ABSTRACT
Notch signalling is a well-conserved signalling pathway that regulates cell fate through cell-cell communication. A typical feature of Notch signalling is ‘lateral inhibition’, whereby two neighbouring cells of equivalent state of differentiation acquire different cell fates. Recently, mathematical and computational approaches have addressed the Notch dynamics in Drosophila neural development. Typical examples of lateral inhibition are observed in the specification of neural stem cells in the embryo and sensory organ precursors in the thorax. In eye disc development, Notch signalling cooperates with other signalling pathways to define the evenly spaced positioning of the photoreceptor cells. The interplay between Notch and epidermal growth factor receptor signalling regulates the timing of neural stem cell differentiation in the optic lobe. In this review, we summarize the theoretical studies that have been conducted to elucidate the Notch dynamics in these systems and discuss the advantages of combining mathematical models with biological experiments.
Introduction
Notch signalling is conserved across metazoans and plays a central role in cell fate decisions [Citation1,Citation2]. Notch was genetically identified in Drosophila with the ‘notching’ mutant phenotype in the wing more than 100 years ago [Citation3]. Subsequent analyses in Drosophila have uncovered the Notch receptor and the other key molecules that constitute the Notch signalling pathway. One of the key processes in Notch signalling is the lateral inhibition system. Typical examples of lateral inhibition have been described in Drosophila neural stem cell (neuroblast, NB) differentiation during embryogenesis and sensory organ precursor (SOP) specification in the thorax [Citation4–6]. The initially uniform field of cells acquires either neuronal or non-neuronal fate through lateral inhibition. In addition to the conserved idea of lateral inhibition, many studies using Drosophila and other animals have revealed diverse functions of Notch signalling in development, tissue homoeostasis, and cancer [Citation7,Citation8].
In this review, we will not mainly focus on the molecular mechanisms of Notch signalling because it has been well documented in several review articles. [Citation9–14]. Here, we simply abstracted the essential factors required for the canonical lateral inhibition system in Drosophila (). Notch is a single-pass transmembrane receptor [Citation15]. There are two membrane-bound Notch ligands in Drosophila: Delta and Serrate [Citation16–18]. Binding of the Notch ligand expressed in neighbouring cells to Notch leads to a conformational change in Notch, and the intracellular domain of Notch (NICD) is cleaved by γ secretase [Citation19]. NICD translocates to the nucleus and activates target gene expression together with Suppressor of Hairless (Su(H)) and Mastermind (Mam) [Citation20–23]. The NICD-Su(H)-Mam complex induces the expression of enhancer of split (E(spl)) complex genes, which encode basic helix-loop-helix (bHLH) transcription factors [Citation20,Citation24,Citation25]. E(spl) transcription factors inhibit the expression of achaete-scute complex (AS-C), which includes achaete (ac), scute (sc), lethal of scute (l’sc), and asense (ase) [Citation26,Citation27]. AS-C genes encode bHLH transcription factors and act as proneural factors. Additionally, AS-C induces Delta expression [Citation28]. The neighbouring cells differentiate into different cell types based on this feedback mechanism. This effect is called as trans-activation. In contrast to trans-activation, cis-inhibition negatively regulates Notch signalling. In cis-inhibition, Notch ligands and the Notch receptor expressed in the same cell bind with each other and inactivate Notch signalling [Citation29–32]. In the wing disc development, high levels of Delta and Serrate inhibit activation of Notch signalling in a cell-autonomous manner through cis-inhibition [Citation32,Citation33]. Although it is unclear whether cis-inhibition is implemented in all biological systems involving Notch signalling, cis-inhibition enhances the effect of trans-activation and contributes to generating mutually exclusive cell states between neighbouring cells during lateral inhibition as explained below.
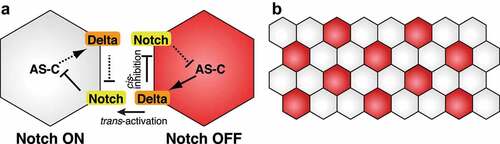
Figure 1. Mathematical modelling of neuroblast and sensory organ precursor differentiation. (a) Notch-mediated lateral inhibition. Delta expression in the right cell activates Notch signalling in the left cell, which leads to downregulation of AS-C expression. Since AS-C induces Delta expression, Delta expression gradually decreases in the left cell. In contrast, Delta expression inhibits Notch signalling in the same cell. This leads to the activation of AS-C and Delta expression in the right cell. (b) The salt-and-pepper-like pattern generated by the Notch function. Neural cells (red) such as embryonic NBs and SOPs are not next to each other
Recently, formulation of mathematical models of Notch signalling to understand its dynamics has become popular [Citation34]. In this review, we focus on the developmental processes of Drosophila, in which Notch signalling plays pivotal roles through lateral inhibition. The basic concept of lateral inhibition was identified by investigating the mechanism of NB formation during embryonic stages and SOP formation in the thorax [Citation4–6].
However, in many other biological systems, Notch signalling cooperates with other signalling pathways and shows complicated interactions. Such examples are observed in eye disc and optic lobe development. In eye disc development, sequential propagation of the morphogenetic furrow induces photoreceptor cell (R cell) differentiation [Citation35,Citation36]. Differentiation of R cells resembles that of embryonic NBs and SOPs in that only a small number of cells are selected as photoreceptor neurons from initially equivalent epithelial cells. Notch signalling, together with the Hedgehog (Hh), Decapentaplegic (Dpp), and epidermal growth factor receptor (EGFR) pathways, defines the evenly spaced positioning of the R cells.
During optic lobe development, a wave of differentiation named ‘proneural wave’ sweeps the surface of the neuroepithelium and determines the timing of NB differentiation [Citation35,Citation37]. In the optic lobe, all neuroepithelial cells (NEs) sequentially differentiate into NBs following the proneural wave. The interaction between Notch-mediated lateral inhibition and EGFR-mediated reaction diffusion determines the speed of proneural wave progression. In these systems, it is difficult to explain Notch dynamics without theoretical approaches because these signalling pathways show complicated interactions. In this article, we introduce molecular bases of these developmental processes and mathematical approaches to decipher diverse Notch functions.
