Abstract
Aminoacyl-tRNA synthetases (aaRSs) ligate amino acids to their cognate tRNAs, allowing them to decode the triplet code during translation. Through different mechanisms aaRSs also perform several non-canonical functions in transcription, translation, apoptosis, angiogenesis and inflammation. Drosophila has become a preferred system to model human diseases caused by mutations in aaRS genes, to dissect effects of reduced translation or non-canonical activities, and to study aminoacylation and translational fidelity. However, the lack of a systematic annotation of this gene family has hampered such studies. Here, we report the identification of the entire set of aaRS genes in the fly genome and we predict their roles based on experimental evidence and/or orthology. Further, we propose a new, systematic and logical nomenclature for aaRSs. We also review the research conducted on Drosophila aaRSs to date. Together, our work provides the foundation for further research in the fly aaRS field.
Introduction
Aminoacyl-tRNA synthetases (aaRSs) constitute an ancient family of enzymes that catalyze aminoacylation reactions by attaching amino acids to cognate tRNAs.Citation1,2 The aminoacylation reaction is a 2-step process (). In the first step, the amino acid is activated by ATP to generate an aminoacyl-adenylate intermediate. In the second step, the activated amino acid is transferred to the 3′ end of the tRNA bearing the appropriate anticodon triplet that recognizes the corresponding codon in the mRNA.Citation3 Such an aminoacylated tRNA is referred to as aa-tRNA and it can now be delivered to the ribosome for nascent polypeptide synthesis. Because aaRSs recognize specific amino acids and the corresponding tRNAs, they translate the nucleic acid language into the amino acid language and thereby decode the “second genetic code”.Citation4 aaRSs are thus fundamental components of the protein synthesis process in all cells of all species in the 3 primary kingdoms of life.
Figure 1. Aminoacyl-tRNA synthetase catalyzes a 2-step aminoacylation reaction. In the first step, the aaRS activates the substrate amino acid. By consuming an ATP it forms an aa-AMP intermediate. In the second step, the aa-AMP is transferred to the acceptor end of the cognate tRNA, generating an aa-tRNA that can be delivered to ribosomes for protein synthesis. aa, amino acid; aaRS, aminoacyl-tRNA synthetase; PPi, pyrophosphate.
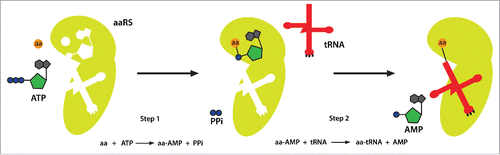
There are 20 standard amino acids, and for each of them cells are expected to express at least one aaRS. Two different criteria may be used to categorize aaRSs. Based on their protein structure, class I aaRSs contain a characteristic Rossman fold catalytic domain and usually function as monomeric or dimeric proteins, while class II aaRSs contain 3 conserved motifs and are usually dimeric or tetrameric.Citation2,5 Alternatively, aaRSs may be classified according to their subcellular sites of action: cytoplasmic, mitochondrial, or both cytoplasmic and mitochondrial (‘dual-localized’).Citation6,7
Interest in aaRSs has grown in recent years for 2 major reasons. First, it has become apparent that aaRSs perform diverse non-canonical functions in addition to their roles in protein synthesis, including roles in regulation of transcription and translation, apoptosis, angiogenesis and inflammation.Citation8,9 These additional functions are mainly achieved by recruitment of other protein complexes, acquisition of additional domains, or generation of novel protein fragments by alternative splicing or proteolysis.Citation8,10,11 Second, genetic studies have revealed that mutations in many aaRS genes are associated with a wide variety of human syndromes and diseases.Citation7,12,13 For example, mutations in 5 genes encoding cytoplasmic or dual-localized aaRSs have been identified in patients with (mainly dominantly inherited) peripheral neuropathies, while 9 mitochondrial aaRS loci have been implicated in heterogeneous recessive disorders.Citation12 In most cases it is not known how aaRS mutations cause the disease phenotypes-whether through reduced translational activity, reduced aminoacylation accuracy or through a defect in a non-canonical function.
The powerful genetic tools available in Drosophila melanogaster offer tremendous potential to explore genotype – phenotype relationships, while the high evolutionary conservation of the tRNA aminoacylation reaction validates the modeling of aaRS-associated diseases in this system.Citation14,15 However, the full complement of Drosophila aaRSs has not been accurately characterized to date, and as a result the annotations in the FlyBase database have been incomplete and in some cases incorrect. For example, prior to our recent study,Citation16 the gene encoding mitochondrial PheRS was named as “Aats-phe, phenylalanyl-tRNA synthetase” (implying a cytoplasmic role) while the genes encoding the 2 subunits of the true cytoplasmic PheRS were unnamed. The lack of a comprehensive and consistent set of aaRS annotations in D. melanogaster potentially hampers understanding and research of these fundamental enzymes in this key model organism.
Here, we report our systematic analysis to identify and classify all aaRSs in D. melanogaster. In so doing, we propose a new nomenclature for Drosophila aaRS genes that is more explicit and consistent with that used in the wider field. In addition, we review the important aaRS studies that have been carried out in flies to date to illustrate how this model organism has already contributed to the field.
