ABSTRACT
The analysis of consequences resulting after experimental elimination of gene function has been and will continue to be an extremely successful strategy in biological research. Mutational elimination of gene function has been widely used in the fly Drosophila melanogaster. RNA interference is used extensively as well. In the fly, exceptionally precise temporal and spatial control over elimination of gene function can be achieved in combination with sophisticated transgenic approaches and clonal analyses. However, the methods that act at the gene and transcript level cannot eliminate protein products which are already present at the time when mutant cells are generated or RNA interference is started. Targeted inducible protein degradation is therefore of considerable interest for controlled rapid elimination of gene function. To this end, a degradation system was developed in yeast exploiting TIR1, a plant F box protein, which can recruit proteins with an auxin-inducible degron to an E3 ubiquitin ligase complex, but only in the presence of the phytohormone auxin. Here we demonstrate that the auxin-inducible degradation system functions efficiently also in Drosophila melanogaster. Neither auxin nor TIR1 expression have obvious toxic effects in this organism, and in combination they result in rapid degradation of a target protein fused to the auxin-inducible degron.
Introduction
The stability of gene products often limits the speed of their experimental depletion. Maternally contributed mRNA and protein can delay and blur the development of abnormal phenotypes in progeny lacking zygotic function of a particular gene. Perdurance of mRNA and protein can also result in gradually changing phenotypes that sometimes impede accurate interpretations after generation of mutant cells by mitotic recombination or comparable approaches in clonal analyses. In case of RNA interference approaches, perdurance of the protein product can mask or delay manifestation of consequences. To circumvent such problems, methods allowing regulated efficient degradation of specific target proteins have been developed.Citation1
In Drosophila, for example, expression of tobacco etch virus protease (TEV) for degradation of a TEV-cleavable Rad21/Vtd variant, a functional cohesin complex subunit, has been shown to result in an apparent null phenotype within the first cell cycle after the onset of TEV expression when expressed instead of endogenous Rad21/Vtd.Citation2 TEV is also exploited in a more versatile strategy involving the N-terminal TIPI tagCitation3 that exposes a destabilizing N-terminus after TEV cleavage followed by degradation via the N-end rule pathway.Citation4 The N end rule pathway recognizes destabilizing N-terminal amino acids by a dedicated ubiquitin ligase, resulting in polyubiquitylation and proteasomal degradation. Interestingly, an N-end rule degron with temperature-sensitive activity has also been developedCitation5 which has been applied in Drosophila to some limited extent.Citation6,7
An elegant method (deGradFP) for targeted degradation of GFP fusion proteins involves expression of a recombinant protein (NSlmb-vhh-GFP4) with an F-box fused to a camelid single chain antibody against GFP.Citation8 F-boxes mediate binding to Skp1, a component of E3 ubiquitin ligase complexes of the Skp1-Cullin-F-box (SCF) type. F-box proteins function as substrate adaptors in these E3 complexes with the help of a second domain allowing specific binding of target proteins. Thereby target proteins are recruited and polyubiquitylated, followed by proteasomal degradation. Expression of NSlmb-vhh-GFP4 generates an SCF ubiquitin ligase complex specific for GFP fusion proteins. This method for specific depletion of GFP fusion proteins has been successfully applied in Drosophila.Citation8-11
While deGradFP acts at the protein level, it is neither particularly fast nor readily reversible. NSlmb-vhh-GFP4 expressed from transgenes needs to accumulate to effective intracellular concentrations that cannot be lowered again rapidly. For the induction of rapid and reversible degradation, activity regulation of degrons by small chemicals is of great interest. A few such controllable degrons have been developed.Citation1 A most successful versionCitation12 is based on the molecular signaling mechanisms of the plant hormone auxin (indole-3-acetic acid, IAA). In plants, auxin acts as molecular glue that mediates specific binding of transcriptional repressors of the AUX/IAA family to the plant-specific F-box protein TIR1. Auxin therefore results in polyubiquitylation and proteasomal degradation of these transcriptional regulators.Citation13-15 The transcriptional repressors, including IAA17, contain an auxin-inducible degron domain (AID) that is required and sufficient for binding to TIR1. Heterologous TIR1 expression in yeast was demonstrated to result in association with endogenous Skp1 and formation of a functional, auxin-dependent SCF ubiquitin ligase.Citation12,16 Fusion proteins containing an AID are rapidly degraded in an auxin-dependent manner. Beyond yeast, the auxin-inducible degradation system has been shown to function in mammalian cells,Citation17 PlasmodiumCitation18,19 and most recently even in the complex metazoan model organism Caenorhabditis elegans.Citation20 Here, we report that this system also functions well in Drosophila melanogaster and describe transgenic strains for its application.