Mathematical models of canonical Notch-mediated lateral inhibition
The basic idea of Notch-dependent lateral inhibition comes from studies of NB differentiation in the embryonic neuroectoderm and SOP selection in the thorax [Citation4–6]. In these biological systems, proneural genes of the AS-C, including ac, sc, and l’sc, are expressed in proneural equivalence groups of neuroectodermal cells, called proneural cluster cells, and have the potential to differentiate into neural cells [Citation38–40]. Among proneural cluster cells, single cells are selected as neural cells, and other cells are fated to be epidermal cells [Citation41,Citation42]. This bistable specification is explained by the Notch-mediated lateral inhibition [Citation9,Citation43]. During the specification step, binding of Delta to the Notch receptor in the neighbouring cell activates Notch signalling and inhibits AS-C and Delta expression (). As a result, these cells do not differentiate into neural cells. In contrast, Delta-expressing cells maintain the expression of AS-C proneural factors to be neural cells. The feedback loop amplifies the slight initial differences in Delta and/or AS-C expression and dictates different cell fates between adjacent cells (). According to the lateral inhibition mechanism, neural cells are not adjacent to each other and show a so-called salt-and-pepper-like pattern ().
To understand the function of lateral inhibition during the specification of embryonic NBs and thoracic SOPs, mathematical and computational approaches have been taken [Citation44–50]. A simple model only considers the differentiation state of the cells; differentiation states of cells were calculated according to the lateral inhibition rule under the assumption that if a cell has acquired a neural fate, then the differentiating cell inhibits the neural differentiation of neighbouring cells. This simple model reproduced the differentiation pattern and the ratio of neural and non-neural cells [Citation44,Citation45]. Collier et al. provided a two-component model to explain the function of Notch signalling in the lateral inhibition mechanism [Citation48]. The model considered the levels of Notch activation (N) and Delta expression (D). N is increased by the effect of trans-activation through D expressed in neighbouring cells and decreased by its degradation. D is decreased by N in the same cell and is regulated by decay. Numerical simulations of the model demonstrated a spatially periodic pattern in which cells with low N are not adjacent to each other and are surrounded by cells with high N, reproducing the salt-and-pepper-like differentiation pattern in vivo. Additionally, mathematical models that include the effect of cis-inhibition efficiently amplified small initial differences between neighbouring cells and accelerated the pattern formation by lateral inhibition [Citation31,Citation51]. These mathematical models and numerical simulations clearly show that Notch-dependent lateral inhibition can determine different cell fates between neighbouring cells.
Recent studies combining mathematical approaches and in vivo analyses have revealed that Notch signalling mediates the tissue-wide patterning of SOPs in the notum. It has been thought that SOP selection includes two distinct steps. First, the expression of proneural factors is regulated by developmental signals that convey positional and temporal information. In the next step, Notch-mediated lateral inhibition singles out SOPs among the proneural cluster cells. In contrast to the two-step model, Notch signalling governs both proneural patterning and SOP selection in the medial thorax [Citation52]. The initial expression pattern of Delta and subsequent self-organized Notch dynamics organizes stereotyped SOP patterning. In addition, Cohen et al. showed that basal actin-based filopodia can transmit intermittent Notch signalling over several cell diameters to adjust the number and spacing of SOPs [Citation53]. These findings suggest that the dynamic action of Notch signalling regulates tissue-wide patterning.
Notch function in eye disc development
The adult eye in Drosophila consists of 750 spatially arranged ommatidia. Each ommatidium comprises eight R cells (R1-8) and several types of accessory cells. Differentiation of R cells starts from the posterior margins of the eye disc and progresses towards the anterior side [Citation35,Citation36,Citation54]. The morphogenetic furrow (MF) is the site where R cell differentiation is initiated () [Citation55]. The posterior to anterior progression of the MF is driven by the secreted Hh protein expressed in the differentiated R cells () [Citation56–58]. In each ommatidium, differentiation of R8 occurs first, followed by R2/R5, R3/R4, and R1/R6 differentiation (). R7 is recruited at the end (). Therefore, patterned differentiation of R8 cells is important for generating a crystalline hexagonal array of the adult eye. Instead of AS-C proneural factors, another bHLH proneural factor, Atonal (Ato), plays a central role in R8 differentiation [Citation59–61]. Ato is expressed in a broad stripe within and just anterior to the MF (). As the MF passes anteriorly, Ato expression is elevated in the regularly spaced intermediate groups. Later, Ato expression is refined to R8 only and shows a more evenly spaced pattern (). Several factors and signalling pathways regulate Ato expression and R8 specification. Dpp signalling, together with Hh signalling, induces Ato expression at the MF (, c) [Citation62–65]. The Notch function in Ato expression is complex [Citation66]. Loss-of-function of Notch does not show elevation of Ato expression, suggesting that Notch signalling is required for establishing a high level of Ato expression in the early step () [Citation66–69]. This function of Notch signalling is called ‘proneural enhancement’ (). Conversely, in the next step, Notch signalling restricts the number of Ato-expressing cells by lateral inhibition (, c) [Citation66,Citation70,Citation71]. The number of Ato-expressing cells is increased when Notch signalling is partially perturbed by using Notchts, a temperature-sensitive mutation of Notch () [Citation72]. A secreted glycoprotein, Scabrous (Sca), regulates R8 specification by activating Notch signalling () [Citation73–76]. Ato induces Sca expression, and the secreted Sca binds to the Notch receptor to inhibit Ato expression [Citation71,Citation72,Citation77,Citation78]. In Sca mutant eye discs, intermediate groups are not formed correctly and more R8s are generated () [Citation71,Citation75]. Since Sca acts as a diffusible protein, it is expected that Sca mediates Notch-mediated lateral inhibition in several cell diameters for regular patterning. In addition to Notch signalling, EGFR signalling also participates in R8 spacing [Citation79,Citation80]. EGFR signalling also activates Hh signalling () [Citation81,Citation82]. Ato induces the expression of a transcription factor, Senseless (Sens), to induce R8 differentiation [Citation83]. Sens, in turn, maintains Ato expression in presumptive R8 cells ().
