ABSTRACT
Polycomb group (PcG) and Trithorax group (TrxG) proteins orchestrate development of a multicellular organism by faithfully maintaining cell fate decisions made early in embryogenesis. An important chromatin mark connected to PcG/TrxG regulation is bivalent domains, the simultaneous presence of H3K27me3 and H3K4me3 on a given locus, originally identified in mammalian embryonic stem cells but considered to be absent in invertebrates. Here, we provide evidence for the existence of bivalency in fly embryos. Using a recently described PcG reporter fly line, we observed a strong reporter inducibility in the embryo and its sharp decrease in larval and adult stages. Analysis of the chromatin landscape of the reporter revealed a strong signal for the repressive PcG mark, H3K27me3, in all three developmental stages and, surprisingly, a strong signal for a transcriptionally activating H3K4me3 mark in the embryo. Using re-chromatin immunoprecipitation experiments, bivalent domains were also uncovered at endogenous PcG targets like the Hox genes.
Introduction
Polycomb group (PcG) proteins together with antagonistically acting Trithorax group (TrxG) proteins are essential epigenetic regulators responsible for the establishment and maintenance of cell fate decisions. Perturbations of PcG and TrxG regulation impact development, neoplastic growth, tissue regeneration and stem cell biology. In Drosophila, perturbations of the PcG repression in larval tissue were recently shown to result in cellular dedifferentiation, a process of reversal to a less differentiated, embryonic-like cell state that precedes tumorigenesis [Citation1]. Transdetermination is a phenomenon where a group of cells in regeneration blastema becomes more plastic and acquires alternative cell identities [Citation2]. In fly and mouse, modulation of PcG function was a prerequisite of transdetermination and reprogramming required for regeneration and wound repair [Citation3,Citation4]. PcG and TrxG proteins function as components of multi-protein complexes. Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) are the most studied of the PcG complexes. PRC2 catalyses the deposition of the repressive H3K27me3 histone mark on transcriptionally inactive PcG target genes [Citation5–Citation7], while PRC1 recognizes and binds the H3K27me3 mark, compacts chromatin and inhibits chromatin remodelling [Citation8–Citation10]. The recruitment of PRC1 and PRC2 to target genes results in their heritable repression. TrxG complexes, on the other hand, associate with transcriptionally active PcG target genes and establish activating H3K4me3 and H3K27ac marks [Citation11,Citation12]. The concerted actions of PcG and TrxG systems guide cells along specific cell lineages and ultimately contribute to the development of various tissues and organs.
While the two systems are generally thought to counteract each other, their synergy results in the establishment of bivalent domains in mammals. Bivalency, defined as the concurrent presence of H3K27me3 and H3K4me3 marks on a given locus, was first discovered in mouse embryonic stem cells [Citation13,Citation14]. Subsequently, bivalency was shown to exist in developing embryo and adult kidney, lung, liver, brain, intestine, blood, skin and sperm [Citation15–Citation18]. While in embryonic stem cells and embryo bivalency is postulated to poise developmentally crucial genes for later activation during differentiation, de novo acquired tissue-restricted bivalency results in repression of affected genes in intestine and expression of these genes in the brain [Citation16,Citation17]. Currently, bivalency is thought to play a vital role in development [Citation19], face morphogenesis [Citation20], female puberty [Citation21], phenotypic differences between species [Citation18] and genetic diversity in neurons [Citation22,Citation23]. Despite its apparent importance, bivalency is functionally and mechanistically not well understood. Studying bivalency in mammals is complicated by the lack of suitable genetic models [Citation24], scarcity of material [Citation15] and redundancy of PcG and TrxG genes [Citation25].