Identification of D. melanogaster aminoacyl-tRNA synthetases
In a typical eukaryotic cell, there are cytoplasmic, mitochondrial, and dual-localized aaRSs.Citation7 Because the number of standard amino acids is 20, the total number of aaRSs is therefore expected to be in the range of 20 to 40. We are aware of one previous study that attempted to identify the D. melanogaster aaRSs17 – this list of 20 different aaRSs, however, comprised a mixture of cytoplasmic and mitochondrial factors.
We began our own study of D. melanogaster aaRSs by searching FlyBaseCitation18 (FB2014_06) for genes with the prefix used in the database for this set of genes, namely ‘Aats-’, for ‘Aminoacyl-tRNA synthetase’. Only 22 aaRS genes were found by this approach (Table S1), suggesting that additional aaRS genes remained to be identified. Furthermore, reference to the location of the enzyme was inconsistent or missing in several gene names: 7 of the 22 named genes encode mitochondrial aaRSs, but this was indicated in only 2 cases; the names of the other 5 mitochondrial aaRS genes did not contain location information and would therefore be wrongly considered to be cytoplasmic (or dual-localized), particularly as the true cytoplasmic form was unnamed in each case (see below).
In order to identify the full complement of fly genes encoding aaRSs, we used the well-characterized set of human aaRS proteins (obtained from the HGNC databaseCitation19) to search for matching D. melanogaster polypeptides in FlyBase (FB2014_06, Dmel Annotation Release 6.03) using BLASTP. We also examined ortholog predictions housed within FlyBase and the HGNC databases, and searched for genes annotated with relevant Gene Ontology terms and InterPro domains. The results are summarized in . We found that 35 genes in the fly nuclear genome encode 34 aaRS enzymes: 15 aaRSs are predicted to act exclusively in the cytoplasm, 15 in the mitochondria, and 4 are dual-localized. The reason for the gene count being one more than the aaRS count is because the cytoplasmic phenylalanyl-tRNA synthetase comprises 2 subunits encoded by 2 separate genes.Citation16 The reason for finding 19 (and not 20) aaRSs that act in the cytoplasm is because the cytoplasmic glutamyl-prolyl-tRNA synthetase (GluProRS) loads both Glu and Pro to their cognate tRNAs.Citation20 Finally the explanation for finding 19 (and not 20) aaRSs that function in mitochondria is that there is no mitochondrial glutaminyl-tRNA synthetase (GlnRS; discussed below). It is also worth noting that CG10802, CG8097 and Slimp (CG31133), encode proteins containing domains associated with alanyl-, arginyl- and seryl-tRNA synthetase activity, respectively (Table S2). However, their overall similarities to the canonical human and Drosophila proteins are relatively low, and it is known that Slimp lacks aminoacylation activity.Citation21 These three genes are therefore not included in and were not considered further.
Table 1. D. melanogaster aminoacyl-tRNA synthetases
We propose a unified Drosophila nomenclature for aaRSs that discriminates between the cytoplasmic and mitochondrial enzymes, rather than one that describes their biochemical and structural properties. This makes sense for work in a system that has a strong emphasis on functional studies. Furthermore, this nomenclature is widely used within the aaRS field. Thus we add a ‘-m’ suffix to the symbols of genes encoding the mitochondrial aaRSs. We also suggest that the ‘Aats-x’ format previously used for aaRS genes in FlyBase is replaced with the more common ‘xRS‘ format, where x indicates the relevant amino acid. Finally, we recommend using the 3-letter, rather than the single letter, amino acid code because this is more explicit and more easily recognized as an amino acid when followed by ‘RS’ in the same word and, again, it is a common convention in the field. With this nomenclature, the gene symbol for the cytoplasmic tyrosyl-tRNA synthetase is ‘TyrRS’, while ‘TyrRS-m’ is the designation for the gene encoding the mitochondrial tyrosyl-tRNA synthetase. While it might also make sense to add a distinguishing suffix to the symbols of the dual-localized enzymes and genes, we have opted to name them the same way as the cytoplasmic ones to keep the symbols simple and short. The symbols and names of all aaRSs following this proposed nomenclature are shown in full in .
In the following parts, we will analyze the 3 groups of aaRSs separately and will review the published work on them.
Cytoplasmic aminoacyl-tRNA synthetases
Cytoplasmic aaRSs charge tRNAs with cognate amino acids in the cytoplasm. Some of these aaRSs are also able to translocate to the nucleus and aminoacylation can also take place in this compartment.Citation22 There are 16 genes encoding 15 cytoplasmic aaRSs in D. melanogaster and these are able to charge 16 different amino acids (). As mentioned, the discrepancies in these figures are explained by the GluProRS gene encoding a protein with 2 enzymatic activities, and by the PheRS enzyme consisting of 2 different subunits encoded by 2 distinct genes. While cytoplasmic aaRS genes generally encode a single, unique polypeptide, it is noteworthy that GluProRS and ValRS, respectively, encode each 2 different polypeptides, generated by alternative promoter usage and alternative splicing, respectively (FlyBase).
