ABSTRACT
Chromatin remodeling, including histone post-translational modifications, during spermatogenesis can affect sperm quality and fertility, and epigenetic marks may therefore be useful for clinical evaluations of sperm. Together with histone hyperacetylation, the dimethylation of histone H3 on lysine K4 (H3K4me2) is also required during protamination. Accordingly, we evaluated the utilization of this epigenetic mark for the identification of sperm with decrease quality and immature chromatin. In this study, 99 semen samples, including 22 normozoospermic (N), 63 asthenozoospermic (A), and 14 oligoasthenozoospermic (OA) samples, were comprehensively analyzed with respect to H3K4me2 levels, DNA damage (DNA fragmentation index, DFI), and sperm immaturity (high DNA stainability, %HDS), as determined by a sperm chromatin structure assay using flow cytometry. We detected a significant relationship between H3K4me2 and %HDS (r = 0.47; p < 0.001). Furthermore, we observed negative correlations between H3K4me2 and sperm concentration, motility, and mitochondrial activity (p < 0.05). The increase in immaturity as semen quality decreased (N > A > OA) indicates the importance of chromatin immaturity and histone code deviations in sperm evaluations. Using various approaches, our study elucidated H3K4me2 as a molecular marker of sperm quality with potential use in reproductive medicine.
Abbreviations: A: asthenozoospermic; AO: acridine orange; ART: assisted reproductive therapy; BWW: Biggers-Whitten Whittingham; DAPI: 4ʹ,6ʹ -diamidino-2-phenylindole; DFI: DNA fragmentation index; H3K4me2: dimethylation of lysine K4 on histones H3; HDS: high DNA stainability; HRP: horseradish peroxidase; MACS: magnetic-activated cell sorting; N: normospermic; NGS: normal goat serum; OA: oligoasthenozoospermic; PTM: post-translational modification; SCSA: sperm chromatin structure assay; SUTI: sperm ubiquitin tag assay; TBS-T: TBS with 0.5% Tween-20
Introduction
To successfully deliver genetic and epigenetic information in the sperm to an embryo, the chromatin has to be hyper-condensed to protect the paternal DNA and epigenome against various external factors (Oliva Citation2006; Carrell et al. Citation2007; Castillo et al. Citation2011). This is ensured by histone replacement with protamines by sperm DNA protamination (Balhorn Citation2007). Despite this, a minute amount of histones retained in the sperm head undergo various post-translational modifications (PTMs) (Rathke et al. Citation2014; Luense et al. Citation2016; Eelaminejad et al. Citation2017). In addition to sperm DNA methylation and non-coding RNA cargo, the histone code contributes to the epigenetic signature of the spermatozoon, to help regulate early post-fertilization events and embryonic development (van der Heijden et al. Citation2008; Hammoud et al. Citation2011; La Spina et al. Citation2014; Sharma et al. Citation2018; Schon et al. Citation2019). Chromatin remodeling during spermatogenesis is quite sensitive to environmental conditions, and exposure to oxidative stress is the most common explanation for decreased sperm quality, as determined by accessibility to DNA fragmentation or apoptosis (Tavalaee et al. Citation2009; Tunc and Tremellen Citation2009; Bahreinian et al. Citation2015). Accordingly, aberrations in sperm chromatin, including histone PTMs, could be related to a decrease in sperm quality, defined by conventional semen parameters or mitochondria activity (Kiani-Esfahani et al. Citation2013). Although the physiological roles of several histone PTMs have been deciphered, a detailed understanding of epigenetic patterns in sperm is lacking (Aoki et al. Citation2005; Siklenka et al. Citation2015; Zhong et al. Citation2015; Pérez-Cerezales et al. Citation2017). Therefore, investigations of the sperm chromatin histone code may lead to improvements in assisted reproductive therapy (ART).
Dimethylation of lysine K4 on histones H3 (H3K4me2) is well-studied in sperm and is a candidate fertility-related histone PTM for several reasons. i) Together with H4 hyperacetylation, H3K4me2 participates in chromatin opening, required for histone-protamine exchange (Rathke et al. Citation2007). ii) The incidence of H3K4me2 is highest in the final stages of spermiogenesis, coinciding with protamination and acrosome formation (Godmann et al. Citation2007). iii) H3K4me2 has been detected at the promoters of transcriptionally active housekeeping genes and genes indispensable for spermatogenesis (Brykczynska et al. Citation2010), and an excess or lack of modification could be responsible for aberrant early embryogenesis (Aoshima et al. Citation2015). iv) Paternal H3K4me2 is involved in the regulation of gene expression during early embryonic development (Teperek et al. Citation2016) and the loss of H3K4me2 is paternally inherited across generations (Siklenka et al. Citation2015).
