ABSTRACT
In mammalian cells, one-third of all polypeptides is transported into or through the ER-membrane via the Sec61-channel. While the Sec61-complex facilitates the transport of all polypeptides with amino-terminal signal peptides (SP) or SP-equivalent transmembrane helices (TMH), the translocating chain-associated membrane protein (now termed TRAM1) was proposed to support transport of a subset of precursors. To identify possible determinants of TRAM1 substrate specificity, we systematically identified TRAM1-dependent precursors by analyzing cellular protein abundance changes upon TRAM1 depletion in HeLa cells using quantitative label-free proteomics. In contrast to previous analysis after TRAP depletion, SP and TMH analysis of TRAM1 clients did not reveal any distinguishing features that could explain its putative substrate specificity. To further address the TRAM1 mechanism, live-cell calcium imaging was carried out after TRAM1 depletion in HeLa cells. In additional contrast to previous analysis after TRAP depletion, TRAM1 depletion did not affect calcium leakage from the ER. Thus, TRAM1 does not appear to act as SP- or TMH-receptor on the ER-membrane’s cytosolic face and does not appear to affect the open probability of the Sec61-channel. It may rather play a supportive role in protein transport, such as making the phospholipid bilayer conducive for accepting SP and TMH in the vicinity of the lateral gate of the Sec61-channel.
Abbreviations: ER, endoplasmic reticulum; OST, oligosaccharyltransferase; RAMP, ribosome-associated membrane protein; SP, signal peptide; SR, SRP-receptor; SRP, signal recognition particle; TMH, signal peptide-equivalent transmembrane helix; TRAM, translocating chain-associated membrane protein; TRAP, translocon-associated protein.
Introduction
In mammalian cells, the endoplasmic reticulum (ER) membrane is a major site for membrane protein biogenesis and for most soluble proteins the entry point into compartments of the secretory pathway [Citation1–Citation5]. Protein transport into the mammalian ER involves various transport components and precursor polypeptides with amino-terminal signal peptides (SP) or SP-equivalent transmembrane helices (TMH) [Citation6–Citation9]. In cotranslational transport, the signal recognition particle (SRP) recognizes SP and TMH of nascent precursor polypeptides emerging from cytosolic ribosomes, and the resulting SRP/ribosome/nascent chain complex is recruited to the ER membrane by the SRP receptor (SR) [Citation10,Citation11]. The precursor polypeptides are next inserted into the Sec61-complex, i.e. the polypeptide-conducting channel of the ER membrane () [Citation12–Citation30]. According to the current view on the opening of the Sec61-channel, SP, or TMH of nascent precursor polypeptides intercalate between the Sec61α transmembrane helices 2 and 7, displace helix 2, and open the “lateral gate” of the Sec61 complex, which is formed by these two transmembrane helices [Citation16,Citation18,Citation23]. Subsequently, the nascent chain can be fully inserted into the Sec61-channel, either in “hairpin” (where the amino-terminus of the SP or TMH stays in the cytosol) or “head-first” configuration (where the amino-terminus of the SP or TMH reaches into the ER lumen), and initiate translocation [Citation27–Citation29]. In either case, the full channel opening may or may not require support from auxiliary transport components (). Elegant in vitro experiments with purified components in proteoliposomes demonstrated that at least for some precursor polypeptides (such as bovine preprolactin) SR and Sec61 complex are sufficient to initiate ER import [Citation13]. In contrast, other precursor polypeptides with inefficient SP or TMH rely on Sec61-auxiliary and -associated membrane components, such as the translocon-associated protein (TRAP) complex [Citation31–Citation38] and/or the translocating chain-associated membrane protein (TRAM) [Citation39–Citation48], which is now termed TRAM1 because of the discovery of close homologs, TRAM1L1 and TRAM2 () [Citation49,Citation50]. Interestingly, TRAM2 has been described to be involved in collagen type I biogenesis [Citation49] and to invert the topology of transmembrane helices that do not promote a specific initial orientation in the membrane [Citation50]. However, TRAM1 function and mechanism as well as its rules of engagement remained largely unknown.
Figure 1. Putative position of TRAM1 in the ER membrane and experimental strategy for the identification of TRAM1 clients and compensatory proteins by TRAM1 depletion in HeLa cells
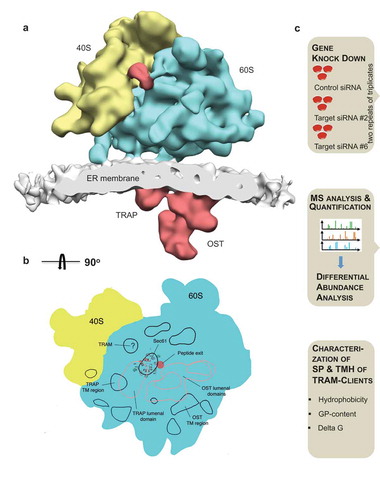
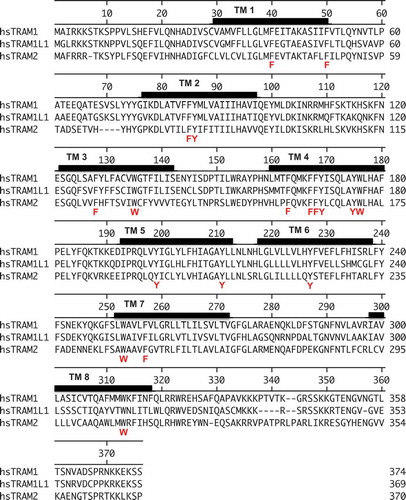
Figure 2. Amino acid sequences of TRAM1, TRAM1L1, and TRAM2
Originally, TRAP complex and TRAM1 were described together with Sec61-complex and the multimeric enzyme oligosaccharyltransferase (OST) as ribosome-associated membrane proteins (RAMP) in canine pancreatic rough microsomes, supporting the notion that TRAP complex and TRAM1 are in close proximity to Sec61 complexes [Citation13]. TRAM1, however, was more easily displaced from the ribosomes as compared to the other three membrane protein complexes, suggesting that it may be more loosely associated with ribosomes and the other three components. In the canine pancreatic rough microsomes TRAM1, TRAP complex, and Sec61 complex were found in approximately stoichiometric amounts [Citation47]. Information on the composition of the native protein transport machinery in the ER membrane came from fluorescence resonance energy transfer (FRET) experiments, which employed fluorescently labeled antibodies against transport components, permeabilized canine cells, and fluorescence microscopy [Citation48]. According to this cell biological approach, the TRAP complex and TRAM1 protein are permanently in close proximity to Sec61 complexes. Notably, a permanent association of ribosome-associated Sec61 complexes with TRAP and OST was also confirmed in the 3D-reconstructions after cryoelectron tomography (CET) of native translocons in rough microsomes, derived from canine pancreas or human cells, and even in intact cells [Citation20–Citation22,Citation25,Citation26] (). However, efforts to pinpoint the position of TRAM1 using 3D-reconstructions after CET of native translocons from TRAM1 depleted versus untreated HeLa cells remained unsuccessful, most likely because it does not comprise lumenal or cytosolic domains large enough for identification by CET [Citation20,Citation21].
TRAM1, an eight-transmembrane domain ER protein, belongs to a protein family, which is characterized by the presence of the TRAM/LAG1/CLN8 homology (TLC) domain and is postulated to bind ceramide or related sphingolipids () [Citation48]. Originally, it was discovered by the crosslinking of nascent presecretory proteins early in their translocation into the ER [Citation39]. Subsequently, it was also found to interact with nascent membrane proteins during their initial integration into the Sec61-channel [Citation40–Citation42,Citation45,Citation46]. A more recent radiolabelling approach employed crosslinking of stalled precursor polypeptide chains in transit through the canine pancreatic ER membrane and subsequent two-dimensional gel electrophoresis plus mass spectrometric analysis [Citation30]. As expected based on both the FRET experiments and previous cross-linking studies with nascent bovine preprolactin chains, preprolactin was found in complex with Sec61 complex, TRAP, TRAM1, and OST also under these conditions.
Traditionally, the substrate specificities of mammalian protein transport components (e.g., the TRAP-complex and TRAM1) have been investigated in cell-free translation reactions in which a small set of model precursor proteins is synthesized one-by-one in the presence of proteoliposomes or in pulse/chase experiments in human cells that overproduce the model precursor of interest [Citation32,Citation36,Citation43]. These approaches are suitable for addressing whether or not a certain component can stimulate ER import of a given precursor polypeptide. However, due to the focus of these experimental strategies on a single precursor, they fail to clearly define the characteristics of precursor polypeptides that lead to TRAM1- or TRAP-dependence and they certainly don´t give insights into whether or not this is relevant under cellular conditions with much faster translation rates and lots of competing precursors with different affinities for the Sec61-channel. Nevertheless, it was suggested that precursor proteins with shorter than average N-regions and shorter than average H-regions in their SP require TRAM1 for efficient insertion into the lateral gate [Citation43].
Here, we identify and characterize the native precursor polypeptides that involve TRAM1 in their biogenesis in human cells under cellular conditions, i.e. at physiological concentrations and in the presence of competing precursors [Citation38]. To this end, we combine the siRNA-mediated gene knock-down of TRAM1 in HeLa cells with label-free quantitative proteomic analysis and differential protein abundance analysis, following the protocol that was established for Sec61 and TRAP depletion [Citation38] (). Furthermore, we employ live-cell calcium imaging in the same cellular system to address the question of whether or not TRAM1 plays a role in Sec61-channel gating. The presented results are discussed in light of the results of both approaches after TRAP depletion and point to a supportive role of TRAM1 in ER protein import and suggest that TRAM1 may affect the phospholipid bilayer in the vicinity of the lateral gate of the Sec61-channel.
Results
The possible client specificity of TRAM1 in human ER protein import
Here, we set out to identify and characterize the native precursor polypeptides that involve TRAM1 in their biogenesis in human cells under cellular conditions. As a positive proof of concept for the proteomic approach, HeLa cells had previously been depleted of the Sec61-complex using two different SEC61A1-targeting siRNAs for 96 h [Citation38]. Then, we assessed the proteomic consequences of this knock-down via label-free quantitative proteomics and differential protein abundance analysis relative to cells treated with non-targeting siRNA. Gene Ontology (GO) terms assigned over 60% of the 482 negatively affected proteins to organelles of the endocytic and exocytic pathways, representing a strong enrichment compared to the value for the total quantified proteome (). We also detected significant enrichment of precursor proteins with SP, N-glycosylated proteins, and membrane proteins. This suggested that the precursors of these proteins are substrates of the Sec61-complex and were degraded by the proteasome upon its depletion, which was experimentally confirmed. According to bioinformatic analysis, ~30% of the total quantified proteome of roughly 7,200 proteins comprises Sec61 substrates. Thus, our experimental approach clearly underestimates the number of different precursor polypeptides that rely on the Sec61-complex. We attribute this to the timing of the experiment, which aims for maximal Sec61 depletion in combination with minimal effects on cell growth and viability, and, therefore, neglects, e.g. proteins with long half-lives or high affinities for Sec61. The positively affected proteins included compensatory components, including the two subunits of the SRP receptor, plus several cytosolic ubiquitin-conjugating enzymes, consistent with cytosolic accumulation of precursors and their proteasomal degradation [Citation38]. Thus, our experimental strategy in human cells was successfully used to analyze the client spectrum of the Sec61-complex-an essential transport component-under physiological conditions. These results further set the stage for subsequent analysis of precursor-specific transport components, such as TRAP complex [Citation38] and TRAM1. The subsequent identification of TRAP-dependent precursors characterized a high glycine and proline content and/or low hydrophobicity of the respective SP as distinguishing features for TRAP dependence [Citation38].
Table 1. Statistics of the identification of TRAM1 clients in comparison to the identification of Sec61α1 and TRAP clients, respectively
Here, we performed similar analyses after TRAM1 depletion using two different TRAM1-targeting siRNAs (TRAM1 #2-UTR siRNA, TRAM1 #6-UTR siRNA) in comparison to the same non-targeting (termed control) siRNA, which had previously been employed in the Sec61 and TRAP depletion experiments [Citation38]. Notably, TRAM1 depletion from HeLa cells for 96 h just began to affect cell growth and viability. The average values for growth and viability were 87.5 ± 4% sem of control siRNA-treated cells (n = 11) and 90 ± 3% sem (n = 11), respectively, and, thus were slightly better than for Sec61 depleted cells and similar to TRAP depleted cells [Citation21]. After TRAM1 depletion in two independent experiments with triplicates for each of the two siRNAs, 7,501.5 ± 223 different proteins were quantitatively characterized by MS, representing approximately 50% of the cellular proteome. Of these proteins, 5,961 were detected in both experiments and were statistically analyzed (, Supplementary Table 1, http://www.ebi.ac.uk/pride/archive/projects/Identifiers PXD008178). They included a good representation of proteins with cleaved SP (7%), N-glycosylated proteins (9%), and membrane proteins (13%), which was comparable to the Sec61 and TRAP experiments () [Citation38]. Applying the same statistical analysis of the ratio changes after TRAM1 depletion as used after SEC61A1 and TRAPB silencing, we found that TRAM1 depletion significantly affected the steady-state levels of 204 proteins: 86 negatively and 118 positively (permutation false discovery rate-adjusted p value < 0.05) (, Supplementary Tables 2 and 3). As expected, TRAM1 itself was negatively affected, which was confirmed by western blot (). Of the other negatively affected proteins, GO terms assigned ~48% to organelles of the secretory pathway, which corresponds to a 1.7-fold enrichment (, large cakes, 47.83% divided by 28.49% = 1.68) and is between the values observed after Sec61 and TRAP depletion (). We also detected for these proteins significant enrichment of proteins with SP (2.3-fold), N-glycosylated proteins (2.2-fold), and membrane proteins (1.8-fold) (, small cakes), which was similar to the TRAP experiment but lower as compared to the Sec61 experiment [Citation38]. Among the proteins negatively affected upon TRAM1 depletion, the precursors are putative clients of the TRAM1-complex (Supplementary Table 2). The identified precursors included 13 proteins with cleavable SP (including 4 membrane proteins with different numbers of transmembrane domains) and 17 membrane proteins with TMH, and represented N-glycosylated proteins (16) and non-glycosylated proteins as well as proteins with low and high cellular concentrations (14) (, and ). The fact that the numbers of negatively affected proteins after TRAM1 depletion are lower as compared to the negatively affected proteins after Sec61 depletion is consistent with TRAM1 being a precursor-specific auxiliary transport component to the Sec61-complex ().
Figure 3. Identification of TRAM1 clients and compensatory proteins by TRAM1 depletion in HeLa cells
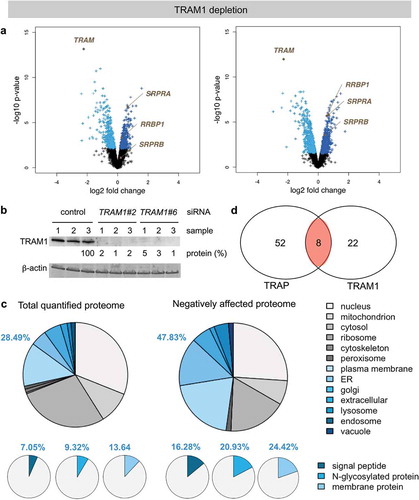
Table 2. Summary of TRAM1 clients and their characteristics in comparison to Sec61α1 and TRAP clients, respectively. Protein abundances in HeLa cells are given in nM and were taken from Hein et al., 2015 [Citation74]. N-glyco, number of N-glycosylation sites; nd, not detected; TMD, number of transmembrane domains, including TMH where applicable
The proteins positively affected by TRAM1 depletion included both SRP receptor subunits (SRPRA, SRPRB) of the ER membrane and two cytosolic ubiquitin-conjugating enzymes (i.e. MYCBP2, UBE2C), which is consistent with accumulation of precursor polypeptides in the cytosol after TRAM1 depletion and reminiscent of the situation after Sec61- and TRAP-complex depletion (, Supplementary Table 3) [Citation38]. There was no indication for activation of the unfolded protein response (UPR) in the course of the 96 h knock-down, which would have been indicative of protein misfolding in the ER and would have resulted in the overproduction of ER-resident molecular chaperones, such as BiP/Grp78 (coded by the HSPA5 gene) or Grp170 (product of the HYOU1 gene) (i.e. would have appeared in Supplementary Table 3). Interestingly, the putative ribosome- and/or mRNA-binding protein in the ER-membrane, termed p180 or RRBP1 [Citation51–Citation56], was positively affected by TRAM1 depletion but not after Sec61- and TRAP-complex depletion [Citation38]. The possible significance of this observation will be addressed in future proteomic analyses after RRBP1 depletion. We note that RRBP1 and SRPRB are type I ER-membrane proteins with TMH and, thus, represent proteins that apparently don´t depend on TRAM1 for membrane insertion and integration (SRPRA is a peripheral ER-membrane protein and recruited to the membrane by SRPRB).