The bifunctionality of GluProRS is unique among all aaRSs and it has been well studied in various systems, including flies.Citation20,23 In bacteria and archaea, 2 distinct genes encode GluRS and ProRS, and it seems that a gene fusion event occurred during the evolution of metazoa.Citation24 The GluProRS protein is composed of 3 domains, the N-terminal domain with Glu-catalyzing activity, the C-terminal domain with Pro-catalyzing activity, and the central domain with repeated motifs. In Drosophila, the GluProRS gene encodes 2 polypeptides, the full-length protein and the C-terminal short protein. Their expression seems to be controlled by different promoters and probably distinct transcriptional regulators. The full-length protein is expressed throughout development, while the C-terminal short protein is especially abundant in 5–10 hours old embryos.Citation23 Interestingly, the C-terminal short protein is functional in Pro-tRNA aminoacylation in vivo,Citation23 thereby providing a second way to generate Pro-tRNA in the cytoplasm.
An interesting feature of cytoplasmic aaRSs in higher eukaryotes (including flies) is that 8 aaRSs, together with 3 non-enzymatic factors, form a ‘multi-synthetase complex’ (MSC).Citation9,20,25 The functional significance of the MSC is unclear, but the auxiliary factors are thought to be responsive to diverse signal transduction pathways and thus provide a mechanism to coordinate protein synthesis with other biological processes.Citation9 In flies, the auxiliary factors are encoded by the CG8235, CG12304, and CG30185 genes, and we propose that these are named AIMP1, AIMP2 and AIMP3 (aminoacyl-tRNA synthetase-interacting multifunctional proteins 1, 2 and 3), respectively, to match the nomenclature used in the wider field (Supplementary Table S2).
Several cytoplasmic aaRSs have been discovered in different genetic screens in flies. Aspartyl-tRNA synthetase (AspRS) was independently identified in screens for Sex-lethal dosage-sensitive modifiersCitation26 and for mutants defective in larval growth.Citation27 Tryptophanyl-tRNA synthetase (TrpRS) was identified in a screen for genes expressed in the embryonic salivary gland,Citation17 while mutations in several different cytoplasmic aaRSs were found to increase lysosomal activity.Citation28 In each case, the specificity and precise function of the aaRS enzyme(s) involved remain to be characterized.
Work from our group has characterized the importance of aminoacylation fidelity in vivo by exploring the ‘double-sieving’ function of PheRS in Drosophila.Citation16 The first sieve — amino-acid recognition — serves to exclude most non-cognate amino acids; the second sieve — amino-acid editing — is capable of correcting aminoacylation errors. Both sieves are important and double-sieving-defective mutations in PheRS result in misacylation by non-cognate Tyr and protein mistranslation, leading to many defects, including ER stress, neuronal cell apoptosis, impaired locomotive performance, reduced lifespan, and decreased organ size. This work demonstrates how malfunctioning of aaRSs at the molecular level can cause a range of phenotypes at the cellular and organismal levels.
Mitochondrial aminoacyl-tRNA synthetases
Mitochondrial aaRSs are required for protein translation in this organelle and are thought to have a bacterial origin. In eukaryotic cells, they are encoded by nuclear genes and, after being expressed, they are imported into mitochondria with the guidance of a mitochondrial targeting sequence (MTSCitation29; ) There are 15 genes coding for 15 mitochondrial aaRSs in D. melanogaster (). In contrast to the situation in the cytoplasm, the mitochondrial PheRS consists of only one polypeptide encoded by a single gene while 2 separate genes code for distinct mitochondrial GluRS and ProRS enzymes. Furthermore, only ProRS-m is annotated to encode more than one polypeptide (FlyBase), though the function of the shorter protein isoform is unknown. We were unable to identify a GlnRS-m gene in our analysis, though this is consistent with a lack of GlnRS activity in all chloroplasts and mitochondria examined.Citation30,31 In these organelles, Gln-tRNA is generated by mischarging a tRNAGln with Glu and converting Glu to Gln via a heterotrimeric Glu-tRNAGln amidotransferase (Gat).Citation31,32 In D. melanogaster, the 3 subunits of this complex are encoded by the GatA,Citation33 CG5463 and CG33649 genes – we propose to name the latter two GatB and GatC, respectively (Table S2).
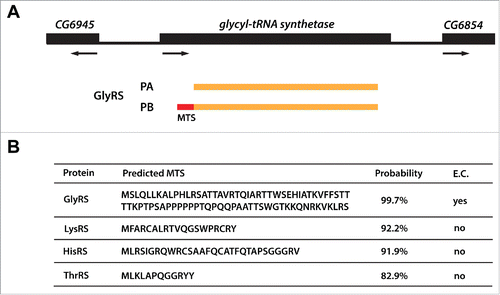
Figure 2. Dual-localized aaRSs in D. melanogaster. (A) GlyRS is shown as an example of dual-localized aaRS. It encodes 2 polypeptides, PA and PB. PB contains an extra N-terminal mitochondrial targeting sequence (MTS; in red) that can be used for its import into mitochondria. (B) The four dual-localized aaRSs with their predicted MTS and probabilities of mitochondrial localization. The analysis was performed by Mitoprot.Citation29 E.C., experimentally confirmed.