Figure 2. Mathematical modelling of the eye disc development. (a) Development of the eye disc. The morphogenetic furrow (MF) progresses in the posterior to the anterior direction (left). Hh is expressed in the differentiating R cells located posterior to the MF. Dpp expression shows a stripe in the MF. Atonal (Ato, green) shows dynamic expression patterns during the eye disc development (right). Notch signalling involves early ‘proneural enhancement’ and late ‘lateral inhibition’ functions. Cells of intermediate groups are circled in red. (b) Recruitment of R cells in the ommatidium. (c) Genetic interaction that regulates Ato expression (modified from Lubensky et al., 2011) [Citation85]. (d) A simplified interaction map of (B) for mathematical modelling of Ato expression (modified from Lubensky et al., 2011) [Citation85]. A, S, H, and N represent activation signals in the cell, delayed positive feedbacks in the cell, long-range cell-non-autonomous activation signals, and cell-non-autonomous inhibitory signals, respectively. (e) Schematics of the Ato expression pattern in Notch−, Notchts, Sca−, and Notchts Sca− mutant conditions
![Figure 2. Mathematical modelling of the eye disc development. (a) Development of the eye disc. The morphogenetic furrow (MF) progresses in the posterior to the anterior direction (left). Hh is expressed in the differentiating R cells located posterior to the MF. Dpp expression shows a stripe in the MF. Atonal (Ato, green) shows dynamic expression patterns during the eye disc development (right). Notch signalling involves early ‘proneural enhancement’ and late ‘lateral inhibition’ functions. Cells of intermediate groups are circled in red. (b) Recruitment of R cells in the ommatidium. (c) Genetic interaction that regulates Ato expression (modified from Lubensky et al., 2011) [Citation85]. (d) A simplified interaction map of (B) for mathematical modelling of Ato expression (modified from Lubensky et al., 2011) [Citation85]. A, S, H, and N represent activation signals in the cell, delayed positive feedbacks in the cell, long-range cell-non-autonomous activation signals, and cell-non-autonomous inhibitory signals, respectively. (e) Schematics of the Ato expression pattern in Notch−, Notchts, Sca−, and Notchts Sca− mutant conditions](/cms/asset/0f23a0bd-32db-4fad-a538-12903914cf47/kfly_a_1953363_f0002_oc.jpg)
Based on the above biological evidence, Pennington and Lubensky developed a mathematical model to understand the mechanism of MF progression and spatial R8 patterning [Citation84]. The authors abstracted molecular interactions controlling Ato expression and set three variables: cell-autonomous activators (A), cell-non-autonomous activators (H), and cell-non-autonomous inhibitors (N). A includes the functions of Ato and Sens. H represents the functions of Hh and Dpp signalling, which drives the MF anteriorly and induces Ato expression. All cell-non-autonomous inhibitory signals, including Notch signalling and Sca functions, were integrated into the variable N. Numerical simulation of the simplified model recapitulated the MF propagation and R8 specification [Citation84]. This result suggests that these interactions among the three components are sufficient for stable and stationary patterning in the eye disc. Lubesnsky et al. revised this model by separating the functions of Ato and Sens (, d) [Citation85]; A, and S represent the function of Ato and Sens, respectively. The revised model also uses the variables H and N, as described in the original model. Simulations based on the four-component model produced continuous stripes of R8s when the effect of lateral inhibition by N was slower than that in wild type situations. Such striped patterns were experimentally reproduced by combining Notchts and Sca mutations () [Citation85]. Zhu et al. established a computational model by further describing the activity of the Hh, Dpp, Notch, and EGFR pathways in R8 specification [Citation86]. Simulations of the computational model showed the robust spatio-temporal order in R8 patterning and revealed that the coupling between long-range inductive signals by Hh and the short-range restrictive signals by Notch and EGFR is important for the accurate spacing of R8s. To elucidate the precise Notch dynamics in eye disc development, the early function of Notch in proneural enhancement and its late function in lateral inhibition need to be separately examined. Future mathematical models that separate these opposing functions of Notch signalling together with in vivo experiments will provide a better understanding of spatio-temporal Notch dynamics in eye disc development.
Notch function in optic lobe development
Another unique function of Notch dynamics is found in the developing optic lobe [Citation35,Citation37,Citation87–92]. The adult optic lobe is composed of four ganglia: the lamina, medulla, lobula, and lobula plate. Among these, the medulla is the largest component, and most of the medulla neurons are generated from NEs in the outer proliferation centre (OPC) during the larval and early pupal stages. During the early stages of larval development, NEs in the OPC proliferate by symmetric cell division () [Citation93]. Later, differentiation from NEs to NBs starts from the medial edge of the optic lobe and progresses in the medial to lateral direction () [Citation94,Citation95]. Proneural factors including Sc, L’sc, and Ase are expressed in wavefront cells and differentiated NBs, and determine the timing of differentiation () [Citation94–96]. Therefore, the wave of differentiation is named as the ‘proneural wave’ [Citation95]. Among the proneural factors, L’sc is transiently expressed in wavefront cells and thus used as a marker for the proneural wave [Citation95]. It has been reported that several conserved signalling pathways, such as Notch, EGFR, JAK/STAT, and Hippo regulate the proneural wave progression. Delta is temporally expressed in the wavefront cells (). Notch signalling shows two activation peaks, once in wavefront cells and again in NBs (). The first Notch activation negatively regulates proneural wave progression () [Citation96–103]. In Su(H) or Delta mutant clones, the proneural wave progression is accelerated [Citation98]. The second Notch activation in NBs regulates the expression of temporal transcription factors in NBs, which defines the neuronal subtype specification [Citation104]. EGFR signalling is activated in the wavefront and induces NB differentiation (, c) [Citation98]. Since Rhomboid (Rho), which is required for EGF ligand secretion, is one of the targets of EGFR signalling, it is speculated that sequential activation of EGFR signalling, which sweeps in a medial to lateral direction, is the driving factor for the progression of the proneural wave [Citation98]. JAK/STAT signalling is activated at lateral NEs and prevents precocious proneural wave progression () [Citation95,Citation101,Citation105]. Hippo signalling restricts NE proliferation and regulates the NE-NB transition () [Citation99,Citation106,Citation107].
Figure 3. Mathematical modelling of the proneural wave progression. (a) Schematic representation of the optic lobe. Neuroepithelial cells (NE), neuroblasts (NB), and the proneural wave (PW) are shown. (b) Development of the optic lobe. The proneural wave progresses in the medial to the lateral direction. EGFR and Notch signalling positively and negatively regulate the wave progression, respectively. (c) Activation of AS-C, EGFR signalling, Notch signalling, JAK/STAT signalling, and Hippo signalling during the proneural wave progression. (d) Genetic interaction among AS-C, EGFR signalling, and Notch signalling, JAK/STAT signalling, and Hippo signalling. (e) The mathematical model of the proneural wave including the interaction among AS-C, EGFR signalling, and Notch signalling (modified from Sato et al., 2016) [Citation111]. (f) Schematics of the simulation results of control (left) and partial EGFR knockdown (right). Activation of A and N is shown. White and black arrowheads represent cells with high A and high N levels, respectively
![Figure 3. Mathematical modelling of the proneural wave progression. (a) Schematic representation of the optic lobe. Neuroepithelial cells (NE), neuroblasts (NB), and the proneural wave (PW) are shown. (b) Development of the optic lobe. The proneural wave progresses in the medial to the lateral direction. EGFR and Notch signalling positively and negatively regulate the wave progression, respectively. (c) Activation of AS-C, EGFR signalling, Notch signalling, JAK/STAT signalling, and Hippo signalling during the proneural wave progression. (d) Genetic interaction among AS-C, EGFR signalling, and Notch signalling, JAK/STAT signalling, and Hippo signalling. (e) The mathematical model of the proneural wave including the interaction among AS-C, EGFR signalling, and Notch signalling (modified from Sato et al., 2016) [Citation111]. (f) Schematics of the simulation results of control (left) and partial EGFR knockdown (right). Activation of A and N is shown. White and black arrowheads represent cells with high A and high N levels, respectively](/cms/asset/2d64671c-55ee-4a3b-8fb7-d8072361fef7/kfly_a_1953363_f0003_oc.jpg)
Although the key components of Notch signalling, such as Delta, Notch, and AS-C are conserved, there are some differences between NB formation in the medulla and that in the embryonic neuroectoderm. First, Delta or AS-C expression does not show any salt-and-pepper pattern during proneural wave progression. Rather, cells at the wavefront show high Notch activity and express L’sc (, f). This is contradictory to the conserved idea of the lateral inhibition mechanism of Notch signalling, which shows a complementary pattern between cells expressing AS-C and cells with high Notch activity. Second, Notch signalling regulates the speed of the proneural wave, and all NEs finally differentiate into NBs following the proneural wave in the optic lobe. This is different from the Notch function in embryonic NB formation, where it regulates binary cell fate choice between neighbouring cells and only a small number of cells differentiate into NBs ().
To elucidate the Notch dynamics in proneural wave progression and interactions among signalling pathways, we and others have developed mathematical models [Citation37,Citation104,Citation108–111]. First, we focused on the interaction between Notch and EGFR signalling pathways because these pathways show transient activation in the transition zone from NEs to NBs and play central roles in proneural wave progression (, e) [Citation98,Citation111]. In the model shown in , e represents the concentration of the EGF ligand and activation of EGFR signalling. The secretion of the EGF ligand is spatially restricted by the function of Rho [Citation111]. Since it is feasible to presume the effect of EGFR signalling as a reaction-diffusion system, E is positively regulated by EGF ligand diffusion (deΔE)
nd negatively regulated by degradation (keE) [Citation112]. Additionally, in vivo experiments showed that activation of EGFR signalling is influenced by AS-C (aeA(A0-A)) () [Citation111]. Ni,j and Di,j show the activity of Notch signalling and the amount of Delta in the i-th and j-th cell, respectively. Since the major components of Notch signalling including Delta, Notch, and AS-C are all involved in proneural wave progression, the effect of Notch signalling is introduced by using a lateral inhibition system into the model. Ni,j is influenced by its degradation (knNi,j), trans-activation by Delta expressed in neighbouring l-th and m-th cells (dtΔl,m∈Λi,jDl,m), and cis-inhibition by Delta expressed in the same i-th and j-th cell (dcDi,j). Di,j is regulated by its degradation (kdDi,j) and receives a positive input from AS-C (adAi,j(A0-Ai,j)). Ai,j reflects the expression of AS-C including Sc, L’sc, and Ase in the i-th and j-th cell because these proneural factors show redundant functions in NB differentiation [Citation95]. Ai,j is used as a variable that represents the level of differentiation. Ai,j = 0 in undifferentiated cells, while Ai,j = A0 in fully differentiated NBs. Ai,j is positively regulated by EGFR signalling (Ei,j), and negatively regulated by Notch signalling (Ni,j) (, e).
Numerical simulations of the model recapitulated the proneural wave progression [Citation111]. Additionally, numerical simulations reproduced loss-of-function phenotypes of EGFR, Notch, or Delta [Citation111]. In the EGFR mutant area, wave progression and NB differentiation did not occur. In contrast, the proneural wave progression was accelerated in the Notch or Delta mutant cells. Therefore, in silico analysis combining Notch-mediated lateral inhibition with EGFR-mediated reaction-diffusion explains the in vivo proneural wave progression.
Moreover, the mathematical model explained why Notch signalling does not show a salt-and-pepper pattern in proneural wave progression. The absence of the salt-and-pepper pattern in the optic lobe can be explained by the diffusion of EGF ligands. If a cell at the wavefront starts expressing AS-C, it leads to the secretion of EGF ligands. Next, diffusible EGF ligands activate EGFR signalling and induce AS-C in neighbouring cells. This function of EGFR signalling may counteract Notch-mediated lateral inhibition and obscure the salt-and-pepper pattern. To test this hypothesis, a computer simulation was performed. In the control condition, A and N were both activated in the same wavefront cells (). However, reduction in EGFR activation showed a complementary pattern of A and N (). This prediction from the simulation was confirmed by the partial knockdown of EGFR activation in vivo. Partial disturbance of EGFR function showed a complementary pattern of L’sc and Notch activation. These results suggest that Notch-mediated lateral inhibition is implemented in proneural wave progression and that EGFR-mediated reaction-diffusion cancels salt-and-pepper pattern formation [Citation111].
