ABSTRACT
To study the relationship between the expression of 10 selected genes in cumulus cells and the corresponding oocyte development competence, and the effect of patient age and body mass index on gene expression of cumulus cells, we collected 354 cumulus cell masses associated with individual oocyte from 48 women. The expression levels of the genes involved in glucose metabolism (PFKP, PKM2, LDHA and GFPT) and expansion (HAS2, VCAN, TNFAIP6, PTGS2, PTX3 and SDC4) in cumulus cells were detected by reverse transcription polymerase chain reaction. These were compared among oocyte maturity, fertilization, embryo morphology and implantation, and analyzed the effect of the subject’s age and body mass index. Cumulus cell PFKP expression from mature oocytes was higher than those from immature oocytes (P = 0.014), and VCAN expression was higher from oocytes that developed into high-quality embryos (P = 0.024). TNFAIP6 expression in cumulus cells from fertilized oocytes was lower than that from unfertilized oocytes (P = 0.044). The levels of VCAN, TNFAIP6, PTX3 and SDC4 were changed significantly as a function of the subject’s age and body mass index. In conclusion, the level of VCAN expression in cumulus cells is positively correlated with the early embryo morphology score, and with further development could perhaps be used to evaluate oocyte developmental competence to complement embryonic morphological assessment.
Abbreviations
CCs: cumulus cells; GDF9: growth differentiation factor 9; BMP15: bone morphogenetic protein 15; PTGS2: prostaglandin synthase 2; HAS2: hyaluronic acid synthase 2; VCAN: versican; GREM1: gremlin 1; PFKP: phosphofructokinase, platelet; PKM2: pyruvate kinase isozyme type M2; LDHA: lactic dehydrogenase; GFPT: glucosaminefructo-6-phosphate transaminase; TNFAIP6: tumor necrosis factor 6 protein; PTX3: penetrin 3; SDC4: syndecan-4; BMI: body mass index; MD: median values; IQR: interquartile range; FSH: follicle-stimulating hormone; LH: luteinizing hormone; HCG: human chorionic gonadotropin; ICSI: intracytoplasmic sperm injection; GnRH: gonadotropin-releasing hormone; hMG: human menopausal gonadotropin; GV: germinal vesicle; M I: metaphase I; M II: metaphase II; cDNA: complementary DNA; SD: standard deviation.
Introduction
Oocyte quality plays a key role in determining embryo development and clinical pregnancy in assisted reproduction, but its evaluation still relies mainly on morphological criterion. Assessment still needs to be improved (Wang and Sun Citation2007; Rienzi et al. Citation2011), suggesting that additional parameters are required.
There is a close interaction between the oocyte and the surrounding cumulus cells (CCs). The hypothesis of bio-directional communication states that the oocyte modulates CCs function using oocyte-secreted factors, and CCs facilitate oocyte development and maturation by providing small molecular nutrients, such as cyclic nucleