ABSTRACT
Epigenetic modifications extensively occur in mammalian embryonic development and cell differentiation process. They play an essential role in the reprogramming of nuclei during somatic cell nuclear transfer (SCNT) and subsequent in vitro embryonic development. Recently, SCNT embryos have been verified to contain a subnormal level of histone H3K4 dimethylation (H3K4me2) in contrast to in vitro fertilized embryos. This finding suggested that increasing H3K4me2 levels may ameliorate the aberrant development of cloned embryos. In this study, we investigated the influence of treating donor cells with trans-2-Phenylcyclopropylamine (2-PCPA), a specific inhibitor of lysine-specific demethylase 1 (LSD1), on embryogenesis, H3K4me2 level, and gene expression in cloned goat embryos. Treated goat fetal fibroblast cells (GFFs) with 2-PCPA served as donor cells for subsequent SCNT. Results showed that H3K4me2 levels in treated GFFs increased gradually with the increasing 2-PCPA concentration (p < 0.05) and had no obvious influence in cell viability. The 2-PCPA-induced up-regulation of H3K4me2 levels led to G0/G1 cell cycle arrest and the difference was significant at 2μM compared with the control group (p < 0.05). Interestingly, the development rate of goat SCNT embryos in vitro was significantly improved and aberrant H3K4me2 levels were effectively corrected in 2-PCPA-treated SCNT embryos in contrast to that in SCNT control embryos. Moreover, 2-PCPA treatment promoted the mRNA expression of key developmental genes Oct4 and Sox2 (p < 0.05) without affecting the expression levels of imprinted genes IGF2R and H19 in goat SCNT embryos. These results indicated that abnormal H3K4me2 status can be corrected and SCNT embryo development can be promoted through treatment of donor cells with 2-PCPA.
Abbreviations: SCNT: somatic cell nuclear transfer; H3K4me2: H3K4 dimethylation; 2-PCPA: trans-2-Phenylcyclopropylamine; LSD1: lysine-specific demethylase 1; GFFs: goat fetal fibroblast cells; IVF: in vitro fertilization; iPS: induced pluripotent stem; PBS: phosphate-buffered saline; IVM: in vitro maturation; RNAPII: RNA polymerase II; HMTs: histone methyltransferase
Introduction
Currently, the application of somatic cell nuclear transfer (SCNT) is limited because of its low cloning efficiency (Vajta et al. Citation2003). Deficient reprogramming of nuclei may be the leading cause of poor clonal embryogenesis, high morbidity, and mortality in cloned individuals (Shao et al. Citation2008). The incomplete erasure of epigenetic modifications in SCNT embryos might result in various abnormalities, such as high levels of DNA methylation and aberrant histone methylation (Humpherys et al. Citation2002). Histone methylation regulates gene expression through complicated mono-, di-, and trimethylation modifications (Santosrosa et al. Citation2002).
During embryonic development, H3K4 dimethylation (H3K4me2) is mainly involved in the transcriptional activation of some genes (Santosrosa et al. Citation2002), playing important roles in embryonic development and cell differentiation. Situated in the gene promoter region, H3K4me2 is an epigenetic marker related to transcriptional activation that is mediated by H3K4 methyltransferases, including Set1 and Set2 (Kim and Buratowski Citation2009). Demethylation of H3K4me2 is implemented mainly with the assistance of lysine acid-specific demethylase 1 (LSD1), which through its action inhibits gene expression (Metzger et al. Citation2005).
During embryo reprogramming, H3K4me2 modifications must be removed, and subsequent embryonic genomic activation must be re-established for the activation of specific genes. In contrast to in vitro fertilized (IVF) embryos, cloned embryos frequently have aberrant levels of modified H3K4me2 (Shao et al. Citation2008). In mice, an oocyte cytoplasm often fails to increase the degree of modification of H3K4me2 levels during reprogramming because of the low levels of H3K4me2 in a donor nucleus. Thus, H3K4me2 levels are unchanged. Clonal embryos undergo a sudden increase in H3K4me2 levels in the 8-cell stage, and this increase may be related to gene activation (Shao et al. Citation2008). Low levels of H3K4me2 in cloned embryos suggest that increasing H3K4me2 levels through the addition of H3K4 demethylation inhibitors may be beneficial to early embryonic development.
Recently, SCNT embryo development have been induced through the use of small molecular inhibitors, such as trichostatin A, UNC0638, and 5-aza-2′-deoxycytidine, which specifically modify histone methylation, DNA methylation, or histone acetylation status (Ding et al. Citation2008; Tsuji and Tsunoda Citation2009; Wang et al. Citation2017). Induced Pluripotent stem (iPS) cells have been obtained through the use of trans-2-Phenylcyclopropylamine (2-PCPA) (Li et al. Citation2009), a H3K4 demethylation inhibitor (Schmidt and Mccafferty Citation2007). The clones of iPS cells can be obtained only with the addition of 2-PCPA combined with six other small molecules and prevents the participation of exogenous factors, such as OCT4 (Hou et al. Citation2013). These studies showed that upregulation of H3K4me2 due to the addition of small molecule compounds reprograms somatic nuclei. In one of our previous studies, an abnormal H3K9me2 level in SCNT embryos was partially corrected after treating donor cells with UNC0638—an inhibitor of histone-lysine methyltransferase G9A, although the in vitro developmental rate was not improved (Wang et al. Citation2017). In the present study, the impact of treating donor cells with 2-PCPA—a specific inhibitor of LSD1—on embryogenesis, H3K4me2 level, and gene expression in cloned goat embryos was investigated.
Results
Effects of 2-PCPA treatment on cell viability and global H3K4me2 levels of goat fetal fibroblast cells (GFFs)
Considering the possible cellular toxicity of 2-PCPA on fibroblasts, we determined the cell viability of the GFFs. After treating the GFFs with increasing concentrations of 2-PCPA up to 10 μM, we did not observe any dramatic change in cell viability (). These results indicated that 2-PCPA has no visible cellular toxicity to fibroblasts at least in the range of 0 μM to 10 μM.
Figure 1. Cell viability and global H3K4me2 level of GFFs treated with different concentrations of 2-PCPA. (A) Cell viability of GFFs treated with different concentrations of 2-PCPA. Cell viability was indicated as the ratio of 2-PCPA treated groups relative to untreated ones. (B) Staining of H3K4me2 (green) and DNA (blue) in GFFs. (C) Relative fluorescence intensity compared with control group (0 μM) was calculated. The ratio of H3K4me2 to DAPI when GFFs were treated with 2-PCPA (0 μM) was set as 1.0. Data are presented as the mean ± SD of three independent experiments. Different superscripts represent significant difference between treated group and control group (p < 0.05). H3K4me2: H3K4 dimethylation; GFFs: goat fetal fibroblast cells; 2-PCPA: trans-2-Phenylcyclopropylamine. Scale bar = 50 μm.
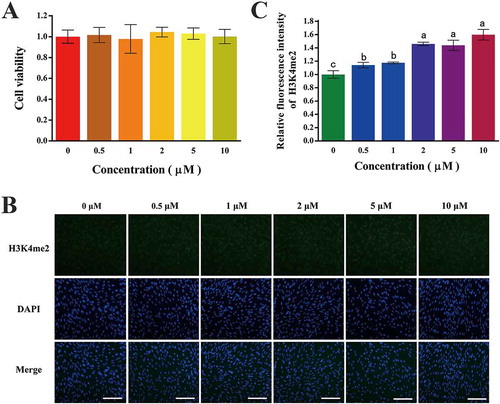
GFFs treated with different concentrations of 2-PCPA were subjected to immunofluorescence staining for the analysis of global H3K4me2 levels (). The quantized fluorescent intensity of the level of H3K4me2 gradually increased with the 2-PCPA enrichment. Specifically, it increased more than 1.4 times when the concentration was above 2 μM with a ceiling value of 1.6 times (10 μM; ; p < 0.05).
G0/G1 cell cycle arrest due to 2-PCPA induction
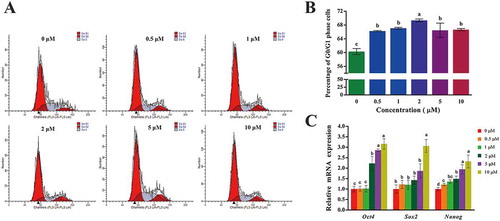
The cell cycle of 2-PCPA-induced cells was measured by flow cytometric analysis (). As the concentration increased, the proportion of cells in the G0/G1 phase showed an upward trend, whereas cells in the Second Gap/Mitosis(G2/M) and Synthesis (S) phases showed a downward trend. Interestingly, when the concentration exceeded 5 μM, the percentage of cells in the G0/G1 phase slightly decreased. Overall, the rate increased after 2-PCPA treatment, and the difference was considerable compared with the control group at 2 μM (, p < 0.05). These data demonstrated that up-regulating the level of H3K4me2 induced by 2-PCPA resulted in G0/G1 cell cycle arrest and suppression of cell proliferation.
Figure 2. Cell cycle and expression of pluripotency genes of GFFs treated with different concentrations of 2-PCPA. (A) Cell cycle analysis using a flow cytometry. (B) The proportion of cells in G0/G1 phase. (C) Expression of key pluripotency genes of GFFs treated with different concentrations of 2-PCPA. Expression of key pluripotency genes in GFFs treated with 2-PCPA (0 μM) was set as 1.0. Data are presented as the mean ± SD of three independent experiments. Different superscripts represent significant difference between treated group and control group (p < 0.05). GFFs: goat fetal fibroblast cells; 2-PCPA: trans-2-Phenylcyclopropylamine.
Effects of 2-PCPA treatment on key pluripotency gene expression in GFFs
Total RNA was extracted from the GFFs treated with different concentrations of 2-PCPA, and the relative expression of key pluripotency genes was obtained by real-time quantitative PCR (qPCR). Statistics indicated that the mRNA expression levels of Oct4, Sox2, and Nanog increased with increasing 2-PCPA concentration. When the concentration exceeded 2 μM, the expression increased, and the peak value was obtained at 10 μM (except for Nanog; ; p < 0.05).
Development of goat SCNT embryos
To investigate the influence of up-regulating H3K4me2 level of GFFs on the development of goat SCNT embryos in vitro in the treated group (SCNT-T group), we treated donor cells with 2 μM 2-PCPA for 48 hours considering the overall results of cell viability, cell cycle, and level of H3K4me2. The control group (SCNT-C group) was untreated, and IVF embryos were set as the standard group (IVF group). As shown in , the developmental rates of early embryos in the SCNT-T and SCNT-C groups were lower than that in the IVF group (p < 0.05) at each stage of development. The developmental rates of early embryos in the SCNT-T group at all stages of development were significantly higher than that in the SCNT-C group (; p < 0.05).
Table 1. Effect of 2-PCPA treatment on the in vitro development of goat SCNT embryos.
Global H3K4me2 level of embryos
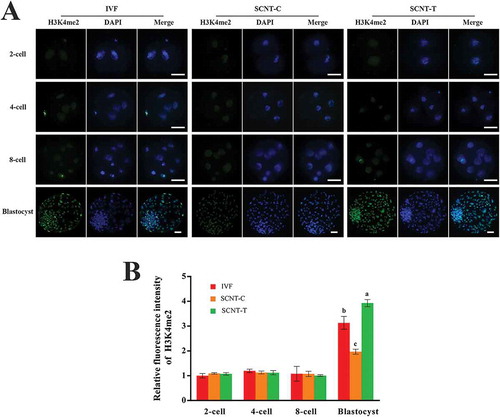
We analyzed the dynamic changes of the level of H3K4me2 in IVF, SCNT-C, and SCNT-T embryos to examine the influence of 2-PCPA treatment on SCNT embryos. As shown in , the three groups all maintained a low level of H3K4me2 in the 2-cell, 4-cell, and 8-cell embryos but showed obvious enhancement at the blastocyst stage. The level of H3K4me2 in the SCNT-C embryos was noticeably lower than in the IVF embryos. After 2-PCPA treatment, the level of H3K4me2 in the SCNT-T embryos increased dramatically compared with those in the SCNT-C embryos (, p < 0.05). Meanwhile, the gap in the level of H3K4me2 between the SCNT-T and IVF blastocysts was also obvious (, p < 0.05). These results suggested that the abnormal H3K4me2 status in cloned goat embryos can be efficiently redressed by treating donor cells with 2-PCPA.
Figure 3. H3K4me2 levels in different stages of IVF, SCNT-C, SCNT-T embryos. (A) H3K4me2 staining of embryos from 2-cell stage to blastocyst stage (green). Nuclei were stained with DAPI (blue). (B) Fluorescence intensity of H3K4me2 was calculated relative to DAPI signal in each group. Ratio of H3K4me2 intensity to DAPI signal when IVF embryos in 2-cell stage were treated with 2-PCPA (2 μM) was set as 1.0. Data are presented as the mean ± SD of three independent experiments. Different superscripts mean significant difference (p < 0.05). H3K4me2: H3K4 dimethylation; IVF: in vitro fertilization; SCNT: somatic cell nuclear transfer; SCNT-T: SCNT treated group; SCNT-C: SCNT control group. Scale bar = 50 μm.
Blastocyst quality assessment and gene expression levels in blastocysts
The influence of 2-PCPA treatment on blastocyst quality was determined by counting the number of blastomeres in blastocysts and obtaining the apoptosis rates of inner cells through the TUNEL assay. As shown in , the mean number of blastomeres in the SCNT-T group was significantly larger than that in the SCNT-C group (p < 0.05). Similarly, no distinct difference in terms of apoptosis rates was observed between the two groups (, p < 0.05).
Figure 4. Blastocyst quality assessment and the mRNA expression of key pluripotency genes and imprinted genes in blastocysts. (A) Total cell number in each blastocyst was confirmed through counting nuclei. (B) Apoptosis rates of inner cells in blastocysts were obtained. (C) Relative mRNA expression of pluripotency genes Oct4, Sox2, and Nanog, imprinted genes H19 and IGF2R in IVF, SCNT-C, and SCNT-T blastocysts. Expression of key pluripotency genes in IVF embryos treated with 2-PCPA (2 μM) was set as 1.0. Data are presented as the mean ± SD of three independent experiments. Different superscripts mean significant difference (p < 0.05). IVF: in vitro fertilization; SCNT: somatic cell nuclear transfer; SCNT-T: SCNT treated group; SCNT-C: SCNT control group.
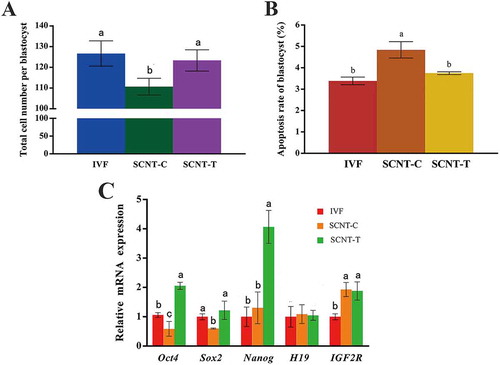
The SCNT-C group presented lower expression levels of Oct4 and Sox2 in their blastocysts than the IVF group, although Nanog expression was slightly higher. After 2-PCPA treatment, the expression levels of the three pluripotency genes were significantly increased in the SCNT-T blastocysts in contrast to those in the SCNT-C blastocysts. However, no obvious difference was observed between the expression levels of the imprinted genes H19 and IGF2R of the two groups. These results indicated that Oct4, Sox2, and Nanog expression levels in cloned goat embryos were up-regulated after the 2-PCPA treatment and this effect had no influence on H19 and IGF2R (; p < 0.05).
Discussion
Histone methylation can lead to gene transcriptional activation and gene transcriptional silencing (Santosrosa et al. Citation2002). The main sites of histone H3 methylation are K4, K36, and K79. Set1 and Set2, which mediate the methylation of both H3K4 and H3K36, directly interact with RNA polymerase II (RNAPII) during the extension phase of mRNAs (Li et al. Citation2003; Robert et al. Citation2003). Histone methyltransferases (HMTs), the enzyme complex that mediates ubiquitination of histone H2B is also associated with RNAP II (Xiao et al. Citation2005). H2B ubiquitination is a prerequisite for the methylation of H3K4 and H3K79, suggesting that H3K79 is also involved in gene transcriptional activation. Furthermore, the association of histone methyltransferases Set1 and Set2 with RNAPII means that the methylation of genes H3K4 and H3K36 is the consequence of gene activation. Methylation of H3K4 or H3K36 can maintain the state of transcriptional activation when some of the transcription factors are down-regulated or absent. The distribution of the H3K4 methylated sites in different organs was analyzed, and H3K4me2 and H3K4me3 both showed high levels of transcriptional activity (Santosrosa et al. Citation2002). However, their distributions were not entirely overlapping. Dimethylation generally occurs at the entire gene segment, while trimethylation occurred at the 5ʹend of these genes (Bernstein Citation2006).
HMTs mediate H3K4 methylation and promote gene activation. H3K4 demethylation results in the decrease in gene transcriptional activity. LSD1 was identified as a demethylase of H3K4me1/2 in 2004 (Shi et al. Citation2004) and the down-regulation of LSD1 was accompanied by an increase in H3K4 methylation levels and up-regulated expression of LSD1 target genes. Associated proteins, such as CoREST, regulate LSD1 by affecting its demethylation (Lee et al. Citation2005, Citation2005). Moreover, LSD1 is incapable of erasing H3K4 trimethylation because of its limited activity. The erasure of H3K4me3 trimethylation is carried out by a number of proteins containing the JmjC domain (Clissold and Ponting Citation2001), and the mechanism is closely related to hydroxylation reaction (Trewick et al. Citation2005).
Consistent with previous reports, our study showed that H3K4me2 levels in the SCNT embryos remained low from the 2-cell stage to the 8-cell stage (Shao et al. Citation2008). The abnormally low H3K4me2 levels in the SCNT embryos may be a continuation of hypomethylation in nuclear donor cells. Nonetheless, the intensity dramatically increased and was accompanied by genome-wide activation, although it remained inferior to the intensity observed in the IVF embryos. The sudden rise of H3K4me2 level during embryonic genome activation indicated the close relationship between H3K4me2 and gene activation. According to previous reports, 2-PCPA can up-regulate H3K4me2 levels by inhibiting LSD1 activity (Schmidt and Mccafferty Citation2007) and has thus been used repeatedly in tumor and stem cell induction studies. Embryos treated with 2-PCPA show an enhanced rate of embryo development in vitro and the expression of key pluripotency genes. Thus, it may be advantageous for the development of cloned embryos in vivo. In the present study, 2-PCPA did not sharply promote the expression of pluripotency genes in the donor cells like that in our previous study (Wang et al. Citation2017). Given the extremely low expression of these genes in GFFs, the actual effect of this inapparent enhancement may be limited.
Recently, Liu et al. reported that overexpression of Kdm5b, a H3K4me3 demethylase, has beneficial effects on cloned embryo development in mice (Liu et al. Citation2016). In addition, Liu et al. have successfully cloned cynomolgus monkeys after the injection of H3K9me3 demethylase Kdm4d mRNA and treatment with histone deacetylase inhibitor trichostatin A at the one-cell stage (Liu et al. Citation2018). These results support that view that histone modifications facilitate the cloned embryo development. Previous studies showed that treatment of reconstructed embryos with low cellular toxicity BIX-01294 and UNC0638 compounds failed to improve the development rates in vitro (Fu et al. Citation2012; Wang et al. Citation2017). Nevertheless, SCNT embryos treated with BIX-01294 or UNC0638 displayed the appropriate patterns of gene expression and epigenetic modifications (Huang et al. Citation2016; Wang et al. Citation2017). Previous studies have shown that SCNT embryos simultaneously suffer from low H3k9 acetylation (Bernstein Citation2006), excessive DNA methylation (Yang et al. Citation2007), and low H3K4 dimethylation (Shao et al. Citation2008). This suggested that multiple epigenetic modifications in SCNT embryos must be simultaneously corrected at the same time to ensure appropriate embryo development.
The H3K4me2 levels in the donor cells increased after the 2-PCPA treatment, which did not have apparent toxicity to GFFs. The up-regulation of the H3K4me2 levels resulted in apparent G0/G1 cell cycle arrest and the suppression of cell proliferation at a 2-PCPA concentration of 2 μM (p < 0.05). Surprisingly, the development rates and qualities of the goat SCNT embryos in vitro were notably improved (p < 0.05), and abnormal H3K4me2 levels were modified effectively in the SCNT-T embryos (p < 0.05). Treatment with 2-PCPA rescued the expression of key pluripotent genes Oct4 and Sox2 (p < 0.05) without disrupting the expression of imprinted genes IGF2R and H19. Our results introduced a promising method for the development of cloned goat embryos. However, the issue of whether a delicate combination between 2-PCPA and other small molecules can correct epigenetic modifications and facilitate the development of SCNT embryos warrants further investigation.
Material and methods
Ethics statements
The entire experimental procedure was approved by the Animal Care and Use Committee of the College of Veterinary Medicine, Northwest A&F University and it was strictly designed under the consideration of animal welfare.
Nuclear donor cells preparation and 2-PCPA treatment
Nuclear donor cells used in the study were derived from a female Saanen Dairy goat fetus. First, the small piece of skin was cut and washed extensively with phosphate-buffered saline (PBS), and then minced into about 1 mm3 pieces and inoculated into 60 mm cell culture dishes. Subsequently, the tissues were cultured in DMEM medium (Gibco, Grand Island, NY, USA). After 1–2 w, the GFFs were passaged when they were amplified up to approximately 90% confluency. Based on a previous report (Li et al. Citation2009), 2-PCPA was diluted into various concentrations (0, 0.5, 1, 2, 5, and 10 μM). Finally, the GFFs were treated with 2-PCPA for 48 h for subsequent cell viability assays, immunostaining, and cell cycle analysis.
Cell viability assay
Cell viability was evaluated with a cell counting kit (Beyotime, China) in accordance with a previous study (Wu et al. Citation2013). Briefly, the GFFs were subcultured in 96-well plates and treated with different concentrations of 2-PCPA for 48 h in an incubator. Then, 10 μL of the CCK-8 solution was added into each well according to protocol. The plates were placed in an incubator for another 2 h. Absorbance was measured at 450 nm with a spectrophotometer (Epoch Biotek, VT, USA). The reference wavelength was 650 nm for the dual wavelength measurement. All the experiments were repeated three times independently.
Immunostaining and quantification of fluorescence intensity
Immunostaining was performed (Wang et al. Citation2017). Cells or embryos were fixed in 4% paraformaldehyde for 30 min, and then permeabilized in 0.1% Triton X-100 (diluted with PBS) for 30 min. All subsequent steps were accomplished at room temperature unless indicated. After every step, an immune staining wash buffer was used for washing the cells or embryos extensively. Subsequently, the samples were blocked with an immune staining blocking solution (Beyotime) at 4°C for 12 h. Then samples were stained with H3K4me2 antibody (Abcam, USA) overnight at 4°C and then incubated in Alexa Fluor 488-labeled goat antirabbit IgG (Beyotime; Diluted 1:500). Finally, the samples were stained with 4,6-diamidino-2-phenylindole (Beyotime) for 5 min. Cells and embryos were both observed under a fluorescent microscope (Olympus, Japan). All experiments were replicated three times independently. In each replication, 15–20 embryos in every group were processed. All images were captured in the same settings without any adjustment of system parameters or brightness. The fluorescent intensity of each image was assessed with Image J software (National Institute of Mental Health, Bethesda, MD, USA). Briefly, images were converted to grayscale. After the calibration of optical density, all individual nuclei in embryos were outlined, and integrated optical density (IOD) and area were measured. The average normalized fluorescence intensity for a single embryo was expressed as sum IOD per sum area. Finally, total DNA contents (DAPI total fluorescence intensities) were divided by H3K4me2 levels for the calculation of normalized H3K4me2 value. For the ease of comparison, the ratio of H3K4me2 intensity (green) to DAPI signal (blue) when the GFFs were treated with 2-PCPA (0 μM) was set as 1.0. For embryos, the ratio of H3K4me2 to DAPI when IVF embryos in the 2-cell stage were treated with 2-PCPA (2 μM) was set as 1.0.
Cell cycle analysis
Cell cycle analysis was confirmed by flow cytometry (Beckman Coulter, CA, USA), which was the same as previous research (Hayes et al. Citation2005). After 2-PCPA treatment and extensive washing, the cells were then trypsinized, centrifugalized, and fixed in 70% pre-cooled ethanol. Then, the GFFs were collected and washed with PBS 1–2 times and then incubated with RNase (Beyotime) for 30 min at 37°C. Finally, the samples were centrifugalized and dyed with 50 μg/mL of propidium iodide (Beyotime) for 30 min at 37°C in the dark. All experiments were replicated three times independently.
Oocyte collection and in vitro maturation (IVM)
Goat ovaries were purchased from a local slaughterhouse and transported to the laboratory in 0.9% NaCl solution at 20–25°C within 2 h. All the cumulus-oocyte complexes (COCs) were obtained by slicing the ovarian surface with a special razor blade and then collected under a stereo microscope (Nikon, Japan). Subsequently, COCs with a diameter of > 150 μm and surrounded by more than three cumulus layers were selected for IVM. The maturation medium used for oocyte IVM was bicarbonate-buffered tissue culture medium-199 (TCM-199, Gibco) supplemented with other accessory factors (Zhang et al. Citation2013). The COCs collected were cultivated in 2 mL of maturation medium in 95% humidified air with 5% CO2 at 38.5°C.
IVF and embryo culture
Fresh semen was collected from a local farmland and stored at 4℃. Consistent with previous reports, matured COCs were then washed in fertilization medium and every group of 20–25 COCs was transferred into 500μL of droplets of fertilization medium under mineral oil (Hajian et al. Citation2017). Matured oocytes and sperm (with a final concentration of about two million sperm per mL) were then co-incubated in 3mL Blacket & Oliphant (BO) media for insemination. The inseminated COCs were incubated in humidified air with 5% CO2 at 38.5°C for 22–24 h. Subsequently, cumulus cells and spermatozoa that attached to oocytes were mechanically removed via mild pipetting.
Nuclear transfer
The nuclear transfer was performed on the basis of a previous study (Liu et al. Citation2011). The first polar body and a small amount of adjacent cytoplasm were removed with a 25 µm diameter pipette. Enucleated oocytes were then checked for the absence of chromosomes under ultraviolet radiation. Subsequently, a round donor cell was injected into the perivitelline space of an oocyte. Later, the couplets were placed between two copper electrodes and then fused by pulse at 32 V for 20 s with an interval of 1 ms. The reconstructed embryos were incubated for 2–3 h in TCM-199 supplemented with 7.5 μg/mL cytochalasin B and 10% FBS. Finally, they were activated in 5 μM ionomycin for 5 min and then 2 mM 6-dimethylaminopurine for 4 h. After extensive washing, the presumptive embryos were cultured in 200 μL mSOF covered with mineral oil.
Apoptosis assays
Apoptosis assays were implemented via a deadend fluorometric TUNEL System (Promega, WI, USA). All steps were accomplished at room temperature unless otherwise stated. Briefly, goat blastocysts were fixed and permeabilized in the same manner as those used in immunostaining. After extensive washes, the samples were equilibrated for 10 min and then incubated with terminal deoxynucleotidyl transferase incubation buffer in the dark at 37℃ for 1 h. Subsequently, the tailing reaction was performed in 2x standard saline citrate for 15 min, and DAPI (Beyotime) was used for DNA staining in the dark for 5 min. Finally, the samples were transferred to a glass slide and dripped with a drop of antifade mounting medium (Beyotime). Goat embryos were observed under a fluorescent microscope (Olympus, Japan). The total cell number in each blastocyst was confirmed through counting nuclei and the number of apoptosis cells in blastocysts was obtained (Su et al. Citation2011). All experiments were replicated three times independently, and 15 to 20 embryos were processed every time.
Total RNA extraction and real-time qPCR
Gene expression was detected by qPCR through StepOne Plus reaction system (ABI, CA, USA). To observe the expression of key pluripotency genes including Oct4, Sox2, and Nanog as well as imprinted genes including H19 and IGF2R, we designed and synthesized six pairs of PCR primers (Supplementary Table S1). According to the manufacturer’s protocol, total RNA was extracted by using trizol-up reagent (TransGen Biotech, China) and inverted to first-stand cDNA with SuperScrpit III CellsDirect cDNA synthesis system kit (Invitrogen, CA, USA). Subsequently, cDNA product was amplified through fluorescence qPCR. The thermal cycling was as follows: pre-denaturation at 95°C for 5 min, 95°C for 15 s, 60°C for 30 s, followed by 40 cycles of 60°C for 30 s with each cycle rising by 0.5°C, until reaching 95°C for 15 s. The relative quantities of the target mRNAs were calculated with SYBR Premix ExTaq II (Takara BIO Inc., CA, USA) and through the Ct method. All the experiments were repeated three times independently.
Statistical analysis
Results were obtained from at least three independent experiments and analyzed using SPSS software (SPSS Inc., Chicago, IL, USA). The differences between means were calculated with one-way analysis of variance (ANOVA) or t-test. Tukey s-b (K) method was used in post-host test. Analysis of percentage values was performed using χ2-test. For all analyses, p < 0.05 was considered statistically significant.
Supplementary_table_2017_258.r2_Supplementary_Materials.doc
Download MS Word (47 KB)Acknowledgments
We sincerely thank our laboratory colleagues for their friendly help in this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary materials
Supplemental data for this article can be accessed here.
Additional information
Funding
Notes on contributors
Yan Luo
Conceived and designed the experiments: YL, YZ; Performed the experiments: TM, CH, RD, BW, PM; Analyzed the data, contributed reagents and materials, and wrote the paper: TM.
References
- Bernstein BE. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125(2):315.
- Clissold PM, Ponting CP. 2001. JmjC: cupin metalloenzyme-like domains in jumonji, hairless and phospholipase A 2 β. Trends Biochem Sci. 26(1):7–9.
- Ding X, Wang Y, Zhang D, Wang Y, Guo Z, Zhang Y. 2008. Increased pre-implantation development of cloned bovine embryos treated with 5-aza-2′-deoxycytidine and trichostatin A. Theriogenology. 70(4):622–630.
- Fu L, Zhang J, Yan FX, Guan H, An XR, Hou J. 2012. Abnormal histone H3K9 dimethylation but normal dimethyltransferase EHMT2 expression in cloned sheep embryos. Theriogenology. 78(9):1929.
- Hajian M, Hosseini SM, Ostadhosseini S, Esfahani MHN. 2017. Comparative stepwise pattern of reactive oxygen species production duringin vitrodevelopment of fertilized and nuclear transferred goat embryos. Int J Fertil Sterility. 11(2):93–98.
- Hayes O, Ramos B, Rodríguez LL, Aguilar A, Badía T, Castro FO. 2005. Cell confluency is as efficient as serum starvation for inducing arrest in the G0/G1 phase of the cell cycle in granulosa and fibroblast cells of cattle. Anim Reprod Sci. 87(3):181–192.
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K. 2013. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 341(6146):651.