PcG and TrxG regulatory systems are highly conserved between fly and mammals, both on the protein level and functionally, and insights initially gained in Drosophila were later validated and expanded in mammalian models [Citation26]. To date, there are four reports discussing bivalency in fly stages and tissues: embryo, larval wing disc and adult testis. These reports analysed heterogeneous cell populations and based their statements on overlapping chromatin immunoprecipitation (ChIP) peaks of H3K27me3 and H3K4me3 signals. Gan et al. evaluated the distribution of H3K4me3 and H3K27me3 marks in male germline stem cells (GSCs) using a Drosophila bag of marbles (bam) mutant strain [Citation27]. Out of 4991 genes that were significantly enriched for either mark, only 91 genes were associated with both H3K27me3 and H3K4me3. However, in addition to enrichment of GSCs, bam mutation results in accumulation of transit-amplifying spermatogonial cells and somatic cells [Citation28,Citation29]. This led the authors to concede that loci associated with both marks were likely monovalently labelled in different cell populations rather than being truly bivalent. Schuettengruber et al. observed a similarly low frequency (2.2%) of loci with overlapping H3K4me3 and H3K27me3 signals in whole fly embryos and attributed this overlap to the mixture of different cell populations in a developing embryo [Citation30]. Kang et al. re-analysed H3K27me3 and H3K4me3 ChIP-seq data from whole embryos generated in the modENCODE project [Citation31,Citation32]. While noting the overlap between the two marks, the authors also commented on the heterogenous nature of embryonic samples and their inability to formally exclude the possibility of these marks occurring in distinct populations of cells. Finally, Schertel et al. performed ChIP-seq assessment of H3K27me3 and H3K4me2-3 (using the antibody that recognized both H3K4me2 and H3K4me3 marks) in imaginal wing discs [Citation33]. The authors identified 241 genes that displayed overlapping signals for both marks but acknowledged non-homogenous cell populations that make up imaginal discs and the resulting ambiguity over the true epigenetic status of these genes. Labelling of loci exhibiting overlap of H3K4me3 and H3K27me3 marks as bivalent by Kang et al. and Schertel et al. generated ambiguity on whether the authors claimed these genes to be truly bivalent. Understandably, in the absence of re-ChIP experiments, these claims failed to convince the broader scientific community, and bivalency is still believed to be absent in Drosophila [Citation34–Citation37].
In this report, we take advantage of the recently introduced PcG reporter construct, PRExpress, to investigate repression of a PcG target gene during development [Citation38]. Experiments with PRExpress showed a progressive decrease of reporter inducibility at later developmental stages. In our attempt to understand epigenetic changes underlying this phenomenon, we discovered the presence of bivalency in the fly embryo and its subsequent decline in larval and adult tissues. In addition to the reporter gene, bivalency could also be demonstrated for endogenous PcG target genes in embryos.
Materials and methods
Transgenic fly lines
no-repeats, PRExpress and PRExpress III fly lines were described in detail previously [Citation38]. Fly lines were reared at 25°C, in 60% humidity and at 13-h light/11-h dark cycles. The reporter gene expression was induced through incubation at 37°C for 1 h followed by a 30-min recovery period at 25°C.
ß-Galactosidase activity assay
Fly embryos were collected overnight (ca. 16 h), dechorionated in 3% hypochlorite solution for 2 min, washed thoroughly with water and frozen in liquid nitrogen. Twenty wandering third instar larvae were collected in 30% (v/v) glycerol solution, washed thoroughly with water and frozen in liquid nitrogen. Three female and three male flies were anaesthetized with CO2 and frozen in liquid nitrogen 3 d post eclosion. After freezing, samples were stored at −80°C. Frozen fly samples were homogenized on ice in the assay buffer (1 mM MgSO4, 2% Triton X-100 (v/v), 100 mM Hepes, pH 8) supplemented with cOmplete™, EDTA-free Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Homogenates were centrifuged twice at 20,000 g for 10 min at 4°C, each time retaining supernatants and discarding the precipitated debris. The protein concentration of homogenates was measured using Pierce BCA protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). One microgram of protein from homogenates was incubated at 37°C for 50 min in 0.1 ml of the assay buffer supplemented with 4-MUG (Sigma-Aldrich, M1633, St. Louis, MO, USA) at 0.9 mM final concentration and with cOmplete™. ß-Galactosidase converts the non-fluorescent substrate 4-MUG into the fluorescent product 4-MU. The 4-MU fluorescence was measured using EnVision Multilabel Reader 2104 (PerkinElmer, Waltham, MA, USA).
Chromatin immunoprecipitation
For ChIP experiment, embryos, larvae and adults were collected similarly as for beta-galactosidase assay but were not frozen before chromatin preparation. Instead, about 600 mg of whole fly embryos, larvae or adults was homogenized at room temperature for 2.5 min in 10 ml of crosslinking buffer (1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 1.8% (v/v) formaldehyde, 50 mM Hepes, pH 8) supplemented with cOmplete™ in a 15-ml glass tissue grinder using the loose-fitting pestle. Glycine was added to the homogenized sample at 0.125 M final concentration, and the sample was rotated at room temperature and 15 rpm for 5 min. From this point on, all steps were performed at 4°C or on ice unless otherwise specified. The sample was centrifuged first at 400 g for 1 min to pellet and discard large debris. The supernatant was centrifuged again at 1,100 g for 10 min. This time, the supernatant was discarded, and the pellet was re-suspended in 10 ml of cell lysis buffer (85 mM KCl, 0.5% (v/v) IGEPAL CA-630, 5 mM Hepes, pH 8) supplemented with cOmplete™ and homogenized in the 15-ml tissue grinder using 20 strokes with the tight-fitting pestle. The sample was then centrifuged at 2,000 g for 4 min to pellet the nuclei. The nuclei were re-suspended in 1 ml of nuclear lysis buffer (10 mM EDTA, 0.5% (w/v) N-lauroylsarcosine, 50 mM Hepes, pH 8) supplemented with cOmplete™ and incubated at room temperature for 20 min. After incubation, another 1 ml of nuclear lysis buffer supplemented with cOmplete™ was added to the sample, and chromatin was fragmented using Bioruptor Plus sonicator to an average 500 bp fragment size. Bioruptor settings: 60 cycles (1 cycle = 30 s on/30 s off), high energy setting, sample briefly vortexed every 10 cycles. Sonicated sample was centrifuged at 20,000 g for 10 min to precipitate debris, and the clear supernatant was frozen in liquid nitrogen and stored at −80°C. Immunoprecipitations (IPs) were performed as described previously [Citation39]. Re-ChIP experiments were performed as described previously [Citation40]. Results of ChIP experiments were analysed using digital droplet PCR (Bio-Rad, Hercules, CA, USA) following the manufacturer’s instructions. Primer pairs and probes for loci evaluated are given in .
Table 1. Primer pairs and probes used in this study.
Antibodies
For ChIP and re-ChiP experiments, antibodies against H3K27me3 (Sigma-Aldrich, St. Louis, MO, USA, 07-449) (), H3K9ac (abcam, Cambridge, UK, ab4441), H3K4me3 (abcam, Cambridge, UK, ab8580), H3K27me3 (abcam, Cambridge, UK, ab6002) (Supplementary Figures S1A, S2; ) and normal rabbit IgG (Santa Cruz, Dallas, TX, USA, sc-2027 X) were used. The first large set of H3K27me3 ChIP experiments in embryo, larva and adult of no-repeats, PRExpress and PRExpress III lines () was performed with the 07-449 antibody due to its good signal-to-noise ratio. As the variability among individual biological replicates in this experiment required normalization to the positive control, a comparison of the results between embryo, larva and adult was not possible. To obtain this important piece of information, additional H3K27me3 ChIP experiments with the ab6002 antibody were performed (Supplementary Figure S1A).
Figure 1. Inducibility of the reporter gene and its chromatin landscape in embryo, larva and adult of no-repeats, PRExpress and PRExpress III lines. (a) Inducibility of lacZ in no-repeats, PRExpress and PRExpress III lines. In each line, inducibility in larva and adult was normalized to inducibility in embryo (n = 3). (b) H3K27 trimethylation at the positive control Ubx, the negative control Act5C and the lacZ reporter in no-repeats, PRExpress and PRExpress III lines. Values for Act5C and lacZ were normalized to Ubx values at each stage and in each line (n = 5). (c) H3K9 acetylation at Ubx, the positive control Act5C and lacZ (n = 5). (d) H3K4 trimethylation at Ubx, the positive control Act5C and lacZ (n = 5). ChIP experiments presented in (b–d) were performed on non-induced fly samples. Data information: Data are represented as mean ± SD.
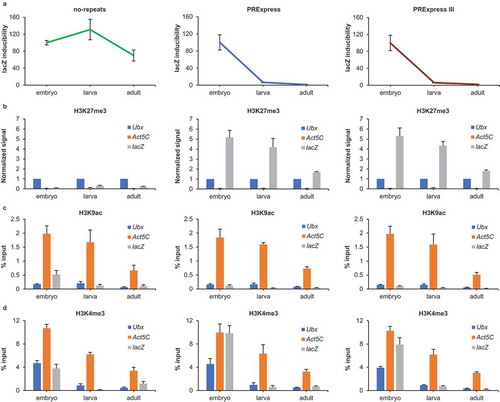
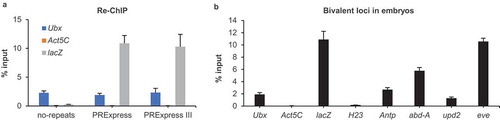
Figure 2. Bivalency in Drosophila embryos. (a) Bivalency at lacZ in no-repeats, PRExpress and PRExpress III embryos (n = 4). (b) Bivalency at additional genomic loci in PRExpress embryos (n = 4). Data information: Re-ChIP experiments were performed using anti-H3K27me3 antibody in the first immunoprecipitation and anti-H3K4me3 in the second immunoprecipitation. Data are represented as mean ± SD.
Statistical analysis
The average values of biological replicates were given as mean ± standard deviation. Significance testing of ChIP results was performed using one-way analysis of variance (ANOVA) test. Statistical significance was defined as P ≤ 0.05. Asterisks indicate statistically significant differences (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001).
Results
Inducibility of a PcG target gene
We recently reported the development of the PRExpress construct, a non-leaky heat-inducible gene expression system for Drosophila [Citation38]. PRExpress consists of a heat-inducible LacZ reporter gene placed downstream of a synthetic PcG response element (PRE), termed poly-PRE. Eight repeats of a 198-bp fragment of the bxd-PRE constitute the poly-PRE sequence. In contrast to a full-length PRE, this short bxd fragment contains no positional information and thus represses the reporter in the entire animal in embryonic, larval and adult stages. Non-leakiness and ubiquitous inducibility of PRExpress rendered it uniquely suitable to assess PcG repression as a function of development in the entire animal. Three previously generated fly lines were used in the present study. PRExpress and PRExpress III lines contain the PRExpress construct integrated into predefined sites on the second and third chromosomes, respectively. Lines with different integration sites furnish an opportunity to exclude the influence of different chromosomal milieus on observed phenotypes. The no-repeats line contains a control construct, identical to PRExpress but lacking the poly-PRE sequence. The no-repeats construct was integrated into the same predefined site on the second chromosome and served as a control for the PRExpress line [Citation38].
Inducibility was defined as a ratio of the ß-galactosidase activity of fly samples in the induced state over the ß-galactosidase activity in the non-induced state (i.e. values of heat-shocked vs non-heat-shocked samples). Analysis of inducibility of the reporter gene in the three lines indicated a steep decrease of inducibility in larva and adult fly in lines containing the poly-PRE sequence by 93% and 97%, respectively, whereas inducibility in no-repeats line remained equally strong throughout development (). This observation suggested an enhancement of PcG repression in advanced developmental stages. Some time ago, we made a comparable observation using a reporter containing the 3.6 kb Fab-7 PRE fragment [Citation41,Citation42]. The activation of the reporter under the control of the Fab-7 PRE was less efficient in larval compared to embryonic tissues. This led to the suggestion that the chromatin of PcG-repressed genes in embryo was more plastic and became progressively more committed as development proceeds [Citation43]. However, elucidation of mechanisms underlying this plasticity was confounded by the leakiness of the Fab-7-reporter construct in embryo and larva in the absence of induction and the lack of appropriate control lines. Leakiness is a manifestation of heterogeneity of the sample and precludes the unequivocal interpretation of ChIP experiments.
H3K27me3, H3K9ac and H3K4me3 marks at the reporter gene
To discern the epigenetic changes that affect the stronger PcG repression in larva and adult, we performed a series of ChIP experiments. All ChIP and re-ChIP experiments were performed on non-induced (i.e. not heat-shocked) fly samples. As expected, the reporter gene in PRExpress and PRExpress III lines was strongly marked with H3K27me3 in embryo, larva and adult (). Correspondingly, the H3K27me3 mark was absent from the reporter in the no-repeats line in all three stages due to the absence of the poly-PRE sequence in this line. Unfortunately, the strong variability among individual biological replicates in this experiment required normalization of results to the positive control, the PcG target gene Ubx. As a result, comparisons among different stages of development were not valid, and it was unclear whether the H3K27me3 mark on the reporter increased or decreased during development, as the H3K27me3 signal at the reference locus Ubx might potentially differ between different developmental stages. To gain this information, we performed another ChIP experiment using an anti-H3K27me3 antibody from a different supplier (see Materials and methods). The H3K27me3 signal at the reporter in PRExpress was similarly strong at all three stages (Supplementary Figure S1A). In contrast, the H3K27me3 signal at Ubx increased twofold in larva compared to embryo and threefold in adult compared to embryo. These results suggested a gradual and significant increase of the H3K27 trimethylation at the endogenous PcG target gene Ubx, while no such increase was observed at the reporter. Hence, changes in H3K27 trimethylation levels could not account for reduced inducibility of the reporter gene at later stages.
The acetylation of H3K9 mediates a switch from transcription initiation to transcription elongation via recruitment of the super elongation complex to chromatin and marks actively transcribed genes [Citation44]. In line with expectations, H3K9ac was strongly present at Act5C, a positive control, in all stages and lines (). The H3K9ac signal was much lower at lacZ compared to Act5C in PRExpress and PRExpress III lines. Interestingly, the H3K9ac signal in no-repeats was higher compared to PRExpress line in all three stages: fourfold higher in embryo, threefold higher in larva and threefold higher in adult (Supplementary Figure S1B). This was consistent with the observed tenfold, twofold and fourfold higher ß-galactosidase activity (i.e. leakiness) in no-repeats compared to PRExpress embryo, larva and adult, respectively [Citation38]. As expected, the H3K9ac signal at Ubx between PRExpress and no-repeats lines was similar in each stage (P > 0.30 for embryo, P > 0.29 for larva and P > 0.13 for adult) (Supplementary Figure S1C). The ability to detect minor leakiness of the reporter gene in no-repeats line underscored the high sensitivity of whole animal ChIP experiments.
The most unexpected result was obtained in ChIP experiments for the H3K4me3 mark (). PRExpress and PRExpress III lines showed a strong H3K4me3 signal at the reporter gene comparable in strength to the signal at Act5C, a positive control, in the embryonic stage. The H3K4me3 signal at lacZ in PRExpress embryo was 2.5-fold higher compared to control no-repeats embryo. Whereas the H3K4me3 signal at the reporter in the no-repeats line could be explained by the leakiness of lacZ in this fly line, this argument is invalid for the PRExpress line where the reporter is completely repressed [Citation38]. Thus, the presence of poly-PRE appeared to result in a stronger H3K4me3 even in the absence of any transcription. At the larval stage, the H3K4me3 level at lacZ in PRExpress line was fourfold higher compared to the H3K4me3 signal at lacZ in no-repeats line. In adult flies, this trend was reversed with lacZ in no-repats line displaying twofold higher H3K4me3 signal compared to the signal at lacZ in PRExpress line. The presence of H3K27me3, H3K4me3 and complete repression of the reporter gene in the entire embryo (i.e. the embryo is a homogenous cell population with respect to the expression of the reporter gene) strongly suggested the existence of bivalency in Drosophila embryos. In agreement with previous reports [Citation30,Citation31], the H3K4me3 signal was also present at the Ubx gene in the embryonic stage in the three lines. However, this could not be assumed to be evidence of bivalency since the gene is expressed in some parasegments of the embryo and repressed in others and would be expected to be marked with H3K4me3 and H3K27me3 in respective parasegments. In larval and adult stages, the H3K4me3 signal at Ubx and lacZ diminished sharply, mirroring the decrease in inducibility of the reporter.
Bivalency in Drosophila embryo
While individual ChIP experiments can hint at the co-occurrence of two histone marks, a re-ChIP (or sequential ChIP) experiment is necessary to confirm the coexistence of both marks on a given locus. In re-ChIP, chromatin is first precipitated using the antibody against H3K27me3 and then using the antibody against H3K4me3. Consequently, enrichment in re-ChIP is indicative of the simultaneous presence of both marks on the same DNA fragment. The reporter gene displayed a strong enrichment in re-ChIP experiment in PRExpress and PRExpress III embryos, while the signal at lacZ in no-repeats embryo was at the background level (). Intriguingly, the Ubx gene exhibited a 40-fold higher enrichment in re-ChIP compared to the negative control, Act5C, suggesting that bivalency was not restricted to the synthetic transgene but was also present at an endogenous PcG target gene. To ensure the re-ChIP signal was specific to anti-H3K27me3 and anti-H3K4me3 antibodies, we performed a control experiment where these antibodies were replaced with normal rabbit IgG in the first and second IPs, respectively. The replacement of anti-H3K27me3 or anti-H3K4me3 antibodies with normal rabbit IgG in re-ChIP resulted in a 1400-fold and 290-fold drop in signal, respectively, indicating high specificity of the technique (Supplementary Figure S2). Bivalency was evaluated at five additional loci in PRExpress embryo (). H23 is a heterochromatin locus of the second chromosome and was included as a negative control in addition to Act5C. Antp and abd-A are canonical Hox genes like Ubx, while eve is a closely related HOX-like homeobox transcription factor. upd2 is a secreted core component of the JAK/STAT signalling pathway. PcG target genes Antp, abd-A, eve and upd2 showed strong bivalency signals, whereas H23 control locus displayed background re-ChIP values. These results suggested that bivalency is a general phenomenon present at different types of PcG target genes in fly embryo.
Discussion
Our results demonstrated the existence of bivalency at PcG target genes in Drosophila melanogaster embryo. As fly developed into larva and adult, the H3K4me3 mark at a PcG target gene declined abruptly, while the H3K27me3 mark remained at similar levels. The downregulation of bivalency at later stages was associated with a dramatic decrease in the gene’s inducibility. In their entirety, our results suggest that bivalency may be a mechanism to keep PcG target genes refractory to subthreshold noise and at the same time responsive to appropriate developmental cues. This function seems to be conserved between fly and mammals. Moreover, bivalency in fly embryos affords an elegant solution to several puzzling observations. PRC2 components Extra sex combs and Enhancer of zeste are expressed most abundantly during embryogenesis and only weakly in post-embryonic stages [Citation45,Citation46]. Polycomblike, another PcG protein required for high levels of H3K27 trimethylation in embryo, associates with PRC2 in embryonic but not larval tissues [Citation47–Citation50]. At the same time, PcG repression appears to be stronger in post-embryonic stages [Citation41–Citation43]. We propose that higher amounts of PcG proteins are necessary to keep bivalent PcG target genes repressed in embryo, whereas less PcG proteins are sufficient to repress target genes in the absence of H3K4me3 at later stages. Additionally, Enhancer of zeste interaction with SIR2, an NAD-dependent histone deacetylase, was reported to be confined to post-embryonic stages of development and was suggested to potentiate fidelity of PcG repression [Citation51]. It is tempting to speculate that SIR2 or other proteins involved in enhancing Polycomb phenotypes (e.g. Enhancer of Polycomb, Suppressor of zeste 2, Histone deacetylase 1) play a role in resolving bivalency in larva and adult fly.
The functional role of bivalent domains in mammalian development has remained enigmatic. The demonstration of bivalency in Drosophila, together with the availability of the bivalency reporter line PRExpress, will allow the application of large-scale high-throughput genetic screens to identify factors involved in establishment or resolution of bivalency and integration of gained insights within a broader context of the development of a multicellular organism.
Author contributions
AA conceived the project, collected samples, designed and performed experiments and wrote the manuscript. MG collected samples and provided critical feedback throughout the project. RP supervised the project. MG and RP provided critical feedback on the manuscript.
Supplemental Material
Download EPS Image (928.7 KB)Supplemental Material
Download MS Word (74.9 KB)Acknowledgements
Work of AA, MG and RP was supported by the ETH Zurich. MG is a member of the Life Science Zurich Graduate School (PhD Program in Molecular Life Sciences) and supported by a fellowship from the German Academic Scholarship Foundation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental material
Supplemental data for this article can be accessed here.
References
- Torres J, Monti R, Moore AL, et al. A switch in transcription and cell fate governs the onset of an epigenetically-deregulated tumor in Drosophila. eLife. 2018;7:e32697.
- Hariharan IK, Serras F. Imaginal disc regeneration takes flight. Curr Opin Cell Biol. 2017;48:10–16.
- Lee N, Maurange C, Ringrose L, et al. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237.
- Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886.
- Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043.
- Czermin B, Melfi R, McCabe D, et al. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196.
- Müller J, Hart CM, Francis NJ, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208.
- Messmer S, Franke A, Paro R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992;6:1241–1254.
- Wang L, Brown JL, Cao R, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646.
- King IFG, Emmons RB, Francis NJ, et al. Analysis of a polycomb group protein defines regions that link repressive activity on nucleosomal templates to in vivo function. Mol Cell Biol. 2005;25:6578–6591.
- Petruk S, Sedkov Y, Smith S, et al. Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science. 2001;294:1331–1334.
- Smith ST, Petruk S, Sedkov Y, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167.
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326.
- Azuara V, Perry P, Sauer S, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538.
- Liu X, Wang C, Liu W, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562.
- Weiner A, Lara-Astiaso D, Krupalnik V, et al. Co-ChIP enables genome-wide mapping of histone mark co-occurrence at single-molecule resolution. Nat Biotechnol. 2016;34:953–961.
- Jadhav U, Nalapareddy K, Saxena M, et al. Acquired tissue-specific promoter bivalency is a basis for PRC2 necessity in adult cells. Cell. 2016;165:1389–1400.
- Lesch BJ, Silber SJ, McCarrey JR, et al. Parallel evolution of male germline epigenetic poising and somatic development in animals. Nat Genet. 2016;48:888–894.
- Mas G, Blanco E, Ballaré C, et al. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat Genet. 2018;50:1452–1462.
- Minoux M, Holwerda S, Vitobello A, et al. Gene bivalency at Polycomb domains regulates cranial neural crest positional identity. Science. 2017;355:eaal2913.