Drosophila mitochondrial aaRSs have received a similar degree of attention in the published literature as their cytoplasmic counterparts. The sequence and structure of the fly PheRS-m was described in a comparative study with the human enzyme.Citation34 ArgRS-m was identified in a genetic screen for nuclear-encoded genes with mitochondrial function.Citation35 Its mitochondrial localization was confirmed in this study by using a GFP fusion protein. MetRS-m was identified in a screen for genes required for neuronal survival and function.Citation36 Mutant flies were characterized and found to exhibit defects in mitochondrial function and cell proliferation. The function of SerRS-m was analyzed through an RNAi approach.Citation37 This was shown to specifically reduce serylation of mitochondrial tRNAs, resulting in defective mitochondrial translation and function. AlaRS-m was studied to address how it distinguishes mitochondrial tRNAAla from the cytoplasmic tRNAAla.Citation38,39 Another series of experiments explored the compatibility between mitochondria-encoded tRNAs and their nucleus-encoded mitochondrial aaRSs.Citation40-42 While individual mutations in a mitochondrial tRNATyr gene and a mitochondrial TyrRS-m showed few phenotypic effects on their own, these mutations caused severe phenotypes coupled with reduced mitochondrial function when combined in the same fly.
Dual-localized aminoacyl-tRNA synthetases
Dual-localized aaRSs are encoded by single genes and perform aminoacylation of tRNAs in the cytoplasmic and the mitochondrial compartments. Our database searches uncovered 4 candidate dual-localized aaRSs in D. melanogaster - GlyRS, LysRS, HisRS, and ThrRS (). Significantly, and in contrast to the genes in the other 2 groups, each of these aaRS genes encodes at least 2 polypeptides. As the mitochondrial version needs an MTS, it is possible that the shorter polypeptide corresponds to the cytoplasmic version and the longer one to the mitochondrial isoform. Indeed, this has been experimentally confirmed for GlyRS ()Citation43 and was suggested for LysRS.Citation44 We analyzed the 2 other aaRSs using Mitoprot, a prediction tool for mitochondrial targeting sequences.Citation29 Indeed, HisRS and ThrRS each encode at least one longer polypeptide with high probability of mitochondrial localization (), strongly suggesting that these enzymes do indeed function as dual-localized aaRSs in Drosophila.
Despite the high conservation of these enzymes and their function through evolution, we noticed that the sets of fly and human dual-localized aaRS genes are not identical. Humans contain only 2 dual-localized aaRS genes, GlyRS and LysRS, while flies additionally have HisRS and ThrRS. This difference needs to be considered when modeling human diseases related to these 2 genes in flies.
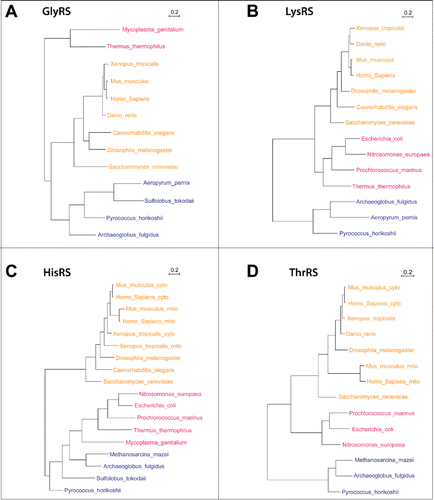
To obtain a better understanding of how these 4 enzymes evolved, we performed phylogenetic analyses with sequences from various eukaryotic species (including flies and human), bacteria and archaea (). All eukaryotic species analyzed contain a single GlyRS gene, which is more closely related to the one from archaea than to the bacterial one, suggesting that it originated from the cytoplasmic aminoacyl-tRNA synthetases. LysRS is also encoded by a single gene. However, the eukaryotic LysRS is closer to the bacterial ones, suggesting that it originated from a mitochondrial gene. Interestingly, lower eukaryotic species contain a single HisRS gene, just like bacteria and archaea, while vertebrates (higher eukaryotes) have 2 separate genes, demonstrating that splitting up their function into 2 separate genes and enzymes was beneficial to vertebrates. For ThrRS the available data suggest that lower eukaryotes may contain only one gene type that was derived from the bacterial/mitochondrial one. The phylogenetic tree then further suggests that there was a subsequent split into 2 types, but that this split was only maintained in some of the analyzed lineages, in the higher vertebrates (mammals). Clearly, more data points are needed to ascertain the apparently rather complex evolution of the ThrRS sequences.
Figure 3. Phylogenetic analysis of 4 dual-localized aaRSs. Protein sequences of common eukaryotes, archaea, and bacteria were obtained from different databases (UniProt, Ensembl, HGNC, FlyBase, Xenbase, WormBase), and also by searching with BLAST. The sequences were aligned using Pagan,Citation54 followed by TrimAl analysis,Citation55 discarding the poorly aligned columns with the threshold of 60%. The treated multiple sequence alignments were used to generate the 4 gene trees using PhyMLCitation56; for topology searches we chose the best out of the NNI and PhyML-Subtree-Pruning-Regrafting (SPR) methods.Citation57,58 All parameters were optimized, i.e., tree topology, branch length and the substitution rate. The number of bootstrap replicates was set to 5. Eukaryotes are shown in yellow, archaea in blue, and bacteria in red. The scale bar stands for the number of substitutions per site.