One may think that Notch activation in the proneural wave progression has a similarity to that in the boundary formation in the Drosophila wing margins and veins. In the wing disc, Notch signalling shows a well-aligned activation pattern that defines sharp and stable boundaries through a process distinct from lateral inhibition [Citation32,Citation113–117]. In contrast, Notch-mediated lateral inhibition mediates desynchronization of NB differentiation between neighbouring cells in the optic lobe. Diffusible action of the EGF ligand cancels the salt-and-pepper pattern and forms the boundary of the proneural wavefront that sweeps across the neuroepithelium. Thus, the roles of Notch signalling in boundary formation are different between the wing and the proneural wave.
In the mathematical model described above, Notch signalling shows a single peak at wavefront [Citation111]. Recently, we reported that a modified mathematical model, formulated by introducing strong non-linearity in cis-inhibition of Notch signalling, reproduced two peaks of Notch activation in silico [Citation104]. in vivo experiments showed that Delta expression induces rapid degradation of the Notch protein in late endosomes, which generates a gap between the two Notch activation peaks. Additionally, we showed that the onset of the second Notch activation in NBs regulates neuronal cell fate decisions. These findings demonstrate the molecular mechanism of cis-inhibition of Notch signalling and the biological importance of the complex activation mechanism of Notch signalling in neurogenesis [Citation104].
In addition to Notch and EGFR signalling, JAK/STAT and Hippo signalling regulate the proneural wave progression [Citation95,Citation99,Citation101,Citation105–107]. It has been shown that JAK/STAT signalling induces the expression of the Notch target genes in the optic lobe () [Citation118]. Simulations from a revised mathematical model of the proneural wave including the interaction between Notch and JAK/STAT predicted that JAK/STAT signalling has a noise-cancelling function to suppress random and spontaneous NB differentiation [Citation110]. This prediction from the mathematical model was confirmed by reducing the JAK/STAT activity in vivo [Citation110]. Hippo signalling acts upstream of EGFR signalling and mediates NB differentiation by regulating Delta expression () [Citation99,Citation106]. Future mathematical studies including the function of Hippo signalling will give us a better understanding of the interaction among these signalling pathways and the progression of the proneural wave. Taken together, the combination of mathematical models with in vivo experiments using the optic lobe as a model has revealed new functions of Notch-mediated lateral inhibition.
Conclusion
In this article, we summarized mathematical approaches to elucidate the dynamics of Notch signalling during Drosophila neural development. The major roles of mathematical modelling in biology are to reproduce in vivo situations and validate the functions of molecules or signalling pathways. In all the cases described above, mathematical and computational simulations accurately recapitulated the in vivo patterning. Another important aspect of mathematical modelling is the prediction of new pathway functions and final differentiation patterns by changing parameters or adding new assumptions. Drosophila genetics using temperature-sensitive alleles or RNAi lines to partially inactivate gene functions in a spatially and/or temporally restricted manner has enabled the confirmation of mathematical predictions in vivo [Citation85,Citation111]. Improving genetic tools to gradually change the activity of target genes and advancing live imaging techniques to acquire quantitative data will be important for validating predictions from mathematical models.
The lateral inhibition mechanism of Notch signalling can be explained by trans-activation and cis-inhibition. However, trans-inhibition and cis-activation of Notch signalling have recently been reported. In mammalian angiogenesis, Delta-like 4 activates Notch signalling in neighbouring cells, while another Notch ligand, Jagged1, plays an opposing role [Citation119]. Single-cell imaging in isolated culture cells shows that cis-binding of the ligand and receptor activates Notch signalling in a cell [Citation120]. These trans-inhibition and cis-activation contradict the conserved idea of the lateral inhibition mechanism in which the signal-sending cells activate Notch signalling in neighbouring cells while Notch signalling is inactivated in the signal-sending cells. These mechanisms may be important for fine-tuning Notch activation. Further studies examining the kinetics of ligand-receptor binding and the molecular mechanisms that distinguish whether the binding activates or inhibits Notch signalling are required. Mathematical approaches will also facilitate our understanding of the complicated trans- and cis- regulations [Citation120–122].
Notch signalling generates different spatial patterns among NBs in the embryo, R8s in the eye disc, and NBs in the optic lobe in Drosophila. Another typical pattern generated by Notch signalling is an oscillatory pattern. In vertebrate somitogenesis, an interplay among Notch, fibroblast growth factor (FGF), and Wnt controls the rhythmic production of somites [Citation123,Citation124]. Interdisciplinary research combining in vivo experiments with mathematical modelling has revealed the molecular mechanism of how Notch signalling synchronizes oscillatory pattern formation [Citation125–127]. In Drosophila eye disc development, decreasing the Notch activity showed a striped differentiation pattern [Citation85]. The numerical simulation of the mathematical model of the proneural wave also showed a striped pattern of NBs when the EGFR activity was decreased, although this has not been reproduced in vivo [Citation111]. It is tempting to speculate that the basic function of Notch signalling is mechanistically well conserved in many biological systems, and only a slight difference in Notch activity or interactions between other signalling pathways can generate diverse outputs. Future interdisciplinary research will elucidate the convergence and divergence of Notch dynamics.
Data availability
No datasets were generated or analyzed during the current study.
Acknowledgments
We would like to thank Koichiro Uriu for his helpful input on Notch signaling in vertebrate segmentation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131(5):965–973.
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284(5415):770–776.
- Morgan TH. The Theory of the Gene. Am Nat. 1917;51(609):513–544.
- Simpson P. Lateral inhibition and the development of the sensory bristles of the adult peripheral nervous system of Drosophila. Development. 1990;109(3):509–519.
- Doe CQ, Goodman CS. Early events in insect neurogenesis. I. Development and segmental differences in the pattern of neuronal precursor cells. Dev Biol. 1985;111(1):193–205.