H3K4me2 has the potential to be a marker of sperm quality, with implications for improving ART. Therefore, we hypothesize that defects in chromatin integrity and pathological sperm quality are associated with an excess of H3K4me2 modification.
Results and discussion
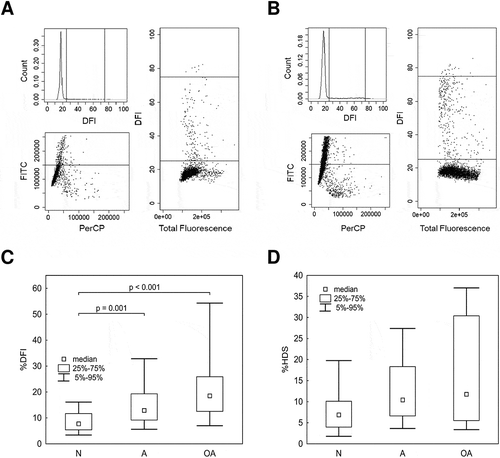
To examine the role of H3K4me2 as an epigenetic marker of human sperm quality, we used sperm samples classified as normospermic (N), asthenozoospermic (A), and oligoasthenozoospermic (OA) according to the WHO (World Health Organization Citation2010) and evaluated the following semen parameters: age, volume, sperm concentration, total and progressive motility, mitochondrial activity/early apoptotic spermatozoa, and chromatin immaturity (expressed as %HDS). As expected, all sperm features other than volume differed significantly among groups (). These parameters also differed when samples were divided into those with low %HDS (HDS ≤ 15) and high %HDS (HDS > 15) () according to Evenson (Citation2011). The concentration of spermatozoa and sperm motility within the HDS < 15 – samples was greater. However, there were no significant differences in mitochondrial activity or, surprisingly, DNA damage (expressed as %DFI) between HDS groups, despite a significant correlation between HDS and DFI (r = 0.200 p = 0.047) ().
Table 1. Overview of sperm parameters according to semen quality.
Table 2. Overview of sperm parameters according to the HDS.
Table 3. Summary of Spearman’s rank correlations between %HDS and other sperm parameters.
Table 4. Summary of Spearman’s rank correlations between H3K4me2 and other sperm parameters.
Immature sperm in the ejaculate is usually explained by the premature release of spermatids from seminiferous tubules in testes before they fully differentiate (Yeung et al. Citation2007; Talebi et al. Citation2008; Elshal et al. Citation2009). Specific treatments for testicular cancer can also increase the presence of immature sperm (Maselli et al. Citation2012), and this may reconcile the residual histones and the abundance of PTMs in semen samples displaying chromatin immaturity. Various PTMs of canonical histones affect residual histones in sperm and have crucial roles with respect to several specific properties of the spermatozoon: the replacement of most core histones via protamination, transcriptional inactivity, and a haploid genome designed for fusion with oocytes. Therefore, the sperm histone code is an important determinant of sperm fertilization ability and the destiny of paternal chromatin in embryos (Nevoral and Sutovsky Citation2017). Moreover, the histone code is sensitive to the environment, which influences sperm quality via epigenetic mechanisms (Delbes et al. Citation2010; Jenkins et al. Citation2017; Gunes et al. Citation2018). Lysine (K) di- and trimethylation of sperm histones are frequent PTMs promoting gene silencing and chromatin protection (van der Heijden et al. Citation2006; Hammoud et al. Citation2009, Citation2011; Brykczynska et al. Citation2010; Siklenka et al. Citation2015). However, the degree of methylation of H3 on lysine K4 has greater effects (i.e., transcriptional repression or activation) than those of the methylation of other lysine residues. Consequently, H3K4me2 that is a coincident marker for transcription factor binding sites was used to evaluate human sperm physiology in this study (visualized in situ on ).
Figure 1. Representative images of the subcellular localization of H3K4me2 in sperm. A) Immunocytochemical localization of H3K4me2 and negative control. B) Difference in H3K4me2 abundance between high and low HDS samples, as determined by western blot densitometry, with an anti-alpha tubulin antibody as a loading control. The data are expressed as means, including min–max whiskers, and different superscripts indicate statistical significance (p < 0.05). HDS, high DNA stainability index.