- Huang J, Zhang H, Yao J, Qin G, Wang F, Wang X, Luo A, Zheng Q, Cao C, Zhao J. 2016. BIX-01294 increases pig cloning efficiency by improving epigenetic reprogramming of somatic cell nuclei. Reproduction. 151(1):39–49.
- Humpherys D, Eggan K, Akutsu H, Friedman A, Hochedlinger K, Yanagimachi R, Lander ES, Golub TR, Jaenisch R. 2002. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc Natl Acad Sci U S A. 99(20):12889–12894.
- Kim T, Buratowski S. 2009. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5ʹ transcribed regions. Cell. 137(2):259–272.
- Lee MG, Wynder C, Cooch N, Shiekhattar R. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 437(7057):432–435.
- Li B, Howe LA, Anderson S, Yates JR, Workman JL. 2003. The set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem. 278(11):8897–8903.
- Li W, Zhou HY, Abujarour R, Zhu S, Jin YJ, Lin T, Hao E, Schöler HR, Hayek A, Ding S. 2009. Generation of human induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 27(12):2992–3000.
- Liu J, Li LL, Du S, Bai XY, Zhang HD, Tang S, Zhao MT, Ma BH, Quan FS, Zhao XE. 2011. Effects of interval between fusion and activation, cytochalasin B treatment, and number of transferred embryos, on cloning efficiency in goats. Theriogenology. 76(6):1076.
- Liu W, Liu X, Wang C, Gao Y, Gao R, Kou X, Zhao Y, Li J, Wu Y, Xiu W. 2016. Identification of key factors conquering developmental arrest of somatic cell cloned embryos by combining embryo biopsy and single-cell sequencing. Cell Discov. 2:16010.
- Liu Z, Cai YJ, Wang Y, Nie YH, Zhang CC, Xu YT, Zhang XT, Lu Y, Wang ZY, Poo MM, et al. 2018. Cloning of macaque monkeys by somatic cell nuclear transfer. Cell. 172:1–7.
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AHFM, Günther T, Buettner R, Schüle R. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 437(7057):436–439.
- Robert F, Ccedil, Hh N. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 11(3):709.
- Santosrosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NCT, Schreiber SL, Mellor J, Kouzarides T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature. 419(6905):407–411.
- Schmidt D, Mccafferty D. 2007. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 46(14):4408.
- Shao GB, Ding HM, Gong AH, Xiao DS. 2008. Inheritance of histone H3 methylation in reprogramming of somatic nuclei following nuclear transfer. J Reprod Dev. 54(3):233–238.
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 119(7):941–953.
- Su J, Wang Y, Li Y, Li R, Li Q, Wu Y, Quan F, Liu J, Guo Z, Zhang Y. 2011. Oxamflatin significantly improves nuclear reprogramming, blastocyst quality, and in vitro development of bovine SCNT embryos. Plos One. 6(8):e23805.
- Trewick SC, McLaughlin PJ, Allshire RC. 2005. Methylation: lost in hydroxylation? EMBO Rep. 6(4):315–320.
- Tsuji Y, Tsunoda YKY. 2009. The developmental potential of mouse somatic cell nuclear-transferred oocytes treated with trichostatin A and 5-aza-2′-deoxycytidine. Zygote. 17(2):109–115.
- Vajta G, Lewis IM, Trounson AO, Purup S, Maddoxhyttel P, Schmidt M, Pedersen HG, Greve T, Callesen H. 2003. Handmade somatic cell cloning in cattle: analysis of factors contributing to high efficiency in vitro1. Biol Reprod. 68(2):571–578.
- Wang Y, Zhang Y, Mao T, Yan B, Deng R, Wei B, Zhang Y, Liu J. 2017. Treatment donor cells with UNC0638 modify the abnormal histone H3K9 dimethylation and gene expression in cloned goat embryos. Small Ruminant Research. 156:27–32.
- Wu S, Ju GQ, Du T, Zhu YJ, Liu GH. 2013. Microvesicles derived from human umbilical Cord Wharton’s jelly mesenchymal stem cells attenuate bladder tumor cell growth in vitro and in vivo. Plos One. 8(4):e61366.
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 25(2):637–651.
- Yang J, Yang S, Beaujean N, Niu Y, He X, Xie Y, Tang X, Wang L, Zhou Q, Ji W. 2007. Epigenetic marks in cloned rhesus monkey embryos: comparison with counterparts produced in vitro1. Biol Reprod. 76(1):36–42.
- Zhang H, Wu B, Liu H, Qiu M, Liu J, Zhang Y, Quan F. 2013. Improving development of cloned goat embryos by supplementing α-lipoic acid to oocyte in vitro maturation medium. Theriogenology. 80(3):228–233.