- Doe CQ, Goodman CS. Early events in insect neurogenesis. II. The role of cell interactions and cell lineage in the determination of neuronal precursor cells. Dev Biol. 1985;111(1):206–219.
- Siebel C, Lendahl U. Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol Rev. 2017;97(4):1235–1294.
- Zhang R, Engler A, Taylor V. Notch: an interactive player in neurogenesis and disease. Cell Tissue Res. 2018;371(1):73–89.
- Bahrampour S, Thor S. The Five Faces of Notch Signalling During Drosophila melanogaster Embryonic CNS Development. Adv Exp Med Biol. 2020;1218:39–58.
- Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13(9):654–666.
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66(10):1631–1646.
- Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–2140.
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233.
- Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722–735.
- Wharton KA, Johansen KM, Xu T, et al. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43(3):567–581.
- Fleming RJ, Scottgale TN, Diederich RJ, et al. The gene Serrate encodes a putative EGF-like transmembrane protein essential for proper ectodermal development in Drosophila melanogaster. Genes Dev. 1990;4(12a):2188–2201.
- Thomas U, Speicher SA, Knust E. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development. 1991;111(3):749–761.
- Vassin H, Campos-Ortega JA. Genetic Analysis of Delta, a Neurogenic Gene of Drosophila melanogaster. Genetics. 1987;116(3):433–445.
- Fortini ME. Gamma-secretase-mediated proteolysis in cell-surface-receptor signalling. Nat Rev Mol Cell Biol. 2002;3(9):673–684.
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9(21):2598–2608.
- Schweisguth F, Posakony JW. Suppressor of Hairless, the Drosophila homolog of the mouse recombination signal-binding protein gene, controls sensory organ cell fates. Cell. 1992;69(7):1199–1212.
- Petcherski AG, Kimble J. Mastermind is a putative activator for Notch. Curr Biol. 2000;10(13):R471–3.
- Wu L, Aster JC, Blacklow SC, et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26(4):484–489.
- Jennings B, Preiss A, Delidakis C, et al. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120(12):3537–3548.
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9(21):2609–2622.
- Nakao K, Campos-Ortega JA. Persistent expression of genes of the enhancer of split complex suppresses neural development in Drosophila. Neuron. 1996;16(2):275–286.
- Oellers N, Dehio M, Knust E. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol Gen Genet. 1994;244(5):465–473.
- Kunisch M, Haenlin M, Campos-Ortega JA. Lateral inhibition mediated by the Drosophila neurogenic gene delta is enhanced by proneural proteins. Proc Natl Acad Sci U S A. 1994;91(21):10139–10143.
- Miller AC, Lyons EL, Herman TG. cis-Inhibition of Notch by endogenous Delta biases the outcome of lateral inhibition. Curr Biol. 2009;19(16):1378–1383.
- Del Alamo D, Schweisguth F. Notch signalling: receptor cis-inhibition to achieve directionality. Curr Biol. 2009;19:R683–4.
- Sprinzak D, Lakhanpal A, Lebon L, et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature. 2010;465(7294):86–90.
- de Celis JF, Bray S. Feed-back mechanisms affecting Notch activation at the dorsoventral boundary in the Drosophila wing. Development. 1997;124(17):3241–3251.
- Micchelli CA, Rulifson EJ, Blair SS. The function and regulation of cut expression on the wing margin of Drosophila: notch, Wingless and a dominant negative role for Delta and Serrate. Development. 1997;124(8):1485–1495.
- Bocci F, Onuchic JN, Jolly MK. Understanding the Principles of Pattern Formation Driven by Notch Signaling by Integrating Experiments and Theoretical Models. Front Physiol. 2020;11:929.
- Sato M, Suzuki T, Nakai Y. Waves of differentiation in the fly visual system. Dev Biol. 2013;380(1):1–11.
- Treisman JE. Retinal differentiation in Drosophila. Wiley Interdiscip Rev Dev Biol. 2013;2(4):545–557.
- Sato M, Yasugi T. Regulation of Proneural Wave Propagation Through a Combination of Notch-Mediated Lateral Inhibition and EGF-Mediated Reaction Diffusion. Adv Exp Med Biol. 2020;1218:77–91.
- Skeath JB, Carroll SB. Regulation of proneural gene expression and cell fate during neuroblast segregation in the Drosophila embryo. Development. 1992;114(4):939–946.
- Cabrera CV, Martinez-Arias A, Bate M. The expression of three members of the achaete-scute gene complex correlates with neuroblast segregation in Drosophila. Cell. 1987;50(3):425–433.
- Martin-Bermudo MD, Martinez C, Rodriguez A, et al. Distribution and function of the lethal of scute gene product during early neurogenesis in Drosophila. Development. 1991;113(2):445–454.
- Artavanis-Tsakonas S, Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991;7(1):403–408.
- Hassan B, Vaessin H. Regulatory interactions during early neurogenesis in Drosophila. Dev Genet. 1996;18(1):18–27.
- Campos-Ortega JA, Jan YN. Genetic and molecular bases of neurogenesis in Drosophila melanogaster. Annu Rev Neurosci. 1991;14(1):399–420.
- Tanemura M, Honda H, Yoshida A. Distribution of differentiated cells in a cell sheet under the lateral inhibition rule of differentiation. J Theor Biol. 1991;153(3):287–300.
- Honda H, Tanemura M, Yoshida A. Estimation of neuroblast numbers in insect neurogenesis using the lateral inhibition hypothesis of cell differentiation. Development. 1990;110(4):1349–1352.
- Marnellos G, Mjolsness E. A gene network approach to modeling early neurogenesis in Drosophila. Pac Symp Biocomput. 1998;30–41.
- Lehotzky D, Zupanc GKH. Cellular Automata Modeling of Stem-Cell-Driven Development of Tissue in the Nervous System. Dev Neurobiol. 2019;79(5):497–517.
- Collier JR, Monk NA, Maini PK, et al. Pattern formation by lateral inhibition with feedback: a mathematical model of delta-notch intercellular signalling. J Theor Biol. 1996;183:429–446.
- Rozante LC, Gubitoso MD, Matioli SR. A framework for modeling of juxtacrine signaling systems. Genet Mol Res. 2007;6:821–845.