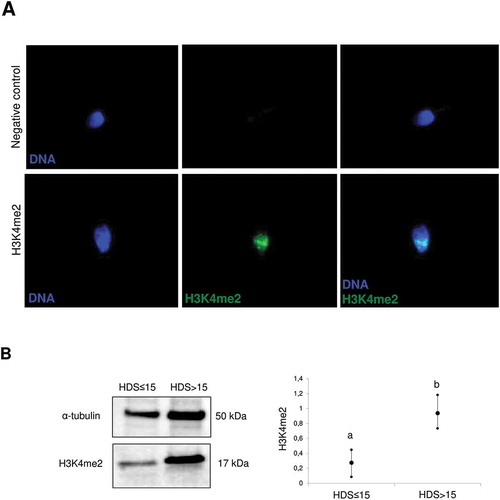
We found a positive association between the H3K4me2 level and sperm chromatin immaturity (%HDS) (r = 0.470; p < 0.001) (). Moreover, there were significant differences between groups classified by %HDS in H3K4me2 levels, as assessed by flow cytometry (,,). The specifity of the H3K4me2 antibody and its localization to the nucleus were confirmed by immunofluorescence and western blotting (,). Increased dimethylated H3K4 occurs in elongating spermatids undergoing nuclear protamination (Godmann et al. Citation2007), and the H3K4me2 modification plays a role in chromatin opening, required for histone replacement by protamines (Rathke et al. Citation2007). Altogether, H3K4me2 in spermatozoa has clinical value and could be used to explain preimplantation failure after ART (Speyer et al. Citation2015), presumably due to the contribution on embryonic gene expression (Hammoud et al. Citation2009, Citation2011; Aoshima et al. Citation2015). Furthermore, H3K4me2 levels were correlated with sperm concentration and motility () and were significantly higher for OA and A samples than for N samples (), emphasizing its value for sperm quality assessment and description.
Figure 2. Relationship between H3K4me2 labeling and semen quality or chromatin maturity. A) Representative images of H3K4me2 histograms generated by flow cytometry, including a normospermic sample with HDS ≤ 15; B) and a sample with pathological semen quality and HDS > 15. C) Comparison of H3K4me2 fluorescence intensity between normozoospermic (N), asthenozoospermic (A), and oligoasthenozoospermic (OA) samples; D) and between samples with HDS ≤ 15 and HDS > 15. The data are expressed as medians and appropriate quartiles, and different superscripts indicate statistical significance (p < 0.05). HDS, high DNA stainability index; DFI, DNA fragmentation index.
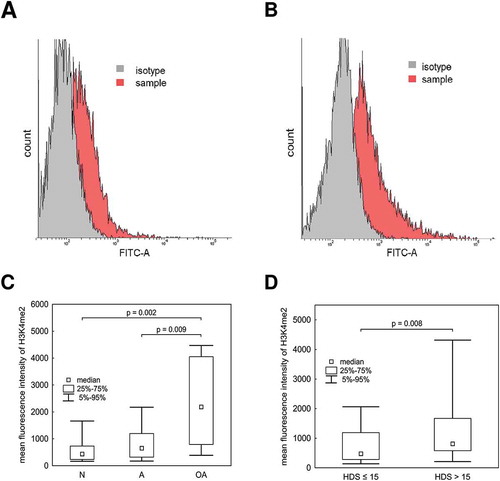
Many studies have found evidence for decreases in concentration and motility in spermatozoa with incomplete protamination (Aoki et al. Citation2005; La Spina et al. Citation2014), consistent with our H3K4me2 results indicating significant correlations with sperm concentration, motility, and %HDS. A correlation was observed between incomplete chromatin condensation and sperm quality parameters, such as concentration, motility, and mitochondrial activity (). Despite a lack of significant differences in %HDS between groups with different semen qualities (), we detected a significant relationship (r = 0.232; p = 0.021) between increasing %HDS and decreasing semen quality (N > A > OA). Indeed, spermatozoa with high chromatin immaturity (%HDS) did not exhibit successful chromatin condensation and are more vulnerable to oxidative stress, DNA fragmentation, and apoptosis (Tunc and Tremellen Citation2009; Bahreinian et al. Citation2015). However, even at levels of high %HDS, an increase in %DFI was not observed (similar to the results of our analysis of HDS groups), but a specific histones were concomitantly up-regulated (Maselli et al. Citation2012). This suggests that, H3K4me2 may be a better indicator than %HDS.
Figure 3. Relationship between semen quality and DNA fragmentation or chromatin maturity measured by SCSA. A) Representative histograms and scatter diagrams of the SCSA results for samples with HDS ≤ 15; B) and HDS > 15; C) Comparison of %DFI; D) and %HDS between normozoospermic (N), asthenozoospermic (A), and oligoasthenozoospermic (OA) samples. The data are expressed as medians and appropriate quartiles, and different superscripts indicate statistical significance (p < 0.05). HDS, high DNA stainability index; DFI, DNA fragmentation index.
Previous studies have shown that incomplete protamination, the protamine ratio, and PTMs of residual histones are reliable prognostic markers for ART success and embryo quality (Carrell et al. Citation1999; Aoki et al. Citation2005; Nasr-Esfahani et al. Citation2007; de Mateo et al. Citation2009; Simon et al. Citation2011). We performed the first comparative analysis of variation in H3K4me2 among semen samples classified by pathological semen quality. H3K4me2 reflects sperm histone code quality as well as protamine features and is advantageous for the assessment of sperm. H3K4me2 levels can partially reflect persisting H3 histones and a shift in the protamine-histone ratio in incompletely protaminated sperm heads (Hamad et al. Citation2014) but the distribution of H3K4me2 did not show an identical pattern to that of H3 in sperm heads (van der Heijden et al. Citation2008). Moreover, our investigation did not reveal strict dependency of the H3K4me2 level to total histone H3 (own unpublished data).
In general, deviations in the sperm histone code have been associated with sperm incompetency and decreased fertility (Gunes et al. Citation2016; Kutchy et al. Citation2017; Rogenhofer et al. Citation2017; Schon et al. Citation2019). The heritability (Jenkins and Carrell Citation2011; Aoshima et al. Citation2015; Colaco and Sakkas Citation2018) of sperm epigenetic marks emphasizes the significance of the H3K4me2 label as an marker of sperm quality. Further studies of the sperm epigenome can improve our understanding of sperm function, explain cases of idiopathic infertility, and improve sperm selection for ART protocols (Castillo et al. Citation2011; Siklenka et al. Citation2015; Gunes et al. Citation2016; Bracke et al. Citation2018). For routine usage, studies of epigenetic marks, selectable sperm labels, and appropriate combinations with noninvasive sperm selection approaches are needed (Ozanon et al. Citation2005; Štiavnická et al. Citation2017). Thus, we expect H3K4me2 may become part of a sperm quality indicator together with previously established noninvasive methods for sperm selection, such as SUTI (sperm ubiquitin tag assay) or MACS (magnetic-activated cell sorting) (Ozanon et al. Citation2005; Nasr-Esfahani et al. Citation2012). Moreover, Raman spectroscopy has potential as a noninvasive method for sperm selection with the ability to distinguish epigenetic differences (Poplineau et al. Citation2011).
By combining three different methods of evaluation, the study supports H3K4me2 as an indicator of aberrant histone-protamine exchange, resulting in improper chromatin condensation. This would also be reflected in the distribution and amount of residual histones and PTMs. Thus, the impact of H3K4me2 on ART success should be assessed as compared to the intragenomic H3K4me2 distribution among spermatozoa with different grades of chromatin maturity.
Materials and methods
Materials
Chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA), unless otherwise stated.
Sperm samples
Human ejaculate samples were obtained after obtaining written consent at the ART center Genetika Pilsen Ltd. (Pilsen, Czech Republic). All subjects were strictly anonymous to the research team. The study was approved by the Ethics committee of Charles University, Faculty of Medicine in Pilsen (238/2016).
Twenty-two sperm samples from healthy donors (normozoospermic, N) and 77 samples from men with pathological semen quality, including 63 presenting with asthenozoospermia (A; progressive motility below 32%) and 14 with oligoasthenozoospermia (OA, concentration below 15 mil/ml and progressive motility under 32%), were assessed. The evaluations of semen concentration, motility, and classification according semen quality were performed in compliance with WHO standards (Citation2010).
Mitochondrial membrane potential and early apoptotic spermatozoa
For the detection of mitochondrial activity and early apoptosis, sperm samples were incubated with 100 nM MitoTracker Deep Red (Thermo Fisher Scientific, Waltham, MA, USA) and 50 nM YO-PRO1 (Thermo Fisher Scientific) in Biggers-Whitten Whittingham medium (BWW) (Biggers et al. Citation1971) for 30 min at room temperature in the dark. Acquisition was performed using a FACSVerse Flow Cytometer (Becton Dickinson, San Diego, CA, USA) containing BD FACSuite software (Becton Dickinson). Cells were run through the instrument at 150 to 300 cells/s and data were collected from 5000 cells. Excitation was performed with a 488-nm laser, except for MitoTracker Deep Red, which was excited at 640 nm. YO-PRO1 green fluorescence was detected with a 537/32BP filter and MitoTracker Deep Red was detected with a 586/42BP filter. MitoTracker+/YO-PRO1- spermatozoa were considered viable with active mitochondria, and MitoTracker-/YO-PRO1+ were identified as early apoptotic spermatozoa with non-functional mitochondria. Flow cytometry data were analyzed using WEASEL Ver. 3 (WEHI, Melbourne, Australia).
Sperm chromatin structure assay (SCSA)
A sperm chromatin structure assay was performed according to the protocol described by Evenson and Jost (Citation2000). This technique is based on the vulnerability of sperm DNA to acid-induced denaturation in situ and subsequent metachromatic staining with acridine orange (AO). The DNA fragmentation index (%DFI) and high DNA stainability (%HDS), indicators of chromatin immaturity (i.e., protamination completeness), were assessed. The samples were evaluated using the FACSVerse Flow Cytometer controlled with BD FACSuite. Data were collected from 5000 events. Excitation of acridine orange was performed with a blue laser (488 nm); red fluorescence was detected with a 700/54BP filter and green fluorescence was detected with a 537/32BP filter. Each sample was run twice and data were analyzed using WEASEL Ver. 3 (WEHI).
Western blot analysis of H3K4me2
Based on the SCSA results, samples with %HDS ≤ 15 and %HDS > 15 (Evenson Citation2011) were compared with respect to H3K4me2 levels by western blotting. Samples were washed twice with PBS and subsequently dissolved in lysis buffer (7 M urea, 2 M thiourea, 4% CHAPS, 120 mM DTT, 40 mM TRIS base) for 30 min on ice. Pooled samples from five patients were prepared and solubilized with the Laemmli buffer (0.003% Triton-X-100 and 0.001% SDS), enriched with Complete Mini Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Thereafter, samples were boiled and subjected to SDS-PAGE on 12.5% separating gels and blotted using the Trans-Blot Turbo Transfer System (BioRad Laboratories, Steenvoorde, France) onto a PVDF membrane (GE Healthcare Life Sciences, Amersham, UK). Approximately 10 million spermatozoa were loaded for each line. Then, the membrane was blocked in 1% BSA in TBS with 0.5% Tween-20 (TBS-T) for 60 min at room temperature. H3K4me2 was detected by rabbit polyclonal anti-H3K4me2 (1:1.000, Abcam, Cambridge, UK), overnight at 4°C. Mouse polyclonal antibody against α-tubulin was used as a loading control (1:1.000, Sigma-Aldrich). Horseradish peroxidase (HRP)-conjugated secondary antibodies, goat anti-mouse, or anti-rabbit IgG (1:15.000; Invitrogen, Waltham, MA, USA), were applied. Target proteins were visualized using ECL Select Western Blotting Detection Reagent (GE Healthcare Life Sciences, Amersham, UK) and the ChemiDoc MP System (Bio-Rad). Densitometry analysis was performed using Image Lab 6.0.1 software (Biorad).
Immunolocalization of H3K4me2
Sperm samples were washed with PBS, placed on microscope slides, and allowed to dry. The spermatozoa on the slides were fixed with 4% paraformaldehyde for 15 min and washed with PBS. They were then permeabilized with 0.5% Triton X-100 and blocked with a solution of 5% normal goat serum (NGS) and 0.1% TritonX-100 in PBS for 60 min at room temperature. Subsequently, the spermatozoa were incubated with a rabbit polyclonal anti-H3K4me2 antibody (1:100; Abcam) for 60 min at room temperature, washed twice with PBS, and incubated with AlexaFluor 488-conjugated goat anti-rabbit secondary antibody (1:200). For negative controls, non-immune rabbit serum with comparable globulin concentrations was used instead of primary antibodies and processed in the same way. Finally, sperm samples were washed twice and mounted onto slides in VectaShield medium with 4ʹ6’-diamidino-2-phenylindole (DAPI; Vector Laboratories Inc., Burlingame, CA, USA). Images were acquired using a 125 spinning disk confocal microscope, Olympus IX83 (Dusseldorf, Germany) and VisiView® (Visitron Systems GmbH, Puchheim, Germany).
H3K4me2 detection by flow cytometry
For H3K4me2 detection, sample preparation and histone analysis by flow cytometry were performed according to the methods of Li et al. (Citation2006). For the detection of H3K4me2, a polyclonal rabbit anti-H3K4me2 antibody (1:100) and Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (1:200; Abcam) were used. Acquisition was performed using the FACSVerse Flow Cytometer and BD FACSuite. Data were collected from 5000 events. A blue laser (488 nm) was used and fluorescence signals were collected using the 537/32BP filter for the excitation of Alexa Fluor 488. An isotype control was evaluated for each sample, and mean fluorescence intensity was measured using the sample and isotype control. Data were analyzed using WEASEL Ver. 3 (WEHI), and the final value for H3K4me2 fluorescence intensity was obtained after subtracting the mean fluorescence intensity of the isotype control from the signal of the sample.
Statistical analysis
Data from all analyses are expressed as the medians with appropriate quantiles and were processed using Statistica Cz 12 (StatSoft, Tulsa, OK, USA). Data were significantly non-normally distributed (according to Shapiro-Wilk tests); accordingly, nonparametric methods were used. Kruskal–Wallis ANOVA (for quantitative variables) was applied for the comparison of all parameters between patient groups with different semen quality. In the case of significant overall findings, pairwise differences between groups were assessed by post-hoc multiple comparisons of mean ranks according to Siegel and Castellan (Citation1988), Mann–Whitney U tests, and a Bonferroni adjustment for multiple testing.
In addition, selected parameters were subjected to Spearman’s rank correlation test. The level of statistical significance was set to p < 0.05, and two-tailed p-values are indicated.