Results and discussion
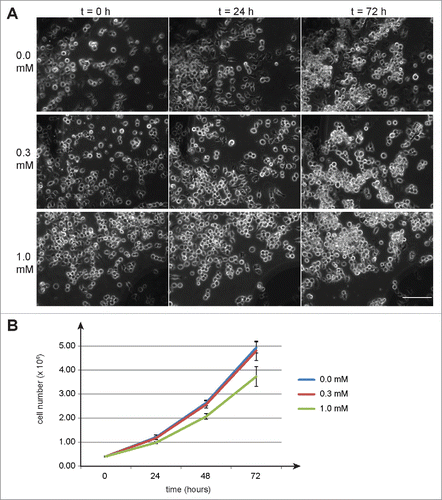
As a first step in our evaluation of the functionality of the auxin-dependent degradation system in Drosophila, we addressed whether auxin might have adverse effects on cultured cells. A suspension of S2R+ cells was distributed into multiwell plates and 24 h later auxin in increasing concentrations was added to the culture medium (final concentration 0, 0.3, 0.6, and 1 mM). Time lapse imaging was used to monitor cells over the next 3 d. Even at the highest concentration we did not observe obvious effects on cell morphology and cell numbers (, data not shown). In a second experiment, cells were harvested from replicate cultures each day after auxin addition and used for determination of cell numbers and viability (). While 0.3 mM auxin did not have a significant effect, 1 mM reduced cell doubling time slightly by about 10% compared to control cells growing in the absence of auxin. Adverse effects on cell viability were not observed even at the highest concentration (1 mM). To evaluate whether auxin might affect Drosophila development, we collected eggs from a w1 strain into vials with food containing auxin at different concentrations (0, 0.3, 0.6, and 1 mM). From all the vials, adult flies were observed to eclose in comparable numbers over a comparable time span. Because of our interest in future applications of the auxin-inducible degradation system during male meiosis, we specifically tested the fertility of males that had developed in the presence of increasing concentrations of auxin. We did not observe any adverse effects of auxin onto male fertility. We conclude that auxin is not acutely toxic for Drosophila melanogaster at the concentrations analyzed. Consistent with our observations, previous analyses have failed to detect mutagenic effects of auxin after feeding Drosophila larvae with concentrations up to 20 mM.Citation21
Figure 1. Auxin effects on Drosophila S2R+ cells. To evaluate potential toxicity of auxin, S2R+ cells were cultured in the presence of auxin at the indicated concentrations (mM). (A) No obvious effects of auxin on cell morphology were observed by imaging defined regions repeatedly at the indicated time points (hours) after auxin addition. Scale bar = 100 µm. (B) Cell counting revealed a slight inhibitory effect of 1 mM auxin on cell proliferation, while 0.3 mM did not appear to have a significant effect. Average cell numbers (+/− s.d., n = 3) at the indicated time points (hours) after addition of auxin (0, 0.3 and 1 mM) are shown.
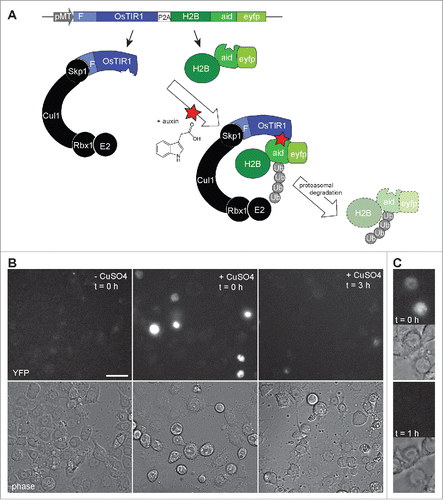
To confirm that auxin actually penetrates into cultured S2R+ cells, we generated a test construct for transient transfection experiments (). This construct (pMT-OsTIR1-P2A-H2B-aid-eyfp) allowed expression of rice TIR1 (OsTIR1) coupled via a “self-cleaving” 2A peptideCitation22 to human histone H2B fused to the auxin-inducible degradation domain (aid) and yellow fluorescent protein (eyfp). After transfection and induction of expression from the construct, strong nuclear YFP signals were observed in transfected cells (). Reassuringly, these signals were no longer observed when cells were fixed 3 h after addition of auxin (1 mM) (). Moreover, time lapse imaging confirmed that the strong nuclear YFP signals vanished within 1 h after auxin addition (). We conclude that auxin penetrates readily into Drosophila cells to reach concentrations capable of inducing degradation of proteins with an auxin-inducible degradation domain.
Figure 2. Auxin induced degradation in Drosophila S2R+ cells. (A) Scheme illustrating the characteristic features of the pMT-OsTIR1-P2A-H2B-aid-eyfp construct and of the auxin-inducible degradation system. The MtnA promoter (pMT) controls expression of an mRNA that includes the sequence of a “self-cleaving” 2A peptide (P2A). Therefore, the mRNA generates 2 distinct proteins. The first is the protein TIR1 from rice (OsTIR1) which includes an F box (F) that allows integration into an SCF ubiquitin ligase complex together with the endogenous Drosophila proteins Skp1, Cul1, Rbx1 and an E2 protein. The second protein is a histone H2B fusion protein with a C-terminal extension that consists of the auxin-inducible degron (aid) followed by EYFP (eyfp). In the presence of auxin, aid-containing proteins are recruited to OsTIR1, resulting in their polyubiquitinylation and proteasomal degradation. (B) The construct illustrated in panel A was transfected into S2R+ cells. Construct expression was either induced (+ CuSO4) or not induced (- CuSO4) before fixation and imaging. The strong nuclear YFP signals, which were observed in transfected and induced cells before addition of auxin (t = 0h), were no longer present when cells were fixed 3 h after addition of auxin (t = 3h). Scale bar = 20 µm. (C) By time lapse imaging the nuclear YFP signals were confirmed to disappear from transfected and induced S2R+ cells within less than an hour after addition of auxin. The upper images (t = 0h) were acquired immediately before and the lower images (t = 1h) one hour after auxin addition.