GlyRS is the only dual-localized aaRS that has been studied experimentally in Drosophila. It was initially identified in a mosaic forward genetic screen for genes having cell-autonomous functions in dendritic and axonal development.Citation43 While the cytoplasmic function of GlyRS was found to be required for terminal arborization of both dendrites and axons during development, the mitochondrial function is preferentially required for the maintenance of dendritic terminals in adults.
Drosophila as a model for aaRS-associated human diseases
Mutations in multiple aaRSs have been implicated in several different human diseases, though the mechanistic details are obscure in most cases.Citation7 Researchers have begun to use the power and efficiency of Drosophila genetics to model some of these diseases, and in so doing more readily investigate their molecular and cellular basis.
Charcot-Marie-Tooth (CMT) neuropathies affect the peripheral nervous system and are associated with axonal degeneration, distal muscle wasting and progressive motor impairment.Citation45 Mutations in the human YARS gene, encoding the cytoplasmic TyrRS, cause dominant-intermediate CMT type C (DI-CMTC).Citation46 Transgenic expression of either human or Drosophila TyrRS bearing disease-associated mutations in flies recapitulated several hallmarks of the human pathology, including progressive decreases in motor performance and axonal degeneration.Citation15,47 By virtue of studying these effects in flies, the authors were able to conclude that the disease phenotypes are not caused by reduced aminoacylation activity, but are more likely due to a gain-of-function alteration of the mutant TyrRS or interference with a non-canonical function.Citation47 This Drosophila disease model was also demonstrated to be a useful and rapid platform for screening the pathogenicity of novel candidate YARS mutations.Citation15
Mutations in a second human aaRS gene, GARS (encoding the dual-localized GlyRS enzyme), cause a different CMT subtype, CMT type 2D (CMT2D).Citation48 An initial study in flies found that loss-of-function mutations in the native Drosophila GlyRS gene resulted in neuronal phenotypes consistent with CMT2D symptoms in humans, although transgenic expression of disease-associated GARS mutations in neuronal clones had no morphological effect.Citation43 A recent study generated a more realistic Drosophila model for CMT2D through ubiquitous or pan-neuronal expression of fly GlyRS transgenes with alterations equivalent to those of pathogenic GARS mutations. These transgenic flies showed both morphological and behavioral phenotypes that recapitulated the human disease.Citation49 Significantly, these phenotypes were observed for disease-associated GlyRS mutants that maintained aminoacylation activity, suggesting that CMT2D is the result of a toxic, neomorphic activity, similar to the conclusion from the DI-CMTC model. A subsequent study confirmed these observations, and further suggested that the gain-of-function effects have a non-cell autonomous contribution.Citation50 Other recent work has generated a complementary Drosophila model of CMT2D in flies through expression of human GARS transgenes harboring disease-associated mutations.Citation51 The disease-relevant phenotypes were again found not to correlate with reduced aminoacylation activity of the enzyme. Nevertheless, a marked decrease in global protein synthesis in motor and sensory neurons was observed in the transgenic flies, suggesting that the mutant enzymes inhibit translation through a cell autonomous mechanism independent of their aminoacylation function. Interestingly, expression of DI-CMTC-associated YARS mutants also resulted in translation inhibition in this assay.Citation51 This finding, together with the facts that the phenotypes of the fly models of both CMT subtypes are similar and share common genetic modifiers,Citation49 suggests that a common mechanism may underlie both YARS- and GARS-associated CMT neuropathies.
Fly MetRS-m was identified in a screen for genes required for neuronal survival and function.Citation36 Mutant flies exhibited defects in mitochondrial function, cell proliferation and age-dependent retinal and muscle degeneration. Remarkably, these findings led to the discovery that mutations in the orthologous human gene, MARS2, are responsible for the neurodegenerative disease Autosomal Recessive Spastic Ataxia with Leukoencephalopathy (ARSAL). Similar to flies, cells from ARSAL patients showed aberrant mitochondrial function and proliferation. This study also reported that treatment with antioxidants could suppress the fly mutant phenotypes, indicating a possible treatment for the human disease.
A different study utilized RNAi to target SerRS-m to produce a fly model of human mitochondrial aminoacylation pathologies in general and mitochondrial serylation defects in particular.Citation37 For example, the fly phenotypes reproduce traits seen in MELAS (mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes) or MERRF (myoclonic epilepsy with ragged red fibers), as well as HUPRA syndrome (hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis), the latter of which is caused by a mutation in the orthologous SARS2 gene. Furthermore, it was found that antioxidant treatment ameliorated the phenotypes resulting from SerRS-m silencing,Citation37 similar to the MetRS-m study.Citation36
Finally, Drosophila TyrRS-m has been studied as a general model for human mitochondrial diseases stemming from an incompatibility between the nuclear-encoded aaRSs and mitochondrially-encoded tRNAs.Citation42 In this model, a mutation in TyrRS-m resulted in defective mitochondrial dysfunction and locomotor defects, though the severity varied across different genetic backgrounds and traits. These context-dependent phenotypes mirror the symptoms of the MLASA syndrome (mitochondrial myopathy, lactic acidosis and sideroblastic anemia) that results from mutations in the orthologous human gene YARS2.