Authors’ contributions
Participated in study design, experiments and manuscript preparation: MŠ, OGA, JN; Participated in experiments: LAP, MD, MK, JM, HŘ, LG, TF; Co-wrote the manuscript and participated in experimental design: PS; Analyzed data: PH; Provided human samples and key information: ZUG, PL (from the Center of Assisted Reproduction Genetika Pilsen Ltd., Pilsen, Czech Republic); Supervised the collaboration with Genetika Ltd. and participated in manuscript preparation: MK.
Acknowledgments
We would like to thank Ms. Kathryn Craighead for manuscript editing. This work was supported by the Czech Health Research Council (NV18-01-00544), Charles University Research Fund (PROGRES Q39), the National Sustainability Programme I (NPU I) Nr LO1503 provided by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR), project no. SVV 02690 provided by MEYS CR, and project no. CZ.02.1.01/0.0/0.0/16_019/0000787 “Fighting Infectious Diseases” awarded by MEYS CR and financed by The European Regional Development Fund. O.GA. was supported by the University of Castilla-La Mancha and Junta de Comunidades de Castilla-La Mancha (SBPLY/17/180501/000470). P.S. was supported by seed funding from the F21C program of the University of Missouri.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Aoki VW, Liu L, Carrell DT. 2005. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum Reprod. 20:1298–1306. doi:10.1093/humrep/deh798
- Aoshima K, Inoue E, Sawa H, Okada Y. 2015. Paternal H3K4 methylation is required for minor zygotic gene activation and early mouse embryonic development. EMBO Rep. 16:803–812.
- Bahreinian M, Tavalaee M, Abbasi H, Kiani-Esfahani A, Shiravi AH, Nasr-Esfahani MH. 2015. DNA hypomethylation predisposes sperm to DNA damage in individuals with varicocele. Syst Biol Reprod Med. 61:179–186. doi:10.3109/19396368.2015.1020116
- Balhorn R. 2007. The protamine family of sperm nuclear proteins. Genome Biol. 8:227. doi:10.1186/gb-2007-8-9-227
- Biggers JD, Whitten WK, Whittingham DG. 1971. The culture of mouse embryos in vitro. In Methods in Mammalian Embryology (ed. JC Daniels, Jr.). San Francisco: Freeman; p. 86–116.
- Bracke A, Peeters K, Punjabi U, Hoogewijs D, Dewilde S. 2018. A search for molecular mechanisms underlying male idiopathic infertility. Reprod Biomed Online. 36:327–339.
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schübeler D, Stadler MB, Peters AHFM. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol. 17:679–687. doi:10.1038/nsmb.1821
- Carrell DT, Emery BR, Hammoud S. 2007. Altered protamine expression and diminished spermatogenesis: what is the link? Hum Reprod Update. 13:313–327. doi:10.1093/humupd/dml057
- Carrell DT, Emery BR, Liu L. 1999. Characterization of aneuploidy rates, protamine levels, ultrastructure, and functional ability of round-headed sperm from two siblings and implications for intracytoplasmic sperm injection. Fertil Steril. 71:511–516.
- Castillo J, Simon L, de Mateo S, Lewis S, Oliva R. 2011. Protamine/DNA ratios and DNA damage in native and density gradient centrifuged sperm from infertile patients. J Androl. 32:324–332. doi:10.2164/jandrol.110.011015
- Colaco S, Sakkas D. 2018. Paternal factors contributing to embryo quality. J Assist Reprod Genet. 35:1953–1968. doi:10.1007/s10815-018-1304-4
- de Mateo S, Gázquez C, Guimerà M, Balasch J, Meistrich ML, Ballescà JL, Oliva R. 2009. Protamine 2 precursors (Pre-P2), protamine 1 to protamine 2 ratio (P1/P2), and assisted reproduction outcome. Fertil Steril. 91:715–722. doi:10.1016/j.fertnstert.2007.12.047
- Delbes G, Hales BF, Robaire B. 2010. Toxicants and human sperm chromatin integrity. Mol Hum Reprod. 16:14–22. doi:10.1093/molehr/gap087
- Eelaminejad Z, Favaedi R, Sodeifi N, Sadighi Gilani MA, Shahhoseini M. 2017. Deficient expression of JMJD1A histone demethylase in patients with round spermatid maturation arrest. Reprod Biomed Online. 34:82–89.
- Elshal MF, El-Sayed IH, Elsaied MA, El-Masry SA, Kumosani TA. 2009. Sperm head defects and disturbances in spermatozoal chromatin and DNA integrities in idiopathic infertile subjects: association with cigarette smoking. Clin Biochem. 42:589–594. doi:10.1016/j.clinbiochem.2008.11.012
- Evenson D, Jost L. 2000. Sperm chromatin structure assay is useful for fertility assessment. Methods Cell Sci. 22:169–189.
