Abstract
Epigenetic regulation of chromatin structure is a fundamental process for eukaryotes. Regulators include DNA methylation, microRNAs and chromatin modifications. Within the chromatin modifiers, one class of enzymes that can functionally bind and modify chromatin, through the removal of methyl marks, is the histone lysine demethylases. Here, we summarize the current findings of the 13 known histone lysine demethylases in Drosophila melanogaster, and discuss the critical role of these histone-modifying enzymes in the maintenance of genomic functions. Additionally, as histone demethylase dysregulation has been identified in cancer, we discuss the advantages for using Drosophila as a model system to study tumorigenesis.
Abbreviations
| JmjC | = | Jumonji-C |
| JmjN | = | Jumonji-N |
| JNK | = | Jun-NH2 terminal kinase |
| K | = | lysine |
| KDM | = | lysine demethylase |
| NAD | = | nicotinamide adenine dinucleotide |
| PHD | = | plant homeo domain |
| R | = | arginine |
| Su(var) | = | suppressor of variegation |
| TPR | = | tetratricopeptide |
Introduction
The basic unit of chromatin is formed by nucleosome building blocks, comprised of 146 base pairs of DNA wrapped around a histone octamer. Assembly of the H2A, H2B, H3, and H4 histone proteins forms the core octamer, and these proteins often undergo modifications on their N-terminal tail. The covalent modification of histones performs essential roles in the organization of chromatin and regulation of genes, and these modifications can be identified by key proteins that regulate chromatin binding and alter functionality.Citation1-3 Histones can be post-translationally modified through covalent addition of a number of distinct chemical moieties such as methylation, acetylation, ubiquitination, phosphorylation, and sumoylation.Citation1-3 Changes on the N-tail of histones can interrupt nucleosomal arrangement and interactions, affecting chromatin packaging into higher order structures. Binding of chromatin modifiers onto the tail also facilitates the function of these proteins.
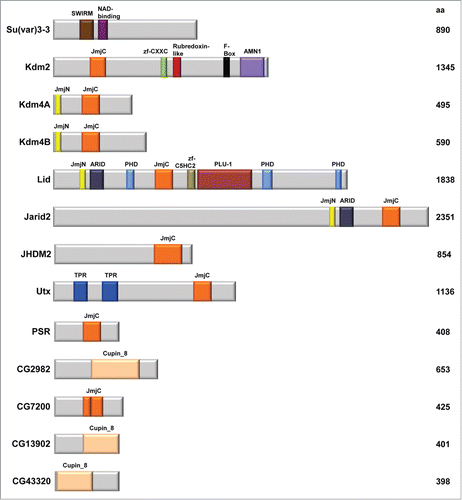
The covalent modification of methylation at particular lysine (K) and arginine (R) residues is the result of the addition or removal of methyl marks by 2 antagonizing enzyme groups, termed “writers” and “erasers.”Citation4 In particular, lysine methylation is substrate specific, and can occur on both the histone H3 and H4 tails. Specific amino acid residues include H3K4, H3K9, H3K27, H3K36, H3K79 and H4K20, where each lysine residue can be un-, mono-, di-, or trimethylated (me0/me1/me2/me3, respectively).Citation1,5,6 Modifications of these residues can be associated with distinct functional outcomes. It has been shown that H3K4, H3K36 and H3K79 marks associate with transcriptional activation, whereas H3K9, H3K27, and H4K20 are linked to transcriptional repression.Citation1,5,7 Lysine methylation has been confirmed to play a critical role in the regulation of a variety of important processes, including gene expression, heterochromatin formation, and dosage compensation.Citation6,8 Here, we review the current findings on the 13 predicted Drosophila histone demethylases (). These proteins are classified as demethylases based on known sequences and domain structure. Further, we address the dysregulation of histone lysine demethylases in cancer and present the advantages for using Drosophila as a model system for studying tumorigenesis.
Amine Oxidase Demethylases
The known or predicted 13 histone lysine demethylases in Drosophila can be subdivided into 2 distinct subgroups based on their mechanism of demethylation. Amine oxidase demethylases, the first group, can oxidize their substrate by FAD to generate an imine that gets hydrolyzed ().Citation5 Suppressor of variegation 3–3, Su(var)3–3, is a histone lysine demethylase that belongs to the flavin monoamine oxidase family, and is the only amine oxidase demethylase in Drosophila.Citation9 Su(var)3–3 is comprised of SWIRM (derived from Swi3p, Rsc8p, and Moira) and nicotinamide adenine dinucleotide-binding (NAD-binding) domains (, ). It is predicted that the SWIRM domain is involved in protein-protein and DNA-protein interactions, in addition to enzyme catalysis.Citation10 Su(var)3–3 has been identified to act as a corepressor by reducing mono- and dimethylated H3K4 (H3K4me1/me2) marks, with no known activity at any other sites of methylation.Citation9,11
Figure 2. Mechanisms of lysine demethylation. (A) Mechanism of LSD1 demethylation via an amine oxidation reaction. (B) Mechanism of JmjC protein demethylation via a hydroxylation reaction. Adapted from Cloos et al.Citation68
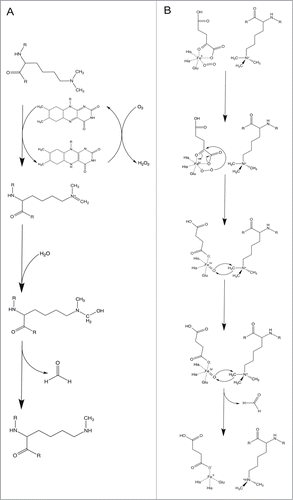
Table 1. Summary of the known 13 histone lysine demethylases in Drosophila melanogaster, including: alternate names, molecular functions, histone demethylase activity, biological processes and lysine demethylase human homologues
Suppressor of variegation, Su(var), genes were originally found to associate with heterochromatin or to prevent heterochromatin spreading into euchromatin.Citation12 While the mechanism to initiate the formation of heterochromatin is unclear, it is noted that a large portion of the eukaryotic genome remains in a heterochromatic, inaccessible state. The role of Su(var)3–3 in heterochromatin formation involves maintenance during embryonic development, and in the establishment of transcriptional silencing in the germline pole cells.Citation11 In that same study, the investigators found that Su(var)3–3 associates with RPD3, heterochromatin protein 1 (HP1) and Su(var)3–9, which controls heterochromatin spreading in position-effect variegation. Further, association of Su(var)3–3 with Su(var)3–9 in this silencing complex established that Su(var)3–3 H3K4 demethylase activity is a requirement for subsequent H3K9 methylation in euchromatic regions.
Su(var)3–3 has also been identified as a protein in the Brahma (Brm) complex, physically associating through its SWIRM domain and functioning as a coregulator.Citation13 The Brm complex is responsible for chromatin remodeling activities during development, involving both gene transcription activation and repression.Citation14 As a member of the Brm complex, Su(var)3–3 represses wing vein formation in inter vein cells.Citation13 Additionally, Su(var)3–3 has been found to functionally interact with the histone demethylase Little imaginal discs (Lid), where together these proteins play a role in Notch pathway-specific control of gene expression in euchromatin.Citation15 Interestingly, while both Su(var)3–3 and Lid demethylate the H3K4 active mark, each is essential for viability (), suggesting that they are not functionally redundant during development. In fact, phenotypes due to mutation of Su(var)3–3 can be suppressed by a mutation in lid, indicating that these enzymes can act in opposition.Citation15 In the Drosophila ovary, Su(var)3–3 is responsible for regulating the size of the germline stem cell (GSC) niche.Citation16 Overall, results from multiple research groups suggest that Su(var)3–3 could regulate gene transcription in a stage or cell-type dependent manner. Data derived from studies on the enzymatic activity have further shown that Su(var)3–3 and its mammalian homolog, LSD1, are functionally conserved demethylases (). However, LSD1, unlike Su(var)3–3, is able to demethylate the repressive H3K9 mark.Citation9,11,17
Table 2. Summary of the phenotypes resulting from mutation of histone lysine demethylases in Drosophila melanogaster
JmjC-Domain Containing Demethylases
The other 12 histone lysine demethylases in Drosophila belong to the JmjC (JumonjiC)-domain containing group. These demethylases are all characterized by a JmjC domain that has defined enzymatic activity, where residues within predicted cofactor binding sites are conserved.Citation5 The mechanism of demethylation requires 5 residues within the JmjC domain that bind to both the α-ketoglutarate (α-KG) and iron cofactors to undergo a hydroxylation reaction, which allow for removal of a methyl mark (). Often, the JmjC domain can be found in combination with other protein domains, which function in substrate specificity.Citation5,8
Lysine-specific demethylase 2, Kdm2, belongs to the group of JmjC-domain containing histone demethylases and is homologous to histone demethylases KDM2 and KDM7 in the mammalian system (). In addition to the JmjC domain, Kdm2 contains zf-CXXC (CXXC-type zinc finger), Rubredoxin-like, F-Box, and AMN1 (Antagonist of mitotic exit network protein 1) domains (). In particular, the F-Box, named for this motif being present in cyclin F, is a protein-protein interaction domain associated with ubiquitylation target selection.Citation18 It was found that Kdm2 is required for efficient H2A ubiquitylation, but has also been shown to mediate demethylation of H3K4me3 and H3K36me2 ().Citation19-21 The observed effects on methylation levels were quite small, suggesting that Kdm2 is redundant with other KDMs. Kdm2 has also been implicated as an enhancer of Polycomb silencing and can act as a suppressor of the Trithorax (Trx) and Absent, small or homeotic discs 1 (Ash1) histone methyltransferases.Citation20 Thus far, Kdm2 has been established to regulate multiple chromatin modifications and linked to factors critical for development.
Histone demethylase 4A, Kdm4A, as well as its homolog Kdm4B, contain both JmjC and JmjN (JumonjiN) domains (). JmjN is a highly conserved domain that is essential for enzymatic activity.Citation5 Proteins that contain both JmjN and JmjC domains, of which there are 4 in Drosophila, serve key roles in regulating development and cell cycle progression. Kdm4A affects the organization of heterochromatin, and overexpression of Kdm4 can induce spreading of HP1 into euchromatin.Citation22 Additionally, it was established that the H3K36me3 activation mark may behave as a barrier to prevent HP1 from spreading into euchromatin, as Kdm4A and HP1 were shown to physically associate.Citation8,22 At particular heterochromatic genes, the Kdm4A-HP1a interaction is required for demethylation of the H3K36me3 mark.Citation23 Kdm4A localizes to sites of euchromatin where it regulates the H3K36 mark, as well as H3K9 to a lesser degree.Citation8,22 Additionally, Kdm4A has been demonstrated to colocalize and physically interact with the Ecdysone Receptor (EcR).Citation24 Those investigators determined that both Kdm4A and Kdm4B regulate ecdysteroid pathway genes through promoter H3K9 demethylation, highlighting overlapping functions between these histone demethylase homologues. Kdm4B, like Kdm4A, demethylates both H3K9 and H3K36 marks.Citation8 Unique to Kdm4B is upregulation of this demethylase by p53 upon ultraviolet irradiation.Citation25 Further, Kdm4B mediates a response to nucleotide excision repair through demethylation of the H3K9 trimethyl mark in heterochromatin. Both Kdm4A and Kdm4B share homology with the mammalian histone demethylase KDM4D (). Mammalian KDM4D, however, is unable to demethylate the H3K36 mark.Citation26
Lid is the third protein that belongs to the subgroup of JmjN and JmjC-domain containing histone demethylases. Lid contains 8 protein domains, which include JmjN, ARID, 3 PHD, JmjC, zf-C5HC2 and PLU-1 (). Lid demethylates the H3K4 mark, specifically H3K4me3.Citation8,27-29 Overexpression of Lid in the wing disc was demonstrated to reduce levels of H3K4me3.Citation27 As mentioned above, Lid activity is not equivalent to that of Su(var)3–3, the other major H3K4 demethylase in flies.Citation15 Lid and KDM2, however, may have some redundant functions as double hypomorph mutants die at an earlier stage in development compared to flies carrying the single mutant alleles.Citation30
Lid localizes at transcription start sites (TSS) of developmental genes.Citation31 Association of Lid with genes that are actively transcribed indicates positive contributions to transcription. In fact, data from recent expression profiling experiments comparing Lid deficient cells to wild type indicate that Lid is required for both gene activation and repression, though the majority of genes were activated by Lid.Citation32 Interestingly, Lid has been found to interact, through its catalytic JmjC domain, with the dMyc oncoprotein, and Lid is required for the expression of a growth regulatory gene.Citation27 dMyc has been shown to inhibit the demethylase activity of Lid. Notably, the interaction of Lid with dMyc is evolutionarily conserved, even in humans.
Additional functions of Lid include its requirement for the correct regulation of homeotic genes during development.Citation8 Lid antagonizes silencing of Ultrabithorax (Ubx) in addition to heterochromatin-dependent gene silencing. It has also been suggested that Lid may function in wing vein development in Drosophila, through interactions with SWI/SNF remodelers.Citation13 Recent genetic and transcriptome studies implicate Lid in appropriate mediation of a response to oxidative stress.Citation32 Lid has also been shown to regulate histone acetylation of polytene chromosomes,Citation8 but not in S2 cells.Citation28 Lid was discovered as a factor in the SIN3 histone deacetylase complex, where it was found to interact with the largest SIN3 isoform, SIN3 220.Citation33,34 KDM5 is the mammalian histone demethylase family with members homologous to Lid ().
Jumonji, AT rich interactive domain 2, Jarid2, is the last JmjN and JmjC-domain containing protein, and is also the largest known histone demethylase in Drosophila. Jarid2 contains JmjN, ARID, and JmjC protein domains (). At present, no demethylase activity on trimethylated H3K4, H3K9, H3K36 and H4K20 have been associated with Jarid2, and speculations have been made that its JmjC domain is not evolutionarily conserved and thus the amino acid changes at the active site may compromise any enzymatic activity.Citation8,35 Instead, it has been suggested that Jarid2 may regulate the presence of the H3K27me3 mark by binding to chromatin.Citation36,37 In addition, Jarid2 interacts with members of the Polycomb repressive complex 2 (PRC2) complex and is thought to control PRC2-dependent transcription.Citation36 Together these proteins may play a role in gene repression as well as function in transcriptional activation of elongation.Citation26,36 Jarid2 also promotes gene activation that is required to block vein differentiation in Drosophila.Citation13 Additional work will be required to fully understand the diverse functional roles of this demethylase. Jarid2 is homologous to the mammalian histone demethylase JARID2 ().
JmjC domain-containing histone demethylase 2, JHDM2, is another known histone lysine demethylase in Drosophila that is homologous to the mammalian KDM3 demethylase (, ). JHDM2 is predicted to play a role in regulating responses to DNA damage stimuli, by contributing to gene expression changes in Chk2 downstream signaling.Citation38 Through overexpression or depletion of this histone demethylase, it has recently been demonstrated to have activity against H3K9 methylation.Citation39 In that study, the investigators found that JHDM2 is a suppressor of position effect variegation, consistent with its role in removal of the repressive H3K9me3 mark.
Histone demethylase UTX, ubiquitously transcribed TPR gene on the X chromosome, contains a JmjC domain as well as 2 Tetratricopeptide repeat (TPR) structural domains, involved in protein-protein interactions (). Similar to the mammalian homolog KDM6, UTX targets the H3K27 repressive mark ().Citation40,41 Initially, UTX was found to demethylate only H3K27me3/me2 marks, with no activity on H3K27me1.Citation40 A subsequent study found that the mono methyl H3K4 mark could also be affected, though the influence there is likely indirect.Citation41 In that study, UTX was found to suppress Notch-dependent and Retinoblastoma-dependent tumors.Citation41 UTX was also found in complex with acetyltransferase CREB-binding protein (CBP) and chromatin remodeler Brm, where they antagonize Polycomb silencing.Citation42 It is interesting to note that Jarid2, which also influences H3K27 methylation levels, copurifies with Polycomb group proteins and localizes to silent chromatin.Citation35 Additional studies are required to dissect the distinct functions of the 2 H3K27 demethylases. Further, UTX was shown to bind to the nuclear hormone receptor EcR complex, and to be recruited to promoter regions for apoptotic and autophagic genes.Citation43 Through mediation of the H3K27 repressive histone mark, UTX was shown to be required for p53-dependent expression of ku80 when Drosophila Kc cells were exposed to ionizing radiation.Citation44 UTX physically associates with p53, but is not a requirement for expression of other genes involved in DNA damage response. UTX was also identified in a genetic screen to find factors necessary during wound healing response to induce a repair transcriptome.Citation45 Together, these studies reveal that UTX plays a critical role to maintain genomic stability.
The phosphatidylserine receptor, PSR, belongs to the JmjC-domain containing subgroup of demethylases (). At present, PSR is predicted to catalyze peptidyl-lysine 5-dioxygenase activity, and is known to play a role in cell competition and negative regulation of both the c-Jun-NH2 terminal kinase, JNK, cascade and apoptotic process ().Citation46,47 PSR plays a role in protection from apoptosis by suppressing the apoptosis regulator Head involution defective, Hid, or Wrinkled. Additionally, activation of the JNK pathway has been shown to suppress phenotypes resulting from PSR overexpression.Citation46 JMJD6 is the mammalian homolog of PSR and a known histone demethylase ().
CG2982, CG7200, CG13902, and CG43320 are all JmjC-domain containing histone demethylases that can be uniquely characterized by a Cupin 8 domain (). Similarities between sequences, secondary structures, and active-sites have led to the identification of JmjC domains as a branch of the cupin family.Citation48 Currently, only CG2982 has predicted histone demethylation, iron ion binding and oxidoreductase activity. NO66, JMJD4, KDM8, and HSPBAP1 are the respective mammalian homologues to these proteins in Drosophila (). The enzymatic activity of these proteins has potential for further studies.