- Plahte E. Pattern formation in discrete cell lattices. J Math Biol. 2001;43(5):411–445.
- Sprinzak D, Lakhanpal A, LeBon L, et al. Mutual inactivation of Notch receptors and ligands facilitates developmental patterning. PLoS Comput Biol. 2011;7(6):e1002069.
- Corson F, Couturier L, Rouault H, et al. Self-organized Notch dynamics generate stereotyped sensory organ patterns in Drosophila. Science. 2017;356.
- Cohen M, Georgiou M, Stevenson NL, et al. Dynamic filopodia transmit intermittent Delta-Notch signaling to drive pattern refinement during lateral inhibition. Dev Cell. 2010;19(1):78–89.
- Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53(2):217–240.
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113(3):841–850.
- Heberlein U, Wolff T, Rubin GM. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75(5):913–926.
- Ma C, Zhou Y, Beachy PA, et al. The segment polarity gene hedgehog is required for progression of the morphogenetic furrow in the developing Drosophila eye. Cell. 1993;75(5):927–938.
- Heberlein U, Moses K. Mechanisms of Drosophila retinal morphogenesis: the virtues of being progressive. Cell. 1995;81(7):987–990.
- Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129(6):1295–1306.
- Jarman AP, Grell EH, Ackerman L, et al. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369(6479):398–400.
- Jarman AP, Sun Y, Jan LY, et al. Role of the proneural gene, atonal, in formation of Drosophila chordotonal organs and photoreceptors. Development. 1995;121:2019–2030.
- Dominguez M. Dual role for Hedgehog in the regulation of the proneural gene atonal during ommatidia development. Development. 1999;126(11):2345–2353.
- Greenwood S, Struhl G. Progression of the morphogenetic furrow in the Drosophila eye: the roles of Hedgehog, Decapentaplegic and the Raf pathway. Development. 1999;126(24):5795–5808.
- Borod ER, Heberlein U. Mutual regulation of decapentaplegic and hedgehog during the initiation of differentiation in the Drosophila retina. Dev Biol. 1998;197(2):187–197.
- Dominguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997;11:3254–3264.
- Baker NE, Yu SY. Proneural function of neurogenic genes in the developing Drosophila eye. Curr Biol. 1997;7(2):122–132.
- Baonza A, Freeman M. Notch signalling and the initiation of neural development in the Drosophila eye. Development. 2001;128(20):3889–3898.
- Ligoxygakis P, Yu SY, Delidakis C, et al. A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development. 1998;125(15):2893–2900.
- Li Y, Baker NE. Proneural enhancement by Notch overcomes Suppressor-of-Hairless repressor function in the developing Drosophila eye. Curr Biol. 2001;11(5):330–338.
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3(8):1099–1112.
- Baker NE, Zitron AE. Drosophila eye development: notch and Delta amplify a neurogenic pattern conferred on the morphogenetic furrow by scabrous. Mech Dev. 1995;49(3):173–189.
- Baker NE, Yu S, Han D. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol. 1996;6(10):1290–1301.
- Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250(4986):1370–1377.
- Mlodzik M, Baker NE, Rubin GM. Isolation and expression of scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 1990;4(11):1848–1861.
- Lee EC, Hu X, Yu SY, et al. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16(3):1179–1188.
- Ellis MC, Weber U, Wiersdorff V, et al. Confrontation of scabrous expressing and non-expressing cells is essential for normal ommatidial spacing in the Drosophila eye. Development. 1994;120(7):1959–1969.
- Powell PA, Wesley C, Spencer S, et al. Scabrous complexes with Notch to mediate boundary formation. Nature. 2001;409(6820):626–630.
- Singson A, Leviten MW, Bang AG, et al. Direct downstream targets of proneural activators in the imaginal disc include genes involved in lateral inhibitory signaling. Genes Dev. 1994;8(17):2058–2071.
- Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr Biol. 2001;11(6):396–404.
- Yang L, Baker NE. Role of the EGFR/Ras/Raf pathway in specification of photoreceptor cells in the Drosophila retina. Development. 2001;128(7):1183–1191.
- Baker NE, Bhattacharya A, Firth LC. Regulation of Hh signal transduction as Drosophila eye differentiation progresses. Dev Biol. 2009;335(2):356–366.
- Rogers EM, Brennan CA, Mortimer NT, et al. Pointed regulates an eye-specific transcriptional enhancer in the Drosophila hedgehog gene, which is required for the movement of the morphogenetic furrow. Development. 2005;132(21):4833–4843.
- Frankfort BJ, Nolo R, Zhang Z, et al. senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron. 2001;32(3):403–414.
- Pennington MW, Lubensky DK. Switch and template pattern formation in a discrete reaction-diffusion system inspired by the Drosophila eye. Eur Phys J E Soft Matter. 2010;33(2):129–148.
- Lubensky DK, Pennington MW, Shraiman BI, et al. A dynamical model of ommatidial crystal formation. Proc Natl Acad Sci U S A. 2011;108(27):11145–11150.
- Zhu H, Owen MR, Mao Y. The spatiotemporal order of signaling events unveils the logic of development signaling. Bioinformatics. 2016;32(15):2313–2320.
- Contreras EG, Sierralta J, Oliva C. Novel Strategies for the Generation of Neuronal Diversity: lessons From the Fly Visual System. Front Mol Neurosci. 2019;12:140.
- Egger B, Gold KS, Brand AH. Regulating the balance between symmetric and asymmetric stem cell division in the developing brain. Fly (Austin). 2011;5(3):237–241.
- Yasugi T, Nishimura T. Temporal regulation of the generation of neuronal diversity in Drosophila. Dev Growth Differ. 2016;58(1):73–87.
- Neriec N, Desplan C. From the Eye to the Brain: development of the Drosophila Visual System. Curr Top Dev Biol. 2016;116:247–271.
- Sato M, Yasugi T, Trush O. Temporal patterning of neurogenesis and neural wiring in the fly visual system. Neurosci Res. 2019;138:49–58.
- Apitz H, Salecker I. A Challenge of Numbers and Diversity: neurogenesis in the Drosophila Optic Lobe. J Neurogenet. 2014;28(3–4):233–249.