In summary, several different aaRS-associated human diseases have so far been effectively modeled in Drosophila using a variety of genetic techniques. These approaches have generated a number of clinically important conclusions, including insights into etiology of CMTCitation15,47,49-51 and the discovery of the underlying cause of ARSAL.Citation36 Moreover, some of these studies have isolated modifiers of the disease model,Citation36,37,49 demonstrating a further advantage of using the Drosophila system.
Conclusion
Most studies of aaRS biology in Drosophila to date have either investigated their canonical functions or have used mutant genotypes to produce models of human diseases linked to aaRS dysfunction. In addition, several aaRSs have been identified in diverse genetic screens, though their precise role in these conditions remains unclear. Notably, the non-canonical roles of aaRSs that have been described in other systems have so far received sparse attention in Drosophila, while many aaRS-associated human diseases have yet to be modeled in flies. Based on these considerations we predict an increase in such studies in the near future. The systematic identification and logical naming of the D. melanogaster aaRSs presented here, together with our literature survey, will aid all these lines of investigation, and thereby facilitate further discoveries into both the normal and aberrant mechanisms of action of these essential and fascinating enzymes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Author Contributions
J.L. and S.J.M. performed the analysis, J.L. and W.H.G. constructed the phylogenetic trees, J.L. prepared the manuscript, S.J.M. and B.S. contributed to and edited the manuscript.
1101196_Supplemental_Material.zip
Download Zip (66 KB)Acknowledgments
We thank Rémy Bruggmann for his support on the bioinformatics side.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
S.J.M. is supported by the FlyBase NIH/NHGRI grant U41HG000739 (W.M. Gelbart, Harvard University, PI; N.H. Brown, University of Cambridge, coPI). J.L. was supported by Cancer Research Switzerland and a SNF grant to B.S., and by a SNSF Early Postdoc Fellowship.
References
- Schimmel PR, Soll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem 1979; 48:601-48; PMID:382994; http://dx.doi.org/10.1146/annurev.bi.48.070179.003125
- Schimmel P. Development of tRNA synthetases and connection to genetic code and disease. Protein Sci 2008; 17:1643-52; PMID:18765819; http://dx.doi.org/10.1110/ps.037242.108
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem 2000; 69:617-50; PMID:10966471; http://dx.doi.org/10.1146/annurev.biochem.69.1.617
- de Duve C. Transfer RNAs: the second genetic code. Nature 1988; 333:117-8; PMID:3367984; http://dx.doi.org/10.1038/333117a0
- Arnez JG, Moras D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem Sci 1997; 22:211-6; PMID:9204708; http://dx.doi.org/10.1016/S0968-0004(97)01052-9
- Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet 2008; 9:87-107; PMID:18767960; http://dx.doi.org/10.1146/annurev.genom.9.081307.164204
- Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med 2013; 5:332-43; PMID:23427196; http://dx.doi.org/10.1002/emmm.201100626
- Brown MV, Reader JS, Tzima E. Mammalian aminoacyl-tRNA synthetases: cell signaling functions of the protein translation machinery. Vasc Pharmacol 2010; 52:21-6; PMID:19962454; http://dx.doi.org/10.1016/j.vph.2009.11.009
- Kim S, You S, Hwang D. Aminoacyl-tRNA synthetases and tumorigenesis: more than housekeeping. Nat Rev Cancer 2011; 11:708-18; PMID:21941282; http://dx.doi.org/10.1038/nrc3124
- Dolde C, Lu J, Suter B. Cross talk between cellular regulatory networks mediated by shared proteins. Adv Biol 2014; 2014: Article ID 274196; http://dx.doi.org/10.1155/2014/274196
- Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol 2010; 11:668-74; PMID:20700144; http://dx.doi.org/10.1038/nrm2956
- Wallen RC, Antonellis A. To charge or not to charge: mechanistic insights into neuropathy-associated tRNA synthetase mutations. Curr Opin Genet Dev 2013; 23:302-9; PMID:23465884; http://dx.doi.org/10.1016/j.gde.2013.02.002
- Konovalova S, Tyynismaa H. Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol Genet Metab 2013; 108:206-11; PMID:23433712; http://dx.doi.org/10.1016/j.ymgme.2013.01.010
- Bilen J, Bonini NM. Drosophila as a model for human neurodegenerative disease. Annu Rev Genet 2005; 39:153-71; PMID:16285856; http://dx.doi.org/10.1146/annurev.genet.39.110304.095804
- Leitao-Goncalves R, Ermanoska B, Jacobs A, De Vriendt E, Timmerman V, Lupski JR, Callaerts P, Jordanova A. Drosophila as a platform to predict the pathogenicity of novel aminoacyl-tRNA synthetase mutations in CMT. Amino Acids 2012; 42:1661-8; PMID:21384131; http://dx.doi.org/10.1007/s00726-011-0868-4