Dysregulation of Histone Lysine Demethylases in Cancer
Histone demethylases play essential roles in dynamically regulating gene expression and chromatin architecture through histone lysine methylation, and are thus implicated in developmental processes, stem cell biology, and tumorigenesis.Citation49-52 Epigenetic alterations, including chromatin modifications, have been shown to play fundamental roles in cancer initiation and progression. Recently, meta-analyses have uncovered that genes encoding histone lysine demethylases have a high frequency of genetic alterations in tumor types, including breast cancer.Citation53,54 Amplification and overexpression of histone demethylases, such as the KDM4 subfamily, has been observed in various tumor tissues,Citation55-58 and histone demethylases, including KDM2A, hold a critical role in preserving aggressive cancer phenotypes.Citation54 A connection has also been established linking histone demethylases, such as KDM5A, to drug resistance in cancer cells.Citation59 Thus, it is critical to understand the mechanisms of how histone lysine demethylases contribute to cancer initiation and progression. The use of a whole organism model system, such as Drosophila melanogaster, in which development and cell and tissue interaction occur, can facilitate and support mechanistic studies of tumorigenesis.
Drosophila Model System in Tumorigenesis
While Drosophila do not typically develop cancer due to their short lifespans, it has been shown that upon perturbation of cancer-related genes, cells can evade apoptosis, sustain proliferative abilities, exhibit metastasis and tissue invasion, have a loss of differentiation, and show genome instability; all proven hallmarks of cancer.Citation60 The reduced redundancy of the fruit fly genome and ability to perform large-scale genetic screens in this organism has contributed to the elucidation and characterization of processes for development and signaling cascades.Citation61 As noted in the sections above and summarized in , mutations in many of the Drosophila KDMs result in distinct developmental phenotypes. Relevant to cancer studies, critical signaling pathways such as Hedgehog and Notch, highly implicated in tumorigenesis,Citation62,63 were first discovered in the fruit fly.Citation60 Additionally, the potential to identify novel tumor suppressors and oncogenes through the use of new molecular technologies, including CRISPR/Cas-9, in this system highlight the promising direction of this field. Thus, there exists potential to expand tumorigenic studies utilizing Drosophila.
One advantage of the Drosophila system is in the ability to model context dependency, as different tissues and organs, including muscle, brain, ovaries, eye discs, wings and hemocytes, can model various cancer hallmarks.Citation60 Flies can allow for in vivo study of cell invasion. Evidence has shown that Drosophila cells can metastasize and spread through epithelial tissue, colonizing in distant new sites of the organism.Citation61 Additionally, studies have been able to utilize mutations and generate neoplastic tumors in the fruit fly. Such tumors can develop quickly and when allografted, can metastasize.Citation64 Thus, there exists the potential to allow for modeling of metastases in Drosophila and expanding tumor mass for further analysis. Additionally, through the capacity to generate clones of mutant tissue for specific genes during development, tumor microenvironment analysis can be facilitated.Citation61
In addition to modeling molecular mechanisms for tumor progression, several findings have begun to show that Drosophila could be used as an in vivo system to screen for cancer drug therapies.Citation65 Advantages to this approach include cost-effectiveness and speed as key advantages for means of drug discovery.Citation61 To date, Drosophila has begun to be utilized to test combination treatments as means for cancer therapy.Citation65-67 For instance, Drosophila were used to perform a small molecule screen to identify drugs that could increase radiation effects to apply these inhibitors to human cancers for novel combination therapy.Citation67 Additional pharmacological approaches to discover cancer drugs took into account Drosophila genetics and profiling of novel compounds to mechanistically investigate drug efficacy.Citation66 These studies highlight the potential and advantages for performing cancer studies using Drosophila as a model system.
Conclusion
Histone lysine demethylases are conserved in organisms from Drosophila to humans (). Modifications of histones on the N-terminal tail can prevent higher order assembly of structures in addition to being binding sites for histone-modifying enzymes. Dysregulation of these histone-modifying enzymes has been implicated in human carcinogenesis. Because histone-modifying enzymes are potential drug targets and epigenetic alterations are reversible, pharmacologic targeting of histone-modifying enzymes is a therapeutic strategy to correct the alteration of histone demethylases, and block cancer progression. Recent evidence has demonstrated that Drosophila can model tumor progression and be screened to identify novel cancer drugs. Taken together, there is promising potential to investigate the dysregulation of histone lysine demethylases in tumorigenesis using the Drosophila model system.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
We thank Dr. Ambikai Gajan for critical reading of the manuscript.
Funding
This work was partially supported by funding from the Cancer Biology Graduate Program, Wayne State University School of Medicine, to AH; by grants from the NIH/NCI (R21CA175244) and the Office of Research on Women's Health (OWRH), the Mary Kay Foundation Cancer Research Grant Program, and the Karmanos Cancer Institute-SRIG to Z-QY; and by a grant from the NIH/NIGMS (R01GM088886–01A2) to LAP.