- Lanet E, Gould AP, Maurange C. Protection of neuronal diversity at the expense of neuronal numbers during nutrient restriction in the Drosophila visual system. Cell Rep. 2013;3(3):587–594.
- Egger B, Boone JQ, Stevens NR, et al. Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2007;2(1):1.
- Yasugi T, Umetsu D, Murakami S, et al. Drosophila optic lobe neuroblasts triggered by a wave of proneural gene expression that is negatively regulated by JAK/STAT. Development. 2008;135(8):1471–1480.
- Orihara-Ono M, Toriya M, Nakao K, et al. Downregulation of Notch mediates the seamless transition of individual Drosophila neuroepithelial progenitors into optic medullar neuroblasts during prolonged G1. Dev Biol. 2011;351(1):163–175.
- Egger B, Gold KS, Brand AH. Notch regulates the switch from symmetric to asymmetric neural stem cell division in the Drosophila optic lobe. Development. 2010;137(18):2981–2987.
- Yasugi T, Sugie A, Umetsu D, et al. Coordinated sequential action of EGFR and Notch signaling pathways regulates proneural wave progression in the Drosophila optic lobe. Development. 2010;137(19):3193–3203.
- Reddy BV, Rauskolb C, Irvine KD. Influence of Fat-Hippo and Notch signaling on the proliferation and differentiation of Drosophila optic neuroepithelia. Development. 2010;137(14):2397–2408.
- Contreras EG, Egger B, Gold KS, et al. Dynamic Notch signalling regulates neural stem cell state progression in the Drosophila optic lobe. Neural Dev. 2018;13(1):25.
- Ngo KT, Wang J, Junker M, et al. Concomitant requirement for Notch and Jak/Stat signaling during neuro-epithelial differentiation in the Drosophila optic lobe. Dev Biol. 2010;346(2):284–295.
- Wang W, Liu W, Wang Y, et al. Notch signaling regulates neuroepithelial stem cell maintenance and neuroblast formation in Drosophila optic lobe development. Dev Biol. 2011;350(2):414–428.
- Weng M, Haenfler JM, Lee CY. Changes in Notch signaling coordinates maintenance and differentiation of the Drosophila larval optic lobe neuroepithelia. Dev Neurobiol. 2012;72(11):1376–1390.
- Wang M, Han X, Liu C, et al. Intracellular trafficking of Notch orchestrates temporal dynamics of Notch activity in the fly brain. Nat Commun. 2021;12(1):2083.
- Wang W, Li Y, Zhou L, et al. Role of JAK/STAT signaling in neuroepithelial stem cell maintenance and proliferation in the Drosophila optic lobe. Biochem Biophys Res Commun. 2011;410(4):714–720.
- Kawamori H, Tai M, Sato M, et al. Fat/Hippo pathway regulates the progress of neural differentiation signaling in the Drosophila optic lobe. Dev Growth Differ. 2011;53(5):653–667.
- Richter C, Oktaba K, Steinmann J, et al. The tumour suppressor L(3)mbt inhibits neuroepithelial proliferation and acts on insulator elements. Nat Cell Biol. 2011;13(9):1029–1039.
- Jorg DJ, Caygill EE, Hakes AE, et al. The proneural wave in the Drosophila optic lobe is driven by an excitable reaction-diffusion mechanism. Elife. 2019;8.
- Ei SI, Ishii H, Sato M, et al. A continuation method for spatially discretized models with nonlocal interactions conserving size and shape of cells and lattices. J Math Biol. 2020;81(4–5):981–1028.
- Tanaka Y, Yasugi T, Nagayama M, et al. JAK/STAT guarantees robust neural stem cell differentiation by shutting off biological noise. Sci Rep. 2018;8(1):12484.
- Sato M, Yasugi T, Minami Y, et al. Notch-mediated lateral inhibition regulates proneural wave propagation when combined with EGF-mediated reaction diffusion. Proc Natl Acad Sci U S A. 2016;113(35):E5153–62.
- Wiley HS, Shvartsman SY, Lauffenburger DA. Computational modeling of the EGF-receptor system: a paradigm for systems biology. Trends Cell Biol. 2003;13(1):43–50.
- de Celis JF, Garcia-Bellido A. Roles of the Notch gene in Drosophila wing morphogenesis. Mech Dev. 1994;46(2):109–122.
- Diaz-Benjumea FJ, Cohen SM. Serrate signals through Notch to establish a Wingless-dependent organizer at the dorsal/ventral compartment boundary of the Drosophila wing. Development. 1995;121(12):4215–4225.
- Kim J, Irvine KD, Carroll SB. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell. 1995;82(5):795–802.
- Rulifson EJ, Blair SS. Notch regulates wingless expression and is not required for reception of the paracrine wingless signal during wing margin neurogenesis in Drosophila. Development. 1995;121(9):2813–2824.
- Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124(17):3283–3291.
- Wang H, Chen X, He T, et al. Evidence for Tissue-Specific JAK/STAT Target Genes in Drosophila Optic Lobe Development. Genetics. 2013;195(4):1291–1306.
- Benedito R, Roca C, Sorensen I, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137(6):1124–1135.
- Nandagopal N, Santat LA, Elowitz MB. Cis-activation in the Notch signaling pathway. Elife. 2019;8.
- Shaya O, Sprinzak D. From Notch signaling to fine-grained patterning: modeling meets experiments. Curr Opin Genet Dev. 2011;21(6):732–739.
- Binshtok U, Sprinzak D. Modeling the Notch Response. Adv Exp Med Biol. 2018;1066:79–98.
- Pourquie O. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell. 2011;145(5):650–663.
- Oates AC, Morelli LG, Ares S. Patterning embryos with oscillations: structure, function and dynamics of the vertebrate segmentation clock. Development. 2012;139(4):625–639.
- Yoshioka-Kobayashi K, Matsumiya M, Niino Y, et al. Coupling delay controls synchronized oscillation in the segmentation clock. Nature. 2020;580(7801):119–123.
- Jiang YJ, Aerne BL, Smithers L, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408(6811):475–479.
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58(2):455–476.