Characteristics of TRAM1 clients
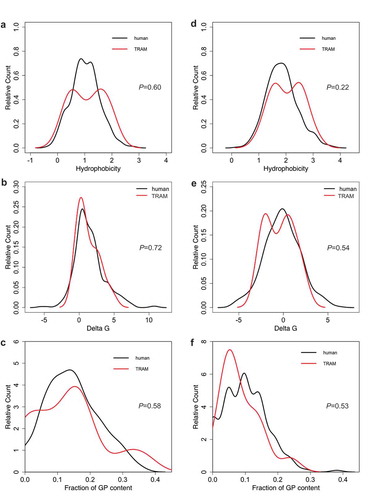
We next addressed the reason for the apparent substrate specificity of TRAM1. Therefore, we analyzed the putative TRAM1 clients with respect to the physicochemical properties of their amino-terminal SP and TMH according to the established procedures [Citation38]. Using custom scripts, we computed the hydrophobicity score and glycine/proline (GP) content of SP and TMH sequences. A peptide’s hydrophobicity score was assigned as the average hydrophobicity of its amino acids according to the Kyte–Doolittle propensity scale (averaged over the sequence length). GP content was calculated as the total fraction of glycine and proline in the respective sequence. ∆Gapp values of SP and TMH were calculated with the ∆Gapp predictor for TM helix insertion (http://dgpred.cbr.su.se). Similar to Sec61 clients, SP and TMH of TRAM1 clients showed average overall hydrophobicities and ∆Gapp values () [Citation38]. Furthermore, TRAM1 clients showed average GP content, i.e. did not share with TRAP the preference for SP with high glycine-plus proline content [Citation38]. Thus, TRAM1 is unlikely to act as a receptor for SP of certain precursor polypeptides on the cytosolic face of the ER membrane, which is consistent with its lack of any soluble cytosolic domains.
Based on in vitro experiments with purified components in proteoliposomes, it had been suggested that precursor proteins with shorter than average N-regions and shorter than average H-regions in their SP require TRAM1 for efficient insertion into the lateral gate [Citation43]. Therefore, SP segmentation into N-, H-, and C-regions was carried out for the 13 TRAM1 clients with SP using the well-established prediction tool Phobius (http://phobius.sbc.su.se). According to these predictions, the lengths of these SP regions are 4.3, 11.1, and 5.2 (n = 10), respectively, for the SP of TRAM1 clients, while the corresponding values for all human SP are 6.5, 11.6, and 5.6 (n = 2876) (S. Schorr, personal communication). Thus, the proteomic data confirmed the tendency toward shorter than average N-regions (Wilcoxon rank test p= 0.22) but not the shorter than average H-regions for the SP of TRAM1 clients under physiological conditions (see Discussion).
TRAM1 does not affect Sec61-channel gating
Finally, we asked if TRAM1 plays a role in Sec61-channel gating. In its open state, the Sec61-channel allows passive Ca2+ efflux from the ER [Citation15,Citation57–Citation60]; therefore, Sec61-channel opening can be monitored in intact HeLa cells via live-cell Ca2+ imaging and identified a role of TRAP in Sec61-channel gating to the open state [Citation38]. Notably, in HeLa cells, the Sec61-channel accounts for roughly 60% of the passive ER Ca2+ leak channels. Thus, in our present work, HeLa cells were depleted of TRAM1 using the same two siRNAs as above, and ER Ca2+ leakage was monitored as an increase of cytosolic Ca2+. Similar to treatment with control siRNA, cellular TRAM1 depletion with one of the two different TRAM1-targeting siRNAs (TRAM1 #2-UTR siRNA, TRAM1 #6-UTR siRNA) did hardly result in altered ER Ca2+ leakage in response to thapsigargin and did hardly affect total cellular Ca2+ content (). These results are drastically different from the TRAP depletion experiments, which revealed a substantially decreased ER Ca2+ leakage in response to thapsigargin [Citation38]. Thus, live-cell calcium imaging did not suggest a Sec61-channel gating activity for TRAM1.
Figure 4. Physico-chemical properties of TRAM1 clients with SP and TMH
Discussion
The Sec61-complex facilitates translocation of all polypeptides with amino-terminal signal peptides (SP) or SP-equivalent transmembrane helices (TMH) into the ER and is permeable to Ca2+ in its open state [Citation4,Citation5,Citation12,Citation13]. The TRAP-complex supports the opening of the Sec61-channel for protein translocation in a substrate-specific manner [Citation32,Citation38]. It was originally termed the signal-sequence receptor (SSR) complex [Citation31], had been crosslinked to nascent polypeptides at late translocation stages [Citation35], and has been demonstrated to physically associate with Sec61 [Citation33,Citation35]. The ribosome-associated Sec61-complex and the TRAP-complex form a stable stoichiometric super-complex, termed translocon, which can be visualized in its native state by CET () [Citation20–Citation22,Citation34]. In vitro transport studies showed that the TRAP-complex stimulates protein translocation depending on the efficiency of the SP in transport initiation and recent studies in intact human cells suggest that TRAP may also affect TMH topology [Citation32,Citation36]. A systematic identification of TRAP-dependent precursors by transient TRAP-depletion in HeLa cells in combination with analysis of cellular protein abundance changes using quantitative label-free proteomics characterized a high glycine and proline content and/or low hydrophobicity of the respective SP as distinguishing features for TRAP dependence [Citation38]. In combination with structural analysis of native translocons by CET [Citation26], these results suggested a scenario, where the TRAP γ-subunit recognizes TRAP-clients on the cytosolic ER-surface and, subsequently, supports Sec61-channel opening by direct interaction of the ER-lumenal domains of the TRAP α- and β-subunits with the ER-lumenal loop 5 of the Sec61 α-subunit, i.e. the crucial “hinge” region between the amino- and carboxy-terminal halves of Sec61α [Citation38]. This function of TRAP is consistent with the observation, that TRAP depletion from HeLa cells results in a reduction in passive calcium leakage from the ER, due to a lower open probability of the calcium-permeable Sec61-channel in the absence of TRAP complex.
So far, efforts to pinpoint the position of TRAM1 using CET of native translocons in ER-membrane vesicles, derived from the canine pancreas or human cells, have not been successful [Citation20,Citation21]. Even though, we suggest that it may represent the density that is consistently found opposite of the “lateral gate” in CET of native translocons [Citation4,Citation5] (). This view is consistent with the above-described i) ribosome-association of TRAM1 [Citation13,Citation48], ii) its stoichiometric association with Sec61 complex and TRAP complex [Citation47,Citation48], and iii) its crosslinking to nascent precursors of soluble as well as membrane proteins at an early stage of their translocation and membrane insertion, respectively [Citation39–Citation42,Citation45,Citation46].
To characterize TRAM1-dependent precursors, we combined siRNA-mediated TRAM1 depletion in human cells, label-free quantitative proteomics, and differential protein abundance analysis, as previously carried out after Sec61 or TRAP depletion. By applying our unbiased approach in living human cells, we identified 30 potential TRAM1 clients that included precursors of soluble and membrane proteins with cleavable SP (13) and membrane proteins without cleavable SP (17), and precursors of both N-glycosylated and non-glycosylated proteins (). Thus, TRAM1 is involved in ER import of these precursors under cellular conditions. However, our experimental approach of gene silencing for 96 h most likely underestimated the number of clients also in the case of TRAM1, i.e. neglected, e.g. proteins with long half-lives or low abundance. Comparison of the distributions of precursor proteins to those found for Sec61 and TRAP clients in similar experiments does not point to a preference of TRAM1 for any type of precursor polypeptides (). Furthermore, analysis of the physicochemical properties of SP and TMH of the TRAM1 clients did not point to a specific feature, i.e. did not identify an obvious substrate specificity, with the possible exception to precursors with short N-regions in their SP. Interestingly, 57% of the TRAM1 clients were also negatively affected by Sec61 depletion and 27% of the TRAM1 clients were also negatively affected by TRAP depletion under similar conditions (, , Venn diagram). This is consistent with the above-described colocalization of TRAM1 with Sec61 and TRAP [Citation13,Citation33,Citation35,Citation48]. Notably, eight of the TRAM1 clients were below the level of detection in HeLa cells in the previous analyses of Sec61 and/or TRAP clients and, therefore, could not be expected in the overlap of clients (). Considering i) the underestimation of clients by our experimental approach for Sec61 as well as TRAM1, ii) the strong overlap in clients of these two transport components (), and iii) TRAM1´s apparent lack of precursor preference () indicates that TRAM1, unlike TRAP, does not act as a receptor for SP or TMH but, rather plays a supportive role in ER protein import for many precursor polypeptides (see below).
Figure 5. TRAM1 depletion plus live-cell Ca2+ imaging does not reveal a TRAM1 function in the Sec61-channel opening
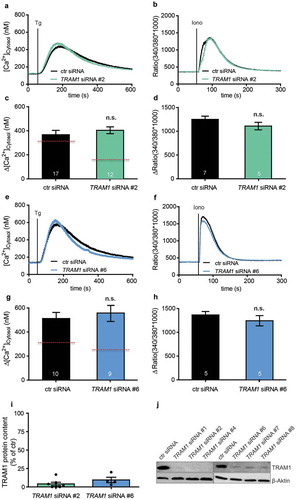
What that role maybe was addressed in live-cell Ca2+ imaging experiments in combination with TRAM1 depletion. In further contrast to TRAP, TRAM1 did not appear to affect the gating of the Sec61-channel. Therefore, the question remains how TRAM1 actually supports ER protein import. We propose that due to its TLC domain TRAM1 may have the ability to interact with ceramides and sphingolipids, or even phospholipids in general, and, therefore, may be able to influence bilayer thickness and/or phospholipid packing in the vicinity of the Sec61-complex, possibly near the lateral gate of the Sec61-channel [Citation50]. In this context, the enrichment of amino acids with aromatic side chains in transmembrane domain 4 may be of particular relevance (). In this domain, there are two rather centrally positioned tyrosines and one tryptophan, which can be expected to align with lipid tails, whereas these residues are typically found at membrane interfaces in other transmembrane domains [Citation61,Citation62]. Reduced bilayer thickness and/or loose phospholipid packing could support the lateral exit of SP and TMH from the Sec61-channel via its lateral gate and this support could be important for different precursors to a different extent. Notably, similar activities have been suggested for YidC and Oxa1 in membrane protein biogenesis in bacteria and mitochondria, respectively [Citation63,Citation64]. Such an activity of TRAM1 would easily explain the observed inhibitory effect of the forced addition of cholesterol into canine pancreatic rough microsomal membranes on ER protein import [Citation65]. The unphysiologically high cholesterol level would counteract the suggested TRAM1 activity on the bilayer. Furthermore, the proposed TRAM1 activity may be able to explain the fact that single-channel recordings from open Sec61-channels routinely showed two pore diameters, one of 0.6-nm width and compatible with the structure of the open Sec61-channel and one of 6-nm width and incompatible with Sec61-channel structure [Citation15]. Therefore, we suggest that the large pore may reflect open Sec61-channels with a distorted lipid bilayer near the open lateral gate of the channel. Consistently, CET of native ER membranes indeed revealed an electron density within the lipid bilayer opposite of the lateral gate of the Sec61-channel and gave the impression that the bilayer in the vicinity of the lateral gate is different as compared to membrane areas distant from ribosomes [Citation5,Citation20–Citation22]. Thus, the phospholipid bilayer in the neighborhood of the lateral gate may indeed be affected by TRAM1 and this may be of different relevance for different precursors, i.e., particularly important for precursors with short N-regions in their SP. This raises the interesting possibility that TRAM1 in human cells could allow regulated access of precursor polypeptides to the ER. Interestingly, TRAM1 was found to be subject to phosphorylation [Citation66]. Thus, this modification could be a candidate for ER protein import-regulation at the level of TRAM1.