To demonstrate the functionality of the auxin-inducible degradation system in the organism, we generated transgenic Drosophila strains. A first construct was used for generation of UASt-OsTIR1 transgenic strains allowing GAL4-dependent expression of OsTIR1. In addition, we generated Ubi-OsTIR1 strains expressing OsTIR1 ubiquitously under control of the Ubi-p63E promoter. Finally, we generated UASt-aid-rux strains allowing GAL4-dependent expression of the auxin-inducible domain fused to Roughex. Drosophila roughex (rux) codes for a Cdk inhibitor.Citation23,24 rux overexpression is known to interfere with normal cell proliferation.Citation23 We observed that the fusion protein Aid-Rux also caused severe developmental abnormalities after expression of UASt-aid-rux with various tissue-specific GAL4 drivers (ey-GAL4, GMR-GAL4, and MS1096) or lethality with more global drivers (Act5C-GAL4 and en-GAL4). To evaluate the auxin-dependent degradation system, we crossed males with UASt-aid-rux and either UASt-OsTIR1 or Ubi-OsTIR1 with en-GAL4 virgin females. From these crosses, eggs were collected into vials with fly food that either did or did not contain 1 mM auxin. After incubation of the vials at 25°C, we counted the number of pupae and adult flies that developed. In the experiments with Ubi-OsTIR1, the number of pupae was lower in the absence of auxin (287 without and 346 with auxin). Moreover, not a single adult fly was observed to eclose in the absence of auxin, while in the presence of auxin 8 flies developed to the adult stage (). However, all these adults displayed morphological abnormalities most prominently in the wings. Often one of the 2 wings was missing. The wings that were present had reduced posterior compartments (). In contrast, the wings of flies with only en-GAL4 or only UASt-aid-rux and Ubi-OsTIR1 were entirely normal (data not shown).
Figure 3. Auxin and OsTIR1 expression suppress Aid-Rux induced lethality during Drosophila development. (A) Schematic illustration of the experimental strategy used for the evaluation of the auxin-inducible degradation system during Drosophila development. Eggs with the indicated genotypes were collected on fly food with or without auxin. While complete developmental lethality was observed in the absence of auxin, the indicated number of adult flies eclosed in the presence of auxin. (B) The posterior wing compartments of en-GAL4>UASt-aid-rux, Ubi-OsTIR1 flies after development in the presence of auxin were observed to be severely reduced. (C) The posterior wing compartments of en-GAL4>UASt-aid-rux, UASt-OsTIR1 flies after development in the presence of auxin were observed to be abnormal as well. Abnormalities were more severe after development in the presence of 0.3 mM compared to 1 mM auxin. While the size of the posterior compartment was usually close to normal, it displayed a multiple wing hair phenotype, as clearly apparent in the high magnification view on the right. Scale bar = 0.4 mm.
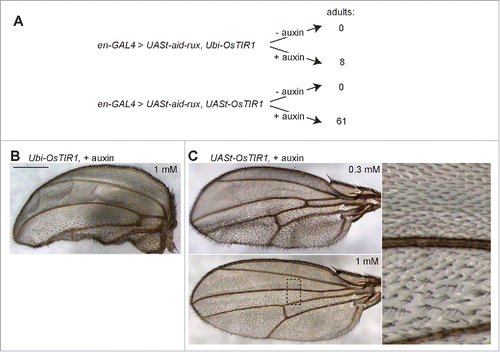
In the experiments with UAS-OsTIR1, the protecting effect of auxin against aid-rux-induced lethality was more prominent. In the presence of auxin, more pupae (294 without and 452 with auxin) and far more adult flies (0 without and 61 with auxin) were obtained compared to absence of auxin (). Those adult flies with UASt-OsTIR1 that were obtained in the presence of auxin were more normal than the few obtained with Ubi-OsTIR1. But also in the UASt-OsTIR1 case, adult wings were not entirely normal (). Abnormalities were more severe after development in lower concentrations of auxin (0.3 mM compared to 1 mM) (). Instead of the regular pattern of wing hairs, a multiple wing hair phenotype was observed in the posterior compartment. During normal development, each cell within the dorsal and ventral wing epithelium produces a single wing hair. We have previously shown that Cdk1 inhibition specifically during the pupal stages disturbs the formation of a single wing hair per cell.Citation25 After Cdk1 inhibition during the pupal stages, wing imaginal disc cells progress through endoreduplication cycles instead of going through the 2 final mitotic cell cycles. The resulting oversized cells produce multiple instead of a single wing hairs during terminal differentiation.Citation25 The presence of a multiple wing hair phenotype in the posterior compartment of en-GAL4>UASt-aid-rux, UASt-OsTIR1 flies indicates that the levels of auxin appear to drop to an ineffective concentration after termination of auxin food uptake at the onset of larval wandering for preparation of pupariation at the end of the third larval instar. In addition, the finding that adult flies were only obtained in the presence of auxin strongly suggested that the auxin-inducible degradation system is functional in Drosophila.
To study the effects of the auxin-induced degradation system more immediately at the cellular level, we analyzed wing imaginal discs. After development of en-GAL4 larvae with UASt-aid-rux and either UASt-OsTIR1 or Ubi-OsTIR1 in the absence of auxin, the posterior compartment was found to be strongly abnormal (). DNA staining revealed the presence of highly endoreduplicated cells within the posterior compartment. Moreover, the overall shape of the disc was distorted to a variable extent. Development in the presence of auxin prevented these abnormalities ().
Figure 4. Aid-Rux depletion by auxin and OsTIR1 expression suppresses endoreduplication in wing discs. (A) Wing imaginal discs from third instar wandering stage larvae with en-GAL4>UASt-aid-rux and either UASt-OsTIR1 or Ubi-OsTIR1, as indicated, were fixed after development in the absence (−) or presence (+) of auxin. DNA staining revealed endoreduplicated nuclei at lower density in the posterior wing disc compartment of larvae grown in the absence of auxin. Boxed regions are shown at higher magnification in the bottom panels. The effects of aid-rux expression in the posterior compartment resulted in variable distortions of the wing imaginal discs, as evident in the example shown in the leftmost panel where the posterior compartment runs oblique across the disc. Scale bar = 50 µm. (B) Larvae with en-GAL4, tubP-GAL80ts>UASt-aid-rux, UASt-OsTIR1 were grown initially at 18°C. GAL4-mediated expression of the UASt transgenes was induced for 24 h at 29°C. Larvae were then transferred into liquid food without (- auxin) or with (+ auxin) auxin for the indicated time (hours). Wing imaginal discs were dissected and stained with anti-Rux, anti-Cyclin A and a DNA stain. Wing pouch regions with anterior and posterior compartment on the left and right side, respectively, are shown. Aid-Rux is degraded more rapidly in the presence of auxin. Scale bar = 10 µm.
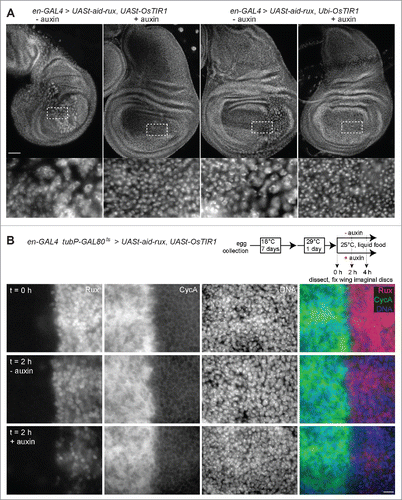
To analyze the dynamics of auxin-induced degradation in further detail, we generated larvae carrying the transgenes en-GAL4, UASt-OsTIR1 and UASt-aid-rux, as well as tubP-GAL80ts (). Growing these larvae initially at 18°C prevented cells within the posterior compartment from becoming highly abnormal in the absence of auxin. Expression of UASt-OsTIR1 and UASt-aid-rux was then induced eventually during 24 h by shifting the larvae to 29°C. Thereafter larvae were transferred to liquid food that either did or did not contain auxin. Wing discs were dissected and fixed at different time points after transfer to liquid food. As expected, immunolabeling with anti-Rux clearly revealed the presence of Aid-Rux in the posterior compartment at the onset of feeding with liquid food (). Moreover, double labeling with anti-Cyclin A revealed that the accumulation of Aid-Rux was paralleled by the disappearance of Cyclin A (), as previously reported.Citation23 Importantly, in the presence of auxin, the intensity of the anti-Rux signals were observed to drop rapidly. Signals were reduced to 22% (± 13 s.d., n = 17 wing discs) and 0.5% (± 0.7 s.d., n = 20) after 2 and 4 h in liquid auxin food, respectively. Presumably because Cyclin A re-accumulation is a comparatively slow process, we did not observe a converse recovery of anti-Cyclin A signal intensities. In addition, somewhat unexpectedly, anti-Rux signal intensities also went down in the absence of auxin () to 41% (± 12 s.d., n = 20) and 15% (± 8 s.d., n = 20) after 2 and 4 h, respectively. However, the reduction in the absence of auxin was significantly less extensive than that in the presence of auxin at both time points (p < 0.0001, t-test). We conclude that Aid-Rux has a limited stability even in the absence of auxin-induced degradation. Moreover, auxin-induced degradation makes it highly unstable.
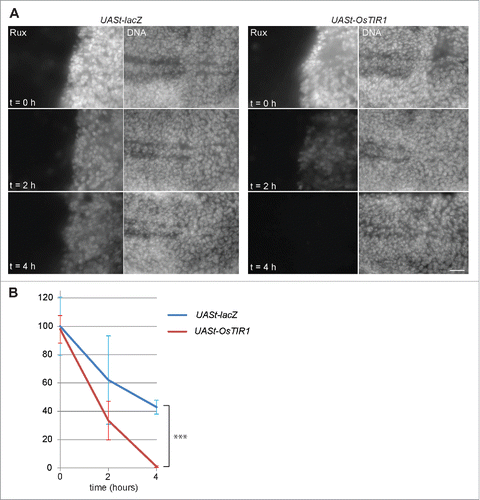
In principle, the limited stability of Aid-Rux in the absence of auxin observed in our experiments might reflect an auxin-independent activity of OsTIR1 in Drosophila. To address whether OsTIR1 might have auxin-independent activity, we performed additional experiments with en-GAL4, tub-GAL80ts, UASt-aid-rux larvae that had either a UASt-OsTIR1 or a UASt-lacZ transgene. After initial development at 18°C, UASt transgene expression was again induced (24 h at 29°C) before transfer to auxin containing liquid food. Signal intensities obtained with anti-Rux at the onset of feeding with liquid food were comparable in the UASt-OsTIR1 and UASt-lacZ wing discs (), indicating that OsTIR1 does not have substantial auxin-independent activity. In addition, analysis of the anti-Rux signal intensities after 2 and 4 h in liquid food provided further confirmation that auxin induces OsTIR1-mediated degradation. The drop in signal intensity was far more drastic in the discs expressing UASt-OsTIR1 compared to those expressing UASt-lacZ (). We conclude that the auxin-dependent degradation system functions also in Drosophila as expected.
Figure 5. OsTIR1 does not cause auxin-independent Aid-Rux degradation. (A) en-GAL4, tubP-GAL80ts>UASt-aid-rux larvae with either UASt-lacZ or UASt-OsTIR1, as indicated, were grown initially at 18°C. GAL4-mediated expression of the UASt transgenes was induced for 24 h at 29°C. Larvae were then transferred for the indicated time (hours) into auxin containing liquid food. Wing imaginal discs were dissected and stained with anti-Rux and a DNA stain. OsTIR1 does not lower Aid-Rux levels before addition of auxin but results in rapid degradation after addition of auxin. Scale bar = 20 µm. (B) Anti-Rux signals in the posterior compartment of wing discs obtained in the experiment illustrated in (A) were quantified. Average signals were normalized with those observed in en-GAL4, tubP-GAL80ts>UASt-aid-rux, UASt-lacZ which were set to 100%; whiskers indicate s.d. (n = 3), *** p = 0.00013 (t-test).
Our work indicates that the auxin-inducible degradation should be an attractive option for spatially and temporally precise depletion of proteins of interest in Drosophila melanogaster. Already the recent evaluation of this approach in Caenorhabditis elegansCitation20 has demonstrated impressively that it functions not only in yeast and cultured cells but also in complex metazoan organism. Moreover, the analyses in the nematode suggest interesting perspectives for further improvements. Instead of the complete AID from IAA17 (229 amino acids) a subregion of only 44 amino acids was shown to function as an efficient auxin-dependent degron in the nematode. Moreover, it is readily conceivable that the TIR1 gene version from Arabidopsis thaliana with 2 point mutations improving affinity and auxin sensitivity, which has been used very successfully in the nematode, might perform better than rice TIR1 that was used in our experiments.
Materials and methods
Plasmids
For the construction of pMT-OsTIR1-P2A-H2B-aid-eyfp we used a plasmid with an insert coding for human histone H2B fused the auxin-inducible degradation domain (aid) and enhanced yellow fluorescent protein (eyfp)Citation17 as well as pNHK36 containing the OsTIR1 coding sequenceCitation12 and pC5Kan-P2A (Addgene #5184).Citation22 Primers AB108 (5′-TGCC AGATCT ATGCCAGAGCCAGCGAAGTC-3′) and AB109 (5′-CGGG ACGCGT TCTAGATTACTTGTACAGCTCGTCCA-3′) were used for enzymatic amplification of the H2B-aid-eyfp fragment. After digestion with BglII and MluI the fragment was inserted into the corresponding restriction sites of pC5-Kan-P2A. Into the Acc65I and SalI sites of the resulting cloning intermediate, the OsTIR1 sequence was inserted after enzymatic amplification with the primers AB110 (5′-ATCC GGTACC ATGACGTACTTCCCGGAGGA-3′) and AB111 (5′-ACCG GTCGAC GCTAGGATTTTAACAAAATTTG-3′) and digestion with the corresponding enzymes. From this second cloning intermediate, the Os-TIR1-P2A-H2B-aid-eyfp fragment was released with KpnI and XbaI, and inserted into pMT between the MtnA promoterCitation26 and the SV40 terminator.
For the generation of pUASt-OsTIR1-K7, the OsTIR1 coding sequence was amplified with the primers CL198 (5′-ACCGG GAATTC AAAATGACGTACTTCCCGGAGGAG-3′) and CL199 (5′-GGCC TCTAGA CTATAGGATTTTAACAAAATTTG-3′). After digestion with EcoRI and XbaI, the fragment was inserted into a modified pUASt vectorCitation27 in which a shortened SV40 terminator (K7) was present instead of the original long SV40 terminator region, which is known to trigger nonsense-mediated mRNA decay and hence reduced expression.Citation28
For the production of pWRpUbi-OsTIR1, the OsTIR1 coding region was amplified with CL191 (5′-CGGA GGTACC AAAATGACGTACTTCCCGGAGGAG-3′) and CL192 (5′-GGCC GAATTC CTATAGGATTTTAACAAAATTTG-3′). After digestion with KpnI and EcoRI, the fragment was inserted into the corresponding sites of pWRpUbiqPE. This places the OsTIR1 coding sequence downstream of the Ubi-p63E promoter and upstream of the rosy+ terminator sequences. Moreover, the resulting construct contains the w+mC marker gene, as well as P element end sequences for Drosophila germline transformation.
For the production of UASt-aid-rux, we amplified the aid coding region from pMT-OsTIR1-P2A-H2B-aid-eyfp with the primers MT55 (5′-TGCC AGATCT ATGGGCAGTGTCGAGCT-3′) (BglII) and MT56 (5′-ACGG ACGCGT AGCTCTGCTCTTGCACTTCTC-3′). After digestions with BglII and MluI, the PCR fragment was cloned into the corresponding restriction sites of pC5Kan-P2A. Into the MluI and NheI sites of the resulting cloning intermediate, a rux cDNA fragment containing the complete coding sequence was inserted. This rux cDNA fragment was amplified first with primers OL5 (5′-AGTAATTATTGAATACAAGAAGAG-3′) and OL6 (5′-GTCCAATTATGTCACACCACAGAA-3′) from genomic DNA isolated from the Drosophila UASt-rux strain,Citation23 followed by re-amplification with primers MT57 (5′-TGCC ACGCGT ATGAGCGCTCCAGAAGAAC-3′) and MT58 (5′-ACGG GCTAGC GCGGCCGC CTAGAAACGCATCCGCC-3′) and digestion with MluI and NheI. BglII and NotI were used for the release of the aid-rux fragment from the second cloning intermediate. The fragment was then inserted into the corresponding restriction sites of pUASt.
Drosophila strains and husbandry
The following GAL4 driver transgenes were used: P{en2.4-GAL4}e16E (en-GAL4),Citation27 P{Act5C-GAL4}25FO1 (Y. Hiromi, unpublished), P{GAL4-ey.H},Citation29 P{GAL4-ninaE.GMR}12 (GMR-GAL4),Citation30 P{GawB}BxMS1096.Citation31 For temperature dependent regulation of en-GAL4 driven expression with the help of P{tubP-GAL80ts}20 (tubP-GAL80ts),Citation32 we used a stock with a recombinant en-GAL4, tubP-GAL80ts chromosome balanced over CyO, P{Dfd-GMR-nvYFP}.Citation33 Larvae lacking the balancer chromosome were selected using a stereomicroscope equipped for fluorescence detection.
ing the UASt-OsTIR1 transgene on either chromosome II (II.1) or chromosome III (III.1 and III.2) were generated with the construct described above. Insertion II.1 was used for the experiments described here.
Strains carrying the Ubi-OsTIR1 transgene on either chromosome II (II.1 and II.2) or III (III.1) were generated with the construct described above. Insertion II.1 was used for the experiments described here.
Strains carrying the UASt-aid-rux gene were generated with the construct described above. Insertion III.1 was used in combination with UASt-OsTIR1(II.1), as well as with Ubi-OsTIR1(II.1). Moreover, for control experiments, UASt-aid-rux (III.1) was combined with UASt-lacZ (II).Citation34
Flies were raised on standard food (100 g yeast, 75 g glucose, 55 g corn meal, 10 g wheat flour, 8 g agar, 250 mg Nipagin, 1 L water). For addition of auxin, fly food was melted in a microwave. After cooling to about 40°C, the required volume of auxin stock solution was added followed by thorough mixing and solidification. The auxin stock solution was generated by dissolving indole-3-acetic acid sodium salt (Sigma-Aldrich, I5148) at a concentration of 1 M in water followed by sterile filtration. The stock solution was frozen in aliquots at −20°C.
Liquid food for Drosophila larvae was prepared essentially as described.Citation35 One liter of liquid food contained 100 g of yeast extract, 100 glucose and 75 g sucrose dissolved in water. The food was sterilized by filtration. Before transfer of larvae into liquid food, eggs were collected from the appropriate crosses in fly bottles during 24 h at 25°C. Parents were discarded and bottles were incubated for 7 d at 18°C. Subsequently, bottles were transferred into a water bath within a 29°C incubator for an additional 24 h. For the isolation of larvae, 20% sucrose in water was added to the bottles containing fly food and larvae. After gentle mixing, larvae floating on top were transferred into a basket with a nylon mesh at the bottom. Excess sucrose solution was washed away with tap water. Baskets with larvae were then transferred into petri dishes containing liquid food (0.95 mL) to which red food color (0.04 mL) had been added just before, as well as auxin stock solution if required. Final concentration of auxin was 1 mM if not specified otherwise. After larval feeding in liquid food for the appropriate time period, the largest larvae with red guts were picked with forceps, followed by dissection of wing imaginal discs in Schneider's tissue culture medium.
Cell culture
S2R+ cell culture, transfection and time lapse imaging were done essentially as previously described.Citation36 To assess auxin effects by time lapse imaging, 75,000 cells/well were plated in a 24 well plate. Auxin was added to the medium one day after plating. For each concentration (0, 0.3, 0.6, and 1 mM), we started 3 replicate cultures. On the bottom of each well, one position was marked. Phase contrast images were acquired next to the mark on consecutive time points (t = 0, 5, 24, 48 and 72 h) with a 20X/0.5 objective on a Zeiss Cell Observer HS wide-field microscope. Replicate cultures displayed identical behavior as illustrated in . For the evaluation of auxin effects on cell numbers and viability, cells were plated into 35 mm dishes. Auxin was again added to the medium one day after plating. Three replicate cultures were started for each time point (0, 24, 48, and 72 h after auxin addition) and auxin concentration (0, 0.3, and 1 mM). Cells were harvested by trypsinization. Trypan Blue was added and the numbers of live and dead cells were determined. For time lapse imaging of H2B-aid-eyfp degradation, S2R+ cells were plated into 35 mm glass bottom dishes and transfected with pMT-OsTIR1-P2A-H2B-aid-eyfp. 0.5 mM CuSO4 was added 40 h after transfection. Auxin (0.5 mM) was added 24 h later. Culture replicates were either fixed at different times after auxin addition or used for time lapse imaging. Images were acquired using a 40x/1.30 oil immersion objective on a Zeiss Cell Observer HS wide-field microscope.
Immunofluorescence
Wing imaginal discs were fixed in 4% formaldehyde for 20 min on a rotating wheel. Subsequent staining was performed as described previously.Citation37 For anti-Rux immunolabeling we used a 1:1 mixture of hybridoma supernatants containing either mouse monoclonal antibody H6 and H9, respectively,Citation23 that was further diluted 1:1.5. Rabbit antiserum against Drosophila Cyclin ACitation38 was used at a dilution of 1:600. Rabbit anti-β-Galactosidase (MP Biomedicals, 0855976) was used at a dilution of 1:2000. For DNA labeling we used Hoechst 33258 at a final concentration of 1 µg/mL. Wing discs of larvae expressing either UASt-lacZ or UASt-OsTIR1 were pooled for fixation, staining and mounting. Image stacks with 3 focal planes spaced by 500 nm were acquired with a 40x/1.30 oil immersion objective on a Zeiss Cell Observer HS microscope. Anti-Rux signals were quantified after maximum intensity projection using Image J. Regions of interest on both sides of the anterior-posterior compartment boundary were selected before determination of average pixel intensity. Signals in the anterior compartment were used for background correction. 17–20 wing discs for each genotype and conditions were quantified in case of the experiments illustrated in , and 3 imaginal discs in case of .
Acknowledgments
We thank M. Kanemaki and E. Caussinus for material and advice, as well as L. Zipursky for monoclonal antibodies against Rux, and S. Moser for technical support.
Funding
This work was supported by the Swiss National Science Foundation (grant 31003A_120276 to C.F.L.).
References
- Kanemaki MT. Frontiers of protein expression control with conditional degrons. Pflugers Arch 2013; 465:419-25; PMID:23271452; http://dx.doi.org/10.1007/s00424-012-1203-y
- Pauli A, Althoff F, Oliveira RA, Heidmann S, Schuldiner O, Lehner CF, Dickson BJ, Nasmyth K. Cell-type-specific TEV protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell 2008; 14:239-51; PMID:18267092; http://dx.doi.org/10.1016/j.devcel.2007.12.009
- Taxis C, Knop M. TIPI: TEV protease-mediated induction of protein instability. Methods Mol Biol 2012; 832:611-26; PMID:22350916; http://dx.doi.org/10.1007/978-1-61779-474-2_43
- Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein science : a publication of the Protein Society 2011; 20:1298-345; PMID:21633985; http://dx.doi.org/10.1002/pro.666
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 1994; 263:1273-6; PMID:8122109; http://dx.doi.org/10.1126/science.8122109
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr Biol 2003; 13:899-910; PMID:12781128; http://dx.doi.org/10.1016/S0960-9822(03)00338-5
- Nicholson SC, Nicolay BN, Frolov MV, Moberg KH. Notch-dependent expression of the archipelago ubiquitin ligase subunit in the Drosophila eye. Development 2011; 138:251-60; PMID:21148181; http://dx.doi.org/10.1242/dev.054429
- Caussinus E, Kanca O, Affolter M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat Struct Mol Biol 2011; 19:117-21; PMID:22157958; http://dx.doi.org/10.1038/nsmb.2180
- Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol 2013; 10:e1001434; PMID:NOT_FOUND; http://dx.doi.org/10.1371/journal.pbio.1001434
- Urban E, Nagarkar-Jaiswal S, Lehner CF, Heidmann SK. The cohesin subunit Rad21 is required for synaptonemal complex maintenance, but not sister chromatid cohesion, during Drosophila female meiosis. PLoS Genet 2014; 10:e1004540; PMID:25101996; http://dx.doi.org/10.1371/journal.pgen.1004540
- Bätz T, Förster D, Luschnig S. The transmembrane protein Macroglobulin complement-related is essential for septate junction formation and epithelial barrier function in Drosophila. Development 2014; 141:899-908; PMID:Can't; http://dx.doi.org/10.1242/dev.102160