- Lomniczi A, Loche A, Castellano JM, et al. Epigenetic control of female puberty. Nat Neurosci. 2013;16:281–289.
- Muotri AR, Chu VT, Marchetto MCN, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910.
- Hoffmann A, Zimmermann CA, Spengler D. Molecular epigenetic switches in neurodevelopment in health and disease. Front Behav Neurosci. 2015;9:Article 120.
- Voigt P, Tee -W-W, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338.
- Denissov S, Hofemeister H, Marks H, et al. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141:526–537.
- Beisel C, Paro R. Silencing chromatin: comparing modes and mechanisms. Nat Rev Genet. 2011;12:123–135.
- Gan Q, Schones DE, Ho Eun S, et al. Monovalent and unpoised status of most genes in undifferentiated cell-enriched Drosophila testis. Genome Biol. 2010;11:R42.
- McKearin DM, Spradling AC. bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 1990;4:2242–2251.
- Gönczy P, Matunis E, DiNardo S. bag-of-marbles and benign gonial cell neoplasm act in the germline to restrict proliferation during Drosophila spermatogenesis. Dev Camb Engl. 1997;124:4361–4371.
- Schuettengruber B, Ganapathi M, Leblanc B, et al. Functional anatomy of polycomb and trithorax chromatin landscapes in drosophila embryos. PLoS Biol. 2009;7:e1000013.
- Kang H, Jung YL, McElroy KA, et al. Bivalent complexes of PRC1 with orthologs of BRD4 and MOZ/MORF target developmental genes in Drosophila. Genes Dev. 2017;31:1988–2002.
- Ho JWK, Jung YL, Liu T, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512:449–452.
- Schertel C, Albarca M, Rockel-Bauer C, et al. A large-scale, in vivo transcription factor screen defines bivalent chromatin as a key property of regulatory factors mediating Drosophila wing development. Genome Res. 2015;25:514–523.
- Schuettengruber B, Bourbon H-M, Di Croce L, et al. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell. 2017;171:34–57.
- Choate LA, Danko CG. Poised for development. Nat Genet. 2016;48:822–823.
- Ringrose L. Noncoding RNAs in Polycomb and Trithorax regulation: a quantitative perspective. Annu Rev Genet. 2017;51:385–411.
- Kassis JA, Kennison JA, Tamkun JW. Polycomb and Trithorax group genes in drosophila. Genetics. 2017;206:1699–1725.
- Akmammedov A, Geigges M, Paro R. Single vector non-leaky gene expression system for Drosophila melanogaster. Sci Rep. 2017;7:6899.
- Sandmann T, Jakobsen JS, Furlong EEM. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc. 2006;1:2839–2855.
- Truax AD, Greer SF. ChIP and Re-ChIP assays: investigating interactions between regulatory proteins, histone modifications, and the DNA sequences to which they bind. In: Vancura A, editor. Transcriptional regulation. New York (NY): Springer New York; 2012. p. 175–188.
- Cavalli G, Paro R. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998;93:505–518.
- Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958.
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22.
- Gates LA, Shi J, Rohira AD, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 2017;292:14456–14472.
- Ng J, Hart CM, Morgan K, et al. A drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078.
- Casas-Vila N, Bluhm A, Sayols S, et al. The developmental proteome of Drosophila melanogaster. Genome Res. 2017;27:1273–1285.
- Nekrasov M, Klymenko T, Fraterman S, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. Embo J. 2007;26:4078–4088.
- Tie F, Prasad-Sinha J, Birve A, et al. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol Cell Biol. 2003;23:3352–3362.
- Savla U, Benes J, Zhang J, et al. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Dev Camb Engl. 2008;135:813–817.
- O’Connell S, Wang L, Robert S, et al. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. J Biol Chem. 2001;276:43065–43073.
- Furuyama T, Banerjee R, Breen TR, et al. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr Biol CB. 2004;14:1812–1821.