Materials & methods
Materials
SuperSignal West Pico Chemiluminescent Substrate (# 34078) was purchased from PierceTM, Thermo Fisher Scientific. ECLTM Plex goat anti-mouse IgG-Cy3 conjugate (PA43009, used dilution 1:2,500) was purchased from GE Healthcare. Horseradish peroxidase coupled anti-rabbit IgG from goat (A 8275, used dilution 1:1,000) was from Sigma-Aldrich. We purchased murine monoclonal antibodies against β-actin also from Sigma (A5441). Rabbit polyclonal antibodies were raised against the carboxy-terminal peptide of canine TRAM1 (12-mer, used dilution 1:500) plus an amino-terminal cysteine. Antibody quality was previously documented [Citation20].
Cell manipulation and analysis
HeLa cells (DSM no. ACC 57) were obtained from the German Collection of Microorganisms and Cell Cultures, routinely tested for mycoplasma contamination by VenorGeM Mycoplasm Detection Kit (Biochrom AG, WVGM), and replaced every five years. They were cultivated at 37°C in a humidified environment with 5% CO2, in DMEM with 10% fetal bovine serum (FBS; Sigma-Aldrich) and 1% penicillin and streptomycin. Cell growth was monitored using the Countess® Automated Cell Counter (Invitrogen) following the manufacturer’s instructions.
For gene silencing, 5.2 × 105 HeLa cells were seeded per 6-cm culture plate, followed by incubation under normal culture conditions. For TRAM1 silencing, the cells were transfected with a final concentration of 10 nM targeting siRNA (TRAM1 #2-UTR siRNA, target sequence: CTGGCATTATACTGAACTATA; TRAM1 #6-UTR siRNA, target sequence: TAGCTTTGGTCCCATCTTTAA) (Qiagen) or 20 nM AllStars Negative Control siRNA (Qiagen) using HiPerFect Reagent (Qiagen) following the manufacturer’s instructions. After 24 h, the medium was changed and the cells were transfected a second time. Silencing efficiencies were evaluated by western blot analysis using the TRAM1 antibody and an anti-β-actin antibody from a mouse for sample comparison. The ß-actin antibodies were visualized with ECLTM Plex goat anti-mouse IgG-Cy3 conjugate using the Typhoon-Trio imaging system combined with Image Quant TL software 7.0 (GE Healthcare). For the detection of TRAM1 antibody, we employed SuperSignal West Pico Chemiluminescent Substrate and the Fusion SL (peqlab) luminescence imaging system with the accompanying software.
Label-free quantitative proteomic analysis
Cells (1 × 106) were harvested, washed twice in PBS, and lysed in buffer containing 6 M GnHCl, 20 mM tris(2-carboxyethyl)phosphine (TCEP; PierceTM, Thermo Fisher Scientific), 40 mM 2-chloroacetamide (CAA; Sigma-Aldrich) in 100 mM Tris, pH 8.0. The lysate was heated to 95°C for 2 min, and then sonicated in a Bioruptor sonicator (Diagenode) at the maximum power setting for 10 cycles of 30 s each. The entire process of heating and sonication was repeated once, and then the sample was diluted 10-fold with digestion buffer (25 mM Tris, pH 8, with 10% acetonitrile). Protein extracts were digested for 4 h with endoproteinase lysC, followed by the addition of trypsin for overnight digestion. After digestion, peptides were purified and loaded for mass spectrometric analysis as previously described [Citation38,Citation67]. Raw data were processed using the MaxQuant computational platform [Citation68]. The peak list was searched against Human Uniprot databases, with an initial precursor and fragment tolerance of 4.5 ppm. The match between the run feature was enabled, and proteins were quantified across samples using the label-free quantification algorithm in MaxQuant as label-free quantification (LFQ) intensities [Citation69].
Data analysis
Each of the two MS experiments provided proteome-wide abundance data as LFQ intensities for three sample groups – one control (non-targeting siRNA treated) and two stimuli (down-regulation by two different targeting siRNAs directed against the same gene) – each having three data points (replicates). Only proteins that were detected in both experiments (repeats) were considered. Missing data points were generated by imputation as previously described [Citation38]. To identify which proteins were affected by TRAM1 knock-down in siRNA-treated cells relative to the non-targeting (control) siRNA-treated sample, we log2-transformed the ratio between siRNA and control siRNA samples and performed two separate unpaired t-tests for each siRNA against the control siRNA sample. The p-values obtained by unpaired t-tests were corrected for multiple testing using a permutation false discovery rate (FDR) test. Proteins with an FDR-adjusted p-value of below 5% were considered significantly affected by knockdown of the targeted proteins. The results from the two unpaired t-tests were then intersected for further analysis meaning that the abundance of all reported candidates was statistically significantly affected in both siRNA silencing experiments. All statistical analyses were performed using the R package SAM (http:www-stat-class.stanford.edu) [Citation70].
Protein annotations of signal peptides, transmembrane regions, and N-glycosylation sites in humans and yeast were extracted from UniProtKB entries using custom scripts [Citation38]. The enrichment of functional Gene Ontology annotations (cellular components and biological processes) among the secondarily affected proteins was computed using the GOrilla package [Citation71]. Using custom scripts, we computed the hydrophobicity score and glycine/proline (GP) content of SP and TMH sequences [Citation38]. A peptide’s hydrophobicity score was assigned as the average hydrophobicity of its amino acids according to the Kyte–Doolittle propensity scale (averaged over the sequence length) [Citation72]. GP content was calculated as the total fraction of glycine and proline in the respective sequence.