References
- Kouzarides T. Chromatin modifications and their function. Cell 2007; 128:693-705; PMID:17320507; http://dx.doi.org/10.1016/j.cell.2007.02.005
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell 2007; 128:707-19; PMID:17320508; http://dx.doi.org/10.1016/j.cell.2007.01.015
- Chen T, Dent SY. Chromatin modifiers and remodellers: regulators of cellular differentiation. Nat Rev Genet 2014; 15:93-106; PMID:24366184; http://dx.doi.org/10.1038/nrg3607
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 2007; 25:15-30; PMID:17218268; http://dx.doi.org/10.1016/j.molcel.2006.12.014
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7:715-27; PMID:16983801; http://dx.doi.org/10.1038/nrg1945
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005; 6:838-49; PMID:16261189; http://dx.doi.org/10.1038/nrm1761
- Zhou X, Ma H. Evolutionary history of histone demethylase families: distinct evolutionary patterns suggest functional divergence. BMC Evol Biol 2008; 8:294; PMID:18950507; http://dx.doi.org/10.1186/1471-2148-8-294
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucleic Acids Res 2008; 36:2852-63; PMID:18375980; http://dx.doi.org/10.1093/nar/gkn098
- Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol 2007; 17:808-12; PMID:17462898; http://dx.doi.org/10.1016/j.cub.2007.03.068
- Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, et al. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure 2006; 14:457-68; PMID:16531230; http://dx.doi.org/10.1016/j.str.2005.12.004
- Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, et al. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell 2007; 26:103-15; PMID:17434130; http://dx.doi.org/10.1016/j.molcel.2007.02.025
- Schotta G, Ebert A, Dorn R, Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin Cell Dev Biol 2003; 14:67-75; PMID:12524009; http://dx.doi.org/10.1016/S1084-9521(02)00138-6
- Curtis BJ, Zraly CB, Marenda DR, Dingwall AK. Histone lysine demethylases function as co-repressors of SWI/SNF remodeling activities during Drosophila wing development. Dev Biol 2011; 350:534-47; PMID:21146519; http://dx.doi.org/10.1016/j.ydbio.2010.12.001
- Tamkun JW. The role of brahma and related proteins in transcription and development. Curr Opin Genet Dev 1995; 5:473-7; PMID:7580139; http://dx.doi.org/10.1016/0959-437X(95)90051-H
- Di Stefano L, Walker JA, Burgio G, Corona DF, Mulligan P, Naar AM, Dyson NJ. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev 2011; 25:17-28; PMID:21205864; http://dx.doi.org/10.1101/gad.1983711
- Eliazer S, Shalaby NA, Buszczak M. Loss of lysine-specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc Natl Acad Sci U S A 2011; 108:7064-9; PMID:21482791; http://dx.doi.org/10.1073/pnas.1015874108
- Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005; 437:436-9; PMID:16079795
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 1996; 86:263-74; PMID:8706131; http://dx.doi.org/10.1016/S0092-8674(00)80098-7
- Kavi HH, Birchler JA. Drosophila KDM2 is a H3K4me3 demethylase regulating nucleolar organization. BMC Res Notes 2009; 2:217; PMID:19852816; http://dx.doi.org/10.1186/1756-0500-2-217
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 2008; 22:2799-810; PMID:18923078; http://dx.doi.org/10.1101/gad.484208
- Zheng Y, Hsu FN, Xu W, Xie XJ, Ren X, Gao X, Ni JQ, Ji JY. A developmental genetic analysis of the lysine demethylase KDM2 mutations in Drosophila melanogaster. Mech Dev 2014; 133:36-53; PMID:25016215; http://dx.doi.org/10.1016/j.mod.2014.06.003
- Lin CH, Li B, Swanson S, Zhang Y, Florens L, Washburn MP, Abmayr SM, Workman JL. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell 2008; 32:696-706; PMID:19061644; http://dx.doi.org/10.1016/j.molcel.2008.11.008
- Lin CH, Paulson A, Abmayr SM, Workman JL. HP1a targets the Drosophila KDM4A demethylase to a subset of heterochromatic genes to regulate H3K36me3 levels. PloS One 2012; 7:e39758; PMID:22761891; http://dx.doi.org/10.1371/journal.pone.0039758
- Tsurumi A, Dutta P, Yan SJ, Sheng R, Li WX. Drosophila Kdm4 demethylases in histone H3 lysine 9 demethylation and ecdysteroid signaling. Sci Rep 2013; 3:2894; PMID:24100631; http://dx.doi.org/10.1038/srep02894