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 2009; 6:917-22; PMID:19915560; http://dx.doi.org/10.1038/nmeth.1401
- Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature 2005; 435:441-5; PMID:15917797; http://dx.doi.org/10.1038/nature03543
- Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005; 435:446-51; PMID:15917798; http://dx.doi.org/10.1038/nature03542
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007; 446:640-5; PMID:17410169; http://dx.doi.org/10.1038/nature05731
- Kanke M, Nishimura K, Kanemaki M, Kakimoto T, Takahashi TS, Nakagawa T, Masukata H. Auxin-inducible protein depletion system in fission yeast. BMC Cell Biol 2011; 12:8; PMID:21314938; http://dx.doi.org/10.1186/1471-2121-12-8
- Holland AJ, Fachinetti D, Han JS, Cleveland DW. Inducible, reversible system for the rapid and complete degradation of proteins in mammalian cells. Proc Natl Acad Sci U S A 2012; 109:E3350-7; PMID:23150568; http://dx.doi.org/10.1073/pnas.1216880109
- Kreidenweiss A, Hopkins AV, Mordmuller B. 2A and the auxin-based degron system facilitate control of protein levels in Plasmodium falciparum. PLoS One 2013; 8:e78661; PMID:24236031; http://dx.doi.org/10.1371/journal.pone.0078661
- Philip N, Waters AP. Conditional Degradation of Plasmodium Calcineurin Reveals Functions in Parasite Colonization of both Host and Vector. Cell Host Microbe 2015; 18:122-31; PMID:26118994; http://dx.doi.org/10.1016/j.chom.2015.05.018
- Zhang L, Ward JD, Cheng Z, Dernburg AF. The auxin-inducible degradation (AID) system enables versatile conditional protein depletion in C. elegans. Development 2015; 142:4374-84; PMID:26552885; http://dx.doi.org/10.1242/dev.129635
- Karadeniz A, Kaya B, Savas B, Topcuoglu SF. Effects of two plant growth regulators, indole-3-acetic acid and beta-naphthoxyacetic acid, on genotoxicity in Drosophila SMART assay and on proliferation and viability of HEK293 cells from the perspective of carcinogenesis. Toxicol Industr Health 2011; 27:840-8; PMID:NOT_FOUND; http://dx.doi.org/10.1177/0748233711399314
- Daniels RW, Rossano AJ, Macleod GT, Ganetzky B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS One 2014; 9:e100637; PMID:24945148; http://dx.doi.org/10.1371/journal.pone.0100637
- Thomas BJ, Zavitz KH, Dong XZ, Lane ME, Weigmann K, Finley RL, Brent R, Lehner CF, Zipursky SL. roughex down-regulates G2 cyclins in G1. Genes Dev 1997; 11:1289-98; PMID:9171373; http://dx.doi.org/10.1101/gad.11.10.1289
- Foley E, O'Farrell PH, Sprenger F. Rux is a cyclin-dependent kinase inhibitor (CKI) specific for mitotic cyclin-Cdk complexes. Curr Biol 1999; 9:1392-402; PMID:10607563; http://dx.doi.org/10.1016/S0960-9822(00)80084-6
- Weigmann K, Cohen SM, Lehner CF. Cell cycle progression, growth and patterning in imaginal discs despite inhibition of cell division after inactivation of Drosophila Cdc2 kinase. Development 1997; 124:3555-63; PMID:9342048
- Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogaster cells. Nucleic Acids Res 1988; 16:1043-61; PMID:3125519; http://dx.doi.org/10.1093/nar/16.3.1043
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993; 118:401-15; PMID:8223268
- Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet 2006; 2:e180; PMID:17196039; http://dx.doi.org/10.1371/journal.pgen.0020180
- Hazelett DJ, Bourouis M, Walldorf U, Treisman JE. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Development 1998; 125:3741-51; PMID:9716539
- Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 1996; 87:651-60; PMID:8929534; http://dx.doi.org/10.1016/S0092-8674(00)81385-9
- Capdevila J, Guerrero I. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J 1994; 13:4459-68; PMID:7925288
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 2003; 302:1765-8; PMID:14657498; http://dx.doi.org/10.1126/science.1089035
- Le T, Liang Z, Patel H, Yu MH, Sivasubramaniam G, Slovitt M, Tanentzapf G, Mohanty N, Paul SM, Wu VM, et al. A new family of Drosophila balancer chromosomes with a w- dfd-GMR yellow fluorescent protein marker. Genetics 2006; 174:2255-7; PMID:17057238; http://dx.doi.org/10.1534/genetics.106.063461
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature 1988; 332:853-6; PMID:3128741; http://dx.doi.org/10.1038/332853a0
- Gasque G, Conway S, Huang J, Rao Y, Vosshall LB. Small molecule drug screening in Drosophila identifies the 5HT2A receptor as a feeding modulation target. Scientific reports 2013; 3:srep02120; PMID:23817146; http://dx.doi.org/10.1038/srep02120
- Lidsky PV, Sprenger F, Lehner CF. Distinct modes of centromere protein dynamics during cell cycle progression in Drosophila S2R+ cells. J Cell Sci 2013; 126:4782-93; PMID:23943877; http://dx.doi.org/10.1242/jcs.134122
- Handke B, Szabad J, Lidsky PV, Hafen E, Lehner CF. Towards long term cultivation of Drosophila wing imaginal discs in vitro. PLoS One 2014; 9:e107333; PMID:25203426; http://dx.doi.org/10.1371/journal.pone.0107333
- Sprenger F, Yakubovich N, O'Farrell PH. S phase function of Drosophila cyclin A and its downregulation in G1 phase. Curr Biol 1997; 7:488-99; PMID:9210381; http://dx.doi.org/10.1016/S0960-9822(06)00220-X