Live-cell Ca2+ imaging
HeLa cells were loaded with 4 µM Fura-2 AM (Molecular Probes, Thermo Fisher Scientific) in DMEM, and incubated for 45 min at 25°C as described [Citation57,Citation58]. Then, the cells were washed twice and incubated at room temperature in Ca2+-free buffer (140 mM NaCl, 5 mM KCl, 1 mM MgCl2, 0.5 mM EGTA, 10 mM glucose in 10 mM HEPES-KOH, pH 7.35). Where indicated, HeLa cells were treated with siRNA for 96 h prior to Ca2+ imaging and were treated with 1 µM Thapsigargin (Molecular Probes, Thermo Fisher Scientific) or 5 µM Ionomycin (Thermo Fisher Scientific). Ratiometric measurements were conducted for 5 or 10 min using an iMIC microscope and the polychromator V (Till Photonics), with alternating excitation at 340 nm and 380 nm and measurement of the fluorescence emitted at 510 nm. The microscope was equipped with a Fluar M27 lens with 20× magnification and 0.75 numerical aperture (Carl Zeiss), and an iXonEM+ camera (Andor Technology). Images containing 50–55 cells/frame were sampled every 3 s using TILLvisION software (Till Photonics). Fura-2 signals were recorded as the F340/F380 ratio, where F340 and F380 correspond to the background-subtracted fluorescence intensities at 340 and 380 nm, respectively. Data were analyzed using Excel 2007. P-values were determined using unpaired Student´s t-test.
Supplemental Material
Download MS Excel (500.6 KB)Supplemental Material
Download MS Excel (22.2 KB)Supplemental Material
Download MS Excel (713.6 KB)Acknowledgments
We thank Dr. Nagarjuna Nagaraj (Max-Planck-Institute of Biochemistry, Biochemistry core facility, Martinsried, Germany) for MS-analyses. Personal communication by Stefan Schorr refers to his unpublished work: Schorr S, Nguyen D, Haßdenteufel S, Nagaraj N, Cavalié A, Greiner M, Weissgerber P, Loi M, Paton AW, Paton JC, Molinari M, Förster F, Dudek J, Lang S, Helms V, and Zimmermann R. Proteomics identifies signal peptide features determining the substrate specificity in human Sec62/Sec63-dependent ER protein import.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [Citation73] partner repository with the dataset identifier: PXD008178 (http://www.proteomexchange.org).
Orbi1931, TRAM experiment 1: Sample 16-18, scr control siRNA; Sample 25-27, TRAM siRNA #2; Sample 28-30, TRAM siRNA #6;
Orbi2085, TRAM experiment 2: Sample 1-3, scr control siRNA; Sample 10-12, TRAM siRNA #2; Sample 13-15, TRAM siRNA #6.
All other data are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358.
- Blobel G, Dobberstein B. Transfer of proteins across membranes: I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–851.
- Blobel G, Dobberstein B. Transfer of proteins across membranes: II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862.
- Pfeffer S, Dudek J, Zimmermann R, et al. Organization of the native ribosome-translocon complex at the mammalian endoplasmic reticulum membrane. Biochim Biophys Acta. 2016;1860:2122–2129.
- Lang S, Pfeffer S, Lee P-H, et al. An update on Sec61-channel function, mechanisms, and related diseases. Front Physiol. 2017;8:887.
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105.
- von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678.
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:564–571.
- Nilsson IM, Lara P, Hessa T, et al. The code for directing proteins for translocation across ER membrane: SRP cotranslationally recognizes specific features of a signal sequence. J Mol Biol. 2015;427:1191–1201.
- Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: structure of the signal recognition particle. Curr Opin Struct Biol. 2005;15:213–220.
- Halic M, Blau M, Becker T, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511.
- Görlich D, Prehn S, Hartmann E, et al. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503.
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630.
- Simon SM, Blobel G. A protein-conducting channel in the endoplasmic reticulum. Cell. 1991;65:371–380.
- Wirth A, Jung M, Bies C, et al. The Sec61p complex is a dynamic precursor activated channel. Mol Cell. 2013;12:261–268.
- van den Berg B, Clemons WM, Collinson I, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44.
- Becker T, Bhushan S, Jarasch A, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373.
- Gumbart J, Trabuco LG, Schreiner E, et al. Regulation of the protein-conducting channel by a bound ribosome. Structure. 2009;17:1453–1464.
- Zhang B, Miller TF III. Long-timescale dynamics and regulation of Sec-facilitated protein translocation. Cell Rep. 2012;2:927–937.
- Pfeffer S, Brandt F, Hrabe T, et al. Structure and 3D arrangement of endoplasmic reticulum membrane-associated ribosomes. Structure. 2012;20:1508–1518.
- Pfeffer S, Dudek J, Gogala M, et al. Structure of the mammalian oligosaccharyl-transferase in the native ER protein translocon. Nat Commun. 2014;5:3072.
- Pfeffer S, Burbaum L, Unverdorben P, et al. Structure of the native Sec61 protein-conducting channel. Nat Commun. 2015;6:8403.
- Voorhees RM, Fernández IS, Scheres SHW, et al. Structure of the mammalian ribosome-Sec61 complex to 3.4 å resolution. Cell. 2014;157:1632–1643.
- Voorhees RM, Hegde RS. Structure of the Sec61 channel opened by a signal sequence. Science. 2016;351:88–91.
- Mahamid J, Pfeffer S, Schaffer M, et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016;351:969–972.
- Pfeffer S, Dudek J, Schaffer M, et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat Commun. 2017;8,14516.
- Devaraneni PK, Conti B, Matsumara Y, et al. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell. 2011;146:134–147.
- Park E, Menetret JF, Gumbart JC, et al. Structure of the SecY channel during initiation of protein translocation. Nature. 2014;506:102–106.
- Vermeire K, Bell TW, Van Puyenbroeck V, et al. Signal peptide-binding drug as a selective inhibitor of co-translational protein translocation. PLoS Biol. 2014;12:e1002011.
- Conti BJ, Devaraneni PK, Yang Z, et al. Cotranslational stabilization of Sec62/63 within the ER Sec61 translocon is controlled by distinct substrate-driven translocation events. Mol Cell. 2015;58:269–283.
- Wiedmann M, Kurzchalia TV, Hartmann E, et al. A signal sequence receptor in the endoplasmic reticulum membrane. Nature. 1987;328:830–833.