- Evenson DP. 2011. Sperm chromatin structure assay (SCSA®): 30 years of experience with the SCSA®. Sperm Chromatin. Springer New York, New York, NY:125–149. doi:10.1007/978-1-4419-6857-9_9
- Godmann M, Auger V, Ferraroni-Aguiar V, Di Sauro A, Sette C, Behr R, Kimmins S. 2007. Dynamic regulation of histone H3 methylation at lysine 4 in mammalian spermatogenesis. Biol Reprod. 77:754–764. doi:10.1095/biolreprod.107.062265
- Gunes S, Arslan MA, Hekim GNT, Asci R. 2016. The role of epigenetics in idiopathic male infertility. J Assist Reprod Genet. 33:553–569. doi:10.1007/s10815-016-0682-8
- Gunes S, Metin Mahmutoglu A, Arslan MA, Henkel R. 2018. Smoking-induced genetic and epigenetic alterations in infertile men. Andrologia. 50:e13124.
- Hamad MF, Shelko N, Kartarius S, Montenarh M, Hammadeh ME. 2014. Impact of cigarette smoking on histone (H2B) to protamine ratio in human spermatozoa and its relation to sperm parameters. Andrology. 2:666–677.
- Hammoud SS, Nix DA, Hammoud AO, Gibson M, Cairns BR, Carrell DT. 2011. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum Reprod. 26:2558–2569. doi:10.1093/humrep/der192
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 460:473–478.
- Jenkins TG, Carrell DT. 2011. The paternal epigenome and embryogenesis: poising mechanisms for development. Asian J Androl. 13:76–80.
- Jenkins TG, James ER, Alonso DF, Hoidal JR, Murphy PJ, Hotaling JM, Cairns BR, Carrell DT, Aston KI. 2017. Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology. 5:1089–1099. doi:10.1111/andr.12416
- Kiani-Esfahani A, Bahrami S, Tavalaee M, Deemeh MR, Mahjour AA, Nasr-Esfahani MH. 2013. Cytosolic and mitochondrial ROS: which one is associated with poor chromatin remodeling? Syst. Biol Reprod Med. 59:352–359. doi:10.3109/19396368.2013.829536
- Kutchy NA, Menezes ESB, Chiappetta A, Tan W, Wills RW, Kaya A, Topper E, Moura AA, Perkins AD, Memili E. 2017. Acetylation and methylation of sperm histone 3 lysine 27 (H3K27ac and H3K27me3) are associated with bull fertility. Andrologia. e12915. doi:10.1111/and.12915
- La Spina FA, Romanato M, Brugo-Olmedo S, De Vincentiis S, Julianelli V, Rivera RM, Buffone MG. 2014. Heterogeneous distribution of histone methylation in mature human sperm. J Assist Reprod Genet. 31:45–49.
- Li Z, Yang J, Huang H. 2006. Oxidative stress induces H2AX phosphorylation in human spermatozoa. FEBS Lett. 580:6161–6168. doi:10.1016/j.febslet.2006.10.016
- Luense LJ, Wang X, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, Berger SL. 2016. Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin. 9:24. doi:10.1186/s13072-016-0072-6
- Maselli J, Hales BF, Chan P, Robaire B. 2012. Exposure to bleomycin, etoposide, and Cis-platinum alters rat sperm chromatin integrity and sperm head protein Profile1. Biol Reprod. 86. doi:10.1095/biolreprod.111.098616.
- Nasr-Esfahani MH, Deemeh MR, Tavalaee M. 2012. New era in sperm selection for ICSI. Int J Androl. 35:475–484. doi:10.1111/j.1365-2605.2011.01227.x
- Nasr-Esfahani MH, Razavi S, Mardani M, Shirazi R, Javanmardi S. 2007. Effects of failed oocyte activation and sperm protamine deficiency on fertilization post-ICSI. Reprod Biomed Online. 14:422–429.
- Nevoral J, Sutovsky P. 2017. Epigenome modification and ubiquitin-dependent proteolysis during pronuclear development of the mammalian zygote: animal models to study pronuclear development. Animal Models and Human Reproduction. New Jersey: Wiley Blackwell; p. 435–466. doi:10.1002/9781118881286.ch17.
- Oliva R. 2006. Protamines and male infertility. Hum Reprod Update. 12:417–435. doi:10.1093/humupd/dml009
- Ozanon C, Chouteau J, Sutovsky P. 2005. Clinical adaptation of the sperm ubuquitin tag immunoassay (SUTI): relationship of sperm ubiquitylation with sperm quality in gradient-purified semen samples from 93 men from a general infertility clinic population. Hum Reprod. 20:2271–2278. doi:10.1093/humrep/dei013
- Pérez-Cerezales S, Ramos-Ibeas P, Lopez-Cardona A, Pericuesta E, Fernandez-Gonzalez R, Pintado B, Gutierrez-Adan A. 2017. Elimination of methylation marks at lysines 4 and 9 of histone 3 (H3K4 and H3K9) of spermatozoa alters offspring phenotype. Reprod Fertil Dev. 29:740. doi:10.1071/RD15349
- Poplineau M, Trussardi-Régnier A, Happillon T, Dufer J, Manfait M, Bernard P, Piot O, Antonicelli F. 2011. Raman microspectroscopy detects epigenetic modifications in living Jurkat leukemic cells. Epigenomics. 3:785–794.
- Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. 2014. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta - Gene Regul Mech. 1839:155–168. doi:10.1016/j.bbagrm.2013.08.004
- Rathke C, Baarends WM, Jayaramaiah-Raja S, Bartkuhn M, Renkawitz R, Renkawitz-Pohl R. 2007. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J Cell Sci. 120:1689–1700. doi:10.1242/jcs.004663
- Rogenhofer N, Ott J, Pilatz A, Wolf J, Thaler CJ, Windischbauer L, Schagdarsurengin U, Steger K, von Schönfeldt V. 2017. Unexplained recurrent miscarriages are associated with an aberrant sperm protamine mRNA content. Hum Reprod. 32:1574–1582. doi:10.1093/humrep/dex224
- Schon SB, Luense LJ, Wang X, Bartolomei MS, Coutifaris C, Garcia BA, Berger SL. 2019. Histone modification signatures in human sperm distinguish clinical abnormalities. J Assist Reprod Genet. 36:267–275.
- Sharma U, Sun F, Conine CC, Reichholf B, Kukreja S, Herzog VA, Ameres SL, Rando OJ. 2018. Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev Cell. 46:481–494.e6. doi:10.1016/j.devcel.2018.06.023
- Siegel S, Castellan NJ. 1988. Non-parametric statistics for the behavioural sciences. New York: MacGraw Hill Int. doi:10.1007/s00464-002-9121-2
- Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, et al. 2015. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science. 350:aab2006. doi:10.1126/science.aab2006
- Simon L, Castillo J, Oliva R, Lewis SEM. 2011. Relationships between human sperm protamines, DNA damage and assisted reproduction outcomes. Reprod Biomed Online. 23:724–734. doi:10.1016/j.rbmo.2011.08.010
- Speyer BE, Pizzey AR, Abramov B, Saab W, Doshi A, Sarna U, Harper JC, Serhal P. 2015. Successful outcomes achieved in assisted reproduction cycles using sperm with high levels of high DNA stainability. Syst Biol Reprod Med. 61:293–299. doi:10.3109/19396368.2015.1033065
- Štiavnická M, Abril-Parreño L, Nevoral J, Králíčková M, García-Álvarez O. 2017. Non-invasive approaches to epigenetic-based sperm selection. Med Sci Monit. 23:4677–4683.
- Talebi AR, Moein MR, Tabibnejad N, Ghasemzadeh J. 2008. Effect of varicocele on chromatin condensation and DNA integrity of ejaculated spermatozoa using cytochemical tests. Andrologia. 40:245–251.
- Tavalaee M, Razavi S, Nasr-Esfahani MH. 2009. Influence of sperm chromatin anomalies on assisted reproductive technology outcome. Fertil Steril. 91:1119–1126. doi:10.1016/j.fertnstert.2008.01.063
- Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, Kwon T, Marcotte EM, Zegerman P, Bradshaw CR, et al. 2016. Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Res. 26:1034–1046. doi:10.1101/gr.201541.115
- Tunc O, Tremellen K. 2009. Oxidative DNA damage impairs global sperm DNA methylation in infertile men. J Assist Reprod Genet. 26:537–544.
- van der Heijden GW, Derijck AAHA, Ramos L, Giele M, van der Vlag J, de Boer P. 2006. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol. 298:458–469. doi:10.1016/j.ydbio.2006.06.051
- van der Heijden GW, Ramos L, Baart EB, van Den Berg IM, Derijck AA, van der Vlag J, Martini E, de Boer P. 2008. Sperm-derived histones contribute to zygotic chromatin in humans. BMC Dev Biol. 8:34.
- World Health Organization. 2010. WHO laboratory manual for the examination and processing of human semen. Geneva: World Health Organization.
- Yeung CH, Beiglböck-Karau L, Tüttelmann F, Nieschlag E. 2007. The presence of germ cells in the semen of azoospermic, cryptozoospermic and severe oligozoospermic patients: stringent flow cytometric analysis and correlations with hormonal status. Clin Endocrinol. 67:767–775.
- Zhong HZ, Lv FT, Deng XL, Hu Y, Xie DN, Lin B, Mo ZN, Lin FQ. 2015. Evaluating ϒH2AX in spermatozoa from male infertility patients. Fertil Steril. 104:574–581. doi:10.1016/j.fertnstert.2015.06.004