- Lu J, Bergert M, Walther A, Suter B. Double-sieving-defective aminoacyl-tRNA synthetase causes protein mistranslation and affects cellular physiology and development. Nat Commun 2014; 5:5650; PMID:25427601; http://dx.doi.org/10.1038/ncomms6650
- Seshaiah P, Andrew DJ. WRS-85D: A tryptophanyl-tRNA synthetase expressed to high levels in the developing Drosophila salivary gland. Mol Biol Cell 1999; 10:1595-608; PMID:10233165; http://dx.doi.org/10.1091/mbc.10.5.1595
- dos Santos G, Schroeder AJ, Goodman JL, Strelets VB, Crosby MA, Thurmond J, Emmert DB, Gelbart WM, FlyBase Consortium. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res 2015; 43:D690-7; PMID:25398896; http://dx.doi.org/10.1093/nar/gku1099
- Gray KA, Yates B, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2015. Nucleic Acids Res 2015; 43:D1079-85; PMID:25361968; http://dx.doi.org/10.1093/nar/gku1071
- Cerini C, Kerjan P, Astier M, Gratecos D, Mirande M, Semeriva M. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J 1991; 10:4267-77; PMID:1756734
- Guitart T, Leon Bernardo T, Sagales J, Stratmann T, Bernues J, Ribas de Pouplana L. New aminoacyl-tRNA synthetase-like protein in insecta with an essential mitochondrial function. J Biol Chem 2010; 285:38157-66; PMID:20870726; http://dx.doi.org/10.1074/jbc.M110.167486
- Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 1998; 282:2082-5; PMID:9851929; http://dx.doi.org/10.1126/science.282.5396.2082
- Cerini C, Semeriva M, Gratecos D. Evolution of the aminoacyl-tRNA synthetase family and the organization of the Drosophila glutamyl-prolyl-tRNA synthetase gene. Intron/exon structure of the gene, control of expression of the two mRNAs, selective advantage of the multienzyme complex. Eur J Biochem 1997; 244:176-85; PMID:9063462; http://dx.doi.org/10.1111/j.1432-1033.1997.00176.x
- Berthonneau E, Mirande M. A gene fusion event in the evolution of aminoacyl-tRNA synthetases. FEBS Lett 2000; 470:300-4; PMID:10745085; http://dx.doi.org/10.1016/S0014-5793(00)01343-0
- Kerjan P, Cerini C, Semeriva M, Mirande M. The multienzyme complex containing nine aminoacyl-tRNA synthetases is ubiquitous from Drosophila to mammals. Biochim Biophys Acta 1994; 1199:293-7; PMID:8161568; http://dx.doi.org/10.1016/0304-4165(94)90009-4
- Stitzinger SM, Pellicena-Palle A, Albrecht EB, Gajewski KM, Beckingham KM, Salz HK. Mutations in the predicted aspartyl tRNA synthetase of Drosophila are lethal and function as dosage-sensitive maternal modifiers of the sex determination gene Sex-lethal. Mol Gen Genet 1999; 261:142-51; PMID:10071220; http://dx.doi.org/10.1007/s004380050951
- Galloni M, Edgar BA. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 1999; 126:2365-75; PMID:10225996
- Arsham AM, Neufeld TP. A genetic screen in Drosophila reveals novel cytoprotective functions of the autophagy-lysosome pathway. PLoS One 2009; 4:e6068; PMID:19562034; http://dx.doi.org/10.1371/journal.pone.0006068
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 1996; 241:779-86; PMID:8944766; http://dx.doi.org/10.1111/j.1432-1033.1996.00779.x
- Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc Natl Acad Sci U S A 1968; 61:229-36; PMID:4972364; http://dx.doi.org/10.1073/pnas.61.1.229
- Schon A, Kannangara CG, Gough S, Soll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature 1988; 331:187-90; PMID:3340166; http://dx.doi.org/10.1038/331187a0
- Nagao A, Suzuki T, Katoh T, Sakaguchi Y, Suzuki T. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc Natl Acad Sci U S A 2009; 106:16209-14; PMID:19805282; http://dx.doi.org/10.1073/pnas.0907602106
- Morris JZ, Bergman L, Kruyer A, Gertsberg M, Guigova A, Arias R, Pogorzelska M. Mutations in the Drosophila mitochondrial tRNA amidotransferase, bene/gatA, cause growth defects in mitotic and endoreplicating tissues. Genetics 2008; 178:979-87; PMID:18245325; http://dx.doi.org/10.1534/genetics.107.084376
- Bullard JM, Cai YC, Demeler B, Spremulli LL. Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol 1999; 288:567-77; PMID:10329163; http://dx.doi.org/10.1006/jmbi.1999.2708
- Liao TS, Call GB, Guptan P, Cespedes A, Marshall J, Yackle K, Owusu-Ansah E, Mandal S, Fang QA, Goodstein GL, et al. An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function. Genetics 2006; 174:525-33; PMID:16849596; http://dx.doi.org/10.1534/genetics.106.061705
- Bayat V, Thiffault I, Jaiswal M, Tetreault M, Donti T, Sasarman F, Bernard G, Demers-Lamarche J, Dicaire MJ, Mathieu J, et al. Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol 2012; 10:e1001288; PMID:22448145; http://dx.doi.org/10.1371/journal.pbio.1001288