- Fons RD, Bogert BA, Hegde RS. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J Cell Biol. 2003;160:529–539.
- Shibatani T, David LL, McCormack AL, et al. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochem. 2005;44:5982–5992.
- Menetret JF, Hegde RS, Aguiar M, et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure. 2008;16:1126–1137.
- Dejgaard K, Theberge J-F, Heath-Engel H, et al. Organization of the Sec61 translocon, studied by high resolution native electrophoresis. J Proteome Res. 2010;9:1763–1771.
- Sommer N, Junne T, Kalies K-U, et al. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim Biophys Acta. 2013;1833:3104–3111.
- Bano-Polo M, Martinez-Garay CA, Grau B, et al. Membrane insertion and topology of the translocon-associated protein (TRAP) gamma subunit. Biochem Biophys Acta. 2017;1859:903–909.
- Nguyen D, Stutz R, Schorr S, et al. Proteomics reveals signal peptide features determining the client specificity in human TRAP-dependent ER protein import. Nat Commun. 2018;9:3765.
- Görlich D, Hartmann E, Prehn S, et al. A protein of the endoplasmic reticulum involved early in polypeptide translocation. Nature. 1992;357:47–52.
- High S, Martoglio B, Görlich D, et al. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993;268:26745–26751.
- Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein translocation through the ER membrane. Embo J. 1994;13:3973–3982.
- Do H, Falcone D, Lin J, et al. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378.
- Voigt S, Jungnickel B, Hartmann E, et al. Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol. 1996;134:25–35.
- Hegde RS, Voigt S, Rapoport TA, et al. TRAM regulates the exposure of nascent secretory proteins to the cytosol during translocation into the endoplasmic reticulum. Cell. 1998;92:621–631.
- Sauri A, McCormick PJ, Johnson AE, et al. Sec61alpha and TRAM are sequentially adjacent to a nascent viral membrane protein during its ER integration. J Mol Biol. 2007;366:366–374.
- McCormick PJ, Miao Y, Shao Y, et al. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol Cell. 2003;12:329–341.
- Snapp EL, Reinhart GA, Bogert BA, et al. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J Cell Biol. 2004;164:997–1007.
- Guth S, Völzing C, Müller A, et al. Protein transport into canine pancreatic microsomes: a quantitative approach. Eur J Biochem. 2004;271:3200–3207.
- Stefanonvic B, Stefanovic L, Schnabl B, et al. TRAM2 protein interacts with endoplasmic reticulum Ca2+ pump Serca2b and is necessary for collagen type I synthesis. Mol Cell Biol. 2004;24:1758–1768.
- Chen Q, Denard B, Lee CE, et al. Inverting the topology of a transmembrane protein by regulating the translocation of the first transmembrane helix. Mol Cell. 2016;63:567–578.
- Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature. 1990;346:540–544.
- Savitz AJ, Meyer DI. 180-kDa ribosome receptor is essential for both ribosome binding and protein translocation. J Cell Biol. 1993;120:853–863.
- Wanker EE, Sun Y, Savitz AJ, et al. Functional characterization of the 180-kD Ribosome Receptor in vivo. J Cell Biol. 1995;130:29–39.
- Cui XA, Zhang H, Palazzo AF. p180 promotes the ribosome-independent localization of a subset of mRNA to the endoplasmic reticulum. PLoS Biol. 2012;10:e1001336.
- Cui XA, Zhang Y, Hong SJ, et al. Identification of a region within the placental alkaline phosphatase mRNA that mediates p180-dependent targeting to the endoplasmic reticulum. J Biol Chem. 2013;288:29633–29641.
- Voigt F, Zhang H, Cui XA, et al. Single-molecule quantification of translation-dependent association of mRNAs with the endoplasmic reticulum. Cell Rep. 2017;21:3740–3753.
- Lang S, Erdmann F, Jung M, et al. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels. 2011;5:228–235.
- Lang S, Schäuble N, Cavalié A, et al. Live cell calcium imaging in combination with siRNA mediated gene silencing identifies Ca2+ leak channels in the ER membrane and their regulatory mechanisms. J Visual Exp. 2011. DOI:10.3791/2730
- Erdmann F, Schäuble N, Lang S, et al. Interaction of calmodulin with Sec61a limits Ca2+ leakage from the endoplasmic reticulum. Embo J. 2011;30:17–31.
- Schäuble N, Lang S, Jung M, et al. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. Embo J. 2012;31:3282–3296.
- MacCallum JL, Bennett WFD, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J. 2008;94:3393–3404.
- Yau WM, Wimley WC, Gawrisch K, et al. The preference of tryptophan for membrane interfaces. Biochem. 1998;37:14713–14718.
- Chen Y, Capponi S, Zu L, et al. YidC insertase of Escherichia coli: water accessibility and membrane shaping. Structure. 2017;25:1403–1414.
- Martin R, Larsen AH, Corey RA, et al. Structure and dynamics of the central lipid pool and proteins of the bacterial holo-translocon. Biophys J. 2019;116:1931–1940.
- Nilsson IM, Ohvo-Rekilä H, Slotte JP, et al. Inhibition of protein translocation across the endoplasmic reticulum membrane by sterols. J Biol Chem. 2001;276:41748–41754.
- Gruss OJ, Feick P, Frank R, et al. Phosphorylation of components of the ER translocation site. Eur J Biochem. 1999;260:785–793.
- Nagaraj N, Kulak NA, Cox J, et al. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol Cell Proteom. 2012;1:111.013722.
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372.
- Cox J, Hein MY, Luber CA, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteom. 2014;13:2513–2526.
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121.
- Eden E, Navon R, Steinfeld I, et al. Gorilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48.
- Kyte J, Doolittle RF. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132.
- Vizcaíno JA, Csordas A, del-Toro N, et al. 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 2016;44:D447–D456.
- Hein MY, Hubner NC, Poser I, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723.