- Palomera-Sanchez Z, Bucio-Mendez A, Valadez-Graham V, Reynaud E, Zurita M. Drosophila p53 is required to increase the levels of the dKDM4B demethylase after UV-induced DNA damage to demethylate histone H3 lysine 9. J Biol Chem 2010; 285:31370-9; PMID:20675387; http://dx.doi.org/10.1074/jbc.M110.128462
- Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006; 125:467-81; PMID:16603238; http://dx.doi.org/10.1016/j.cell.2006.03.028
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes Dev 2007; 21:537-51; PMID:17311883; http://dx.doi.org/10.1101/gad.1523007
- Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase. Nat Struct Mol Biol 2007; 14:344-6; PMID:17351630; http://dx.doi.org/10.1038/nsmb1217
- Lee N, Zhang J, Klose RJ, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. The trithorax-group protein Lid is a histone H3 trimethyl-Lys4 demethylase. Nat Struct Mol Biol 2007; 14:341-3; PMID:17351631; http://dx.doi.org/10.1038/nsmb1216
- Li L, Greer C, Eisenman RN, Secombe J. Essential functions of the histone demethylase lid. PLoS Genet 2010; 6:e1001221
- Lloret-Llinares M, Perez-Lluch S, Rossell D, Moran T, Ponsa-Cobas J, Auer H, Corominas M, Azorín F. dKDM5/LID regulates H3K4me3 dynamics at the transcription-start site (TSS) of actively transcribed developmental genes. Nucleic Acids Res 2012; 40:9493-505; PMID:22904080; http://dx.doi.org/10.1093/nar/gks773
- Liu X, Greer C, Secombe J. KDM5 interacts with Foxo to modulate cellular levels of oxidative stress. PLoS Genet 2014; 10:e1004676; PMID:25329053; http://dx.doi.org/10.1371/journal.pgen.1004676
- Spain MM, Caruso JA, Swaminathan A, Pile LA. Drosophila SIN3 isoforms interact with distinct proteins and have unique biological functions. J Biol Chem 2010; 285:27457-67; PMID:20566628; http://dx.doi.org/10.1074/jbc.M110.130245
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell 2009; 35:782-93; PMID:19782028; http://dx.doi.org/10.1016/j.molcel.2009.07.020
- Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M. The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. FEBS J 2007; 274:6139-51; PMID:17970746; http://dx.doi.org/10.1111/j.1742-4658.2007.06135.x
- Herz HM, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, et al. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol Cell Biol 2012; 32:1683-93; PMID:22354997; http://dx.doi.org/10.1128/MCB.06503-11
- Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Müller J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 2014; 21:569-71; PMID:24837194; http://dx.doi.org/10.1038/nsmb.2833
- Park SY, Song YH. Genetic screen for genes involved in Chk2 signaling in Drosophila. Mol Cells 2008; 26:350-5; PMID:18612247
- Herz HM, Morgan M, Gao X, Jackson J, Rickels R, Swanson SK, Florens L, Washburn MP, Eissenberg JC, Shilatifard A. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science 2014; 345:1065-70; PMID:25170156; http://dx.doi.org/10.1126/science.1255104
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 2008; 28:1041-6; PMID:18039863; http://dx.doi.org/10.1128/MCB.01504-07
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 2010; 30:2485-97; PMID:20212086; http://dx.doi.org/10.1128/MCB.01633-09
- Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ. Histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 2012; 32:2323-34; PMID:22493065; http://dx.doi.org/10.1128/MCB.06392-11
- Denton D, Aung-Htut MT, Lorensuhewa N, Nicolson S, Zhu W, Mills K, Cakouros D, Bergmann A, Kumar S. UTX coordinates steroid hormone-mediated autophagy and cell death. Nat Commun 2013; 4:2916; PMID:24336022; http://dx.doi.org/10.1038/ncomms3916
- Zhang C, Hong Z, Ma W, Ma D, Qian Y, Xie W, Tie F, Fang M. Drosophila UTX coordinates with p53 to regulate ku80 expression in response to DNA damage. PloS One 2013; 8:e78652; PMID:24265704; http://dx.doi.org/10.1371/journal.pone.0078652
- Campos I, Geiger JA, Santos AC, Carlos V, Jacinto A. Genetic screen in Drosophila melanogaster uncovers a novel set of genes required for embryonic epithelial repair. Genetics 2010; 184:129-40; PMID:19884309; http://dx.doi.org/10.1534/genetics.109.110288
