;469(659–664):659–664.
- Nakamura F, Stossel TP, Hartwig JH. The filamins: organizers of cell structure and function. Cell Adh Migr. 2011;5(160–169):160–169.
- Zhang K, Zhu T, Gao D, et al. Filamin A expression correlates with proliferation and invasive properties of human metastatic melanoma tumors: implications for survival in patients. J Cancer Res Clin Oncol. 2014;140(1913–1926):1913–1926.
- Uramoto H, Akyurek LM, Hanagiri T. A positive relationship between filamin and VEGF in patients with lung cancer. Anticancer Res. 2010;30(10):3939–3944.
- Kircher P, Hermanns C, Nossek M, et al. Filamin A interacts with the coactivator MKL1 to promote the activity of the transcription factor SRF and cell migration. Sci Signal. 2015;8(ra112). DOI:10.1126/scisignal.aad2959
- Xu Y, Bismar TA, Su J, et al. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med. 2010;207(2421–2437):2421–2437.
- Dai W, Zhou F, Tang D, et al. Single-cell transcriptional profiling reveals the heterogenicity in colorectal cancer. Medicine (Baltimore). 2019;98(e16916):e16916.
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17(528–542):528–542.
- Akin D, Wang SK, Habibzadegah-Tari P, et al. A novel ATG4B antagonist inhibits autophagy and has a negative impact on osteosarcoma tumors. Autophagy. 2014;10(2021–2035):2021–2035.
- Xie Q, Liu Y, Li X. The interaction mechanism between autophagy and apoptosis in colon cancer. Transl Oncol. 2020;13(100871):100871.
- Chen X, Mao R, Su W, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p -STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16(659–671):659–671.
- Jiang F, Zhou JY, Zhang D, et al. Artesunate induces apoptosis and autophagy in HCT116 colon cancer cells, and autophagy inhibition enhances the artesunate induced apoptosis. Int J Mol Med. 2018;42(1295–1304):1295–1304.
- Lin Z, Niu Y, Wan A, et al. RNA m 6 A methylation regulates sorafenib resistance in liver cancer through FOXO 3-mediated autophagy. EMBO J. 2020;39(e103181). DOI:10.15252/embj.2019103181
- Wei L, Chen Z, Cheng N, et al. MicroRNA-126 Inhibit Viability of Colorectal Cancer Cell by Repressing mTOR Induced Apoptosis and Autophagy. Onco Targets Ther. 2020;13(2459–2468):2459–2468.
- Lu S, Yao Y, Xu G, et al. CD24 regulates sorafenib resistance via activating autophagy in hepatocellular carcinoma. Cell Death Dis. 2018;9(646). DOI:10.1038/s41419-018-0681-z