- Guitart T, Picchioni D, Pineyro D, Ribas de Pouplana L. Human mitochondrial disease-like symptoms caused by a reduced tRNA aminoacylation activity in flies. Nucleic Acids Res 2013; 41:6595-608; PMID:23677612; http://dx.doi.org/10.1093/nar/gkt402
- Lovato MA, Chihade JW, Schimmel P. Translocation within the acceptor helix of a major tRNA identity determinant. EMBO J 2001; 20:4846-53; PMID:11532948; http://dx.doi.org/10.1093/emboj/20.17.4846
- Lovato MA, Swairjo MA, Schimmel P. Positional recognition of a tRNA determinant dependent on a peptide insertion. Mol Cell 2004; 13:843-51; PMID:15053877; http://dx.doi.org/10.1016/S1097-2765(04)00125-X
- Hoekstra LA, Siddiq MA, Montooth KL. Pleiotropic effects of a mitochondrial-nuclear incompatibility depend upon the accelerating effect of temperature in Drosophila. Genetics 2013; 195:1129-39; PMID:24026098; http://dx.doi.org/10.1534/genetics.113.154914
- Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL. An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet 2013; 9:e1003238; PMID:23382693; http://dx.doi.org/10.1371/journal.pgen.1003238
- Holmbeck MA, Donner JR, Villa-Cuesta E, Rand DM. A Drosophila model for mito-nuclear diseases generated by an incompatible interaction between tRNA and tRNA synthetase. Dis Models Mech 2015; 8:843-54; PMID:26035388; http://dx.doi.org/10.1242/dmm.019323
- Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci 2007; 10:828-37; PMID:17529987; http://dx.doi.org/10.1038/nn1910
- Tolkunova E, Park H, Xia J, King MP, Davidson E. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J Biol Chem 2000; 275:35063-9; PMID:10952987; http://dx.doi.org/10.1074/jbc.M006265200
- Reilly MM, Murphy SM, Laura M. Charcot-Marie-Tooth disease. J Peripher Nerv Syst 2011; 16:1-14; PMID:21504497; http://dx.doi.org/10.1111/j.1529-8027.2011.00324.x
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, Dierick I, Jacobs A, De Vriendt E, Guergueltcheva V, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet 2006; 38:197-202; PMID:16429158; http://dx.doi.org/10.1038/ng1727
- Storkebaum E, Leitao-Goncalves R, Godenschwege T, Nangle L, Mejia M, Bosmans I, Ooms T, Jacobs A, Van Dijck P, Yang XL, et al. Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc Natl Acad Sci U S A 2009; 106:11782-7; PMID:19561293; http://dx.doi.org/10.1073/pnas.0905339106
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 2003; 72:1293-9; PMID:12690580; http://dx.doi.org/10.1086/375039
- Ermanoska B, Motley WW, Leitao-Goncalves R, Asselbergh B, Lee LH, De Rijk P, Sleegers K, Ooms T, Godenschwege TA, Timmerman V, et al. CMT-associated mutations in glycyl- and tyrosyl-tRNA synthetases exhibit similar pattern of toxicity and share common genetic modifiers in Drosophila. Neurobiol Dis 2014; 68:180-9; PMID:24807208; http://dx.doi.org/10.1016/j.nbd.2014.04.020
- Grice SJ, Sleigh JN, Motley WW, Liu JL, Burgess RW, Talbot K, Cader MZ. Dominant, toxic gain-of-function mutations in gars lead to non-cell autonomous neuropathology. Hum Mol Genet 2015; 24:4397-406; PMID:25972375; http://dx.doi.org/10.1093/hmg/ddv176
- Niehues S, Bussmann J, Steffes G, Erdmann I, Kohrer C, Sun L, Wagner M, Schäfer K, Wang G, Koerdt SN, et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat Commun 2015; 6:7520; PMID:26138142; http://dx.doi.org/10.1038/ncomms8520
- Otani A, Slike BM, Dorrell MI, Hood J, Kinder K, Ewalt KL, Cheresh D, Schimmel P, Friedlander M. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci U S A 2002; 99:178-83; PMID:11773625; http://dx.doi.org/10.1073/pnas.012601899
- Cheng G, Zhang H, Yang X, Tzima E, Ewalt KL, Schimmel P, Faber JE. Effect of mini-tyrosyl-tRNA synthetase on ischemic angiogenesis, leukocyte recruitment, and vascular permeability. Am J Physiol Regul Integr Comp Physiol 2008; 295:R1138-46; PMID:18753262; http://dx.doi.org/10.1152/ajpregu.90519.2008
- Loytynoja A, Vilella AJ, Goldman N. Accurate extension of multiple sequence alignments using a phylogeny-aware graph algorithm. Bioinformatics 2012; 28:1684-91; PMID:22531217; http://dx.doi.org/10.1093/bioinformatics/bts198
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009; 25:1972-3; PMID:19505945; http://dx.doi.org/10.1093/bioinformatics/btp348
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010; 59:307-21; PMID:20525638; http://dx.doi.org/10.1093/sysbio/syq010
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003; 52:696-704; PMID:14530136; http://dx.doi.org/10.1080/10635150390235520
- Hordijk W, Gascuel O. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 2005; 21:4338-47; PMID:16234323; http://dx.doi.org/10.1093/bioinformatics/bti713