- Krieser RJ, Moore FE, Dresnek D, Pellock BJ, Patel R, Huang A, Brachmann C, White K. The Drosophila homolog of the putative phosphatidylserine receptor functions to inhibit apoptosis. Development 2007; 134:2407-14; PMID:17522160; http://dx.doi.org/10.1242/dev.02860
- Li W, Baker NE. Engulfment is required for cell competition. Cell 2007; 129:1215-25; PMID:17574031; http://dx.doi.org/10.1016/j.cell.2007.03.054
- Clissold PM, Ponting CP. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A2beta. Trends Biochem Sci 2001; 26:7-9; PMID:11165500; http://dx.doi.org/10.1016/S0968-0004(00)01700-X
- Varier RA, Timmers HT. Histone lysine methylation and demethylation pathways in cancer. Biochim Biophys Acta 2011; 1815:75-89; PMID:20951770
- Kampranis SC, Tsichlis PN. Histone demethylases and cancer. Adv Cancer Res 2009; 102:103-69; PMID:19595308; http://dx.doi.org/10.1016/S0065-230X(09)02004-1
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13:297-311; PMID:22473470
- Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 2012; 13:343-57; PMID:22473383; http://dx.doi.org/10.1038/nrg3173
- Ye Q, Holowatyj A, Wu J, Liu H, Zhang L, Suzuki T, Yang ZQ. Genetic alterations of KDM4 subfamily and therapeutic effect of novel demethylase inhibitor in breast cancer. Am J Cancer Res 2015; 5(4):1519-30
- Liu H, Liu L, Holowatyj A, Jiang Y, Yang ZQ. Integrated genomic and functional analyses of histone demethylases identify oncogenic KDM2A isoform in breast cancer. Molecular Carcinogenesis Forthcoming 2015; PMID: 26207617.
- Liu G, Bollig-Fischer A, Kreike B, van de Vijver MJ, Abrams J, Ethier SP, Yang ZQ. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene 2009; 28:4491-500; PMID:19784073; http://dx.doi.org/10.1038/onc.2009.297
- Holowatyj A, Yang ZQ. The Role of Histone Demethylase GASC1 in Cancer and its Therapeutic Potential. Curr Cancer Ther Rev 2013; 9:78-85
- Labbe RM, Holowatyj A, Yang ZQ. Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. Am J Transl Res 2013; 6:1-15; PMID:24349617
- Ye Q, Holowatyj A, Wu J, Liu H, Zhang L, Suzuki T, Yang ZQ. Genetic alterations of KDM4 subfamily and therapeutic effect of novel demethylase inhibitor in breast cancer. Am J Cancer Res 2015; 5:1519-30; PMID:26101715
- Hou J, Wu J, Dombkowski A, Zhang K, Holowatyj A, Boerner JL, Yang ZQ. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. Am J Transl Res 2012; 4:247-56; PMID:22937203
- Tipping M, Perrimon N. Drosophila as a model for context-dependent tumorigenesis. J Cell Physiol 2014; 229:27-33; PMID:23836429
- Miles WO, Dyson NJ, Walker JA. Modeling tumor invasion and metastasis in Drosophila. Dis Models Mech 2011; 4:753-61; PMID:21979943; http://dx.doi.org/10.1242/dmm.006908
- Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic overgrowth in Drosophila. EMBO J 2003; 22:5769-79; PMID:14592975; http://dx.doi.org/10.1093/emboj/cdg548
- Ogden SK, Ascano M, Jr., Stegman MA, Robbins DJ. Regulation of Hedgehog signaling: a complex story. Biochem Pharmacol 2004; 67:805-14; PMID:15104233; http://dx.doi.org/10.1016/j.bcp.2004.01.002
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 2010; 330:1824-7; PMID:21205669; http://dx.doi.org/10.1126/science.1195481
- Gladstone M, Su TT. Chemical genetics and drug screening in Drosophila cancer models. J Genet Genomics 2011; 38:497-504; PMID:22035870; http://dx.doi.org/10.1016/j.jgg.2011.09.003
- Dar AC, Das TK, Shokat KM, Cagan RL. Chemical genetic discovery of targets and anti-targets for cancer polypharmacology. Nature 2012; 486:80-4; PMID:22678283; http://dx.doi.org/10.1038/nature11127
- Edwards A, Gladstone M, Yoon P, Raben D, Frederick B, Su TT. Combinatorial effect of maytansinol and radiation in Drosophila and human cancer cells. Dis Models Mech 2011; 4:496-503; PMID:21504911; http://dx.doi.org/10.1242/dmm.006486
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 2008; 22:1115-40; PMID:18451103; http://dx.doi.org/10.1101/gad.1652908
- McQuilton P, St Pierre SE, Thurmond J, FlyBase C. FlyBase 101–the basics of navigating FlyBase. Nucleic Acids Res 2012; 40:D706-14; PMID:22127867; http://dx.doi.org/10.1093/nar/gkr1030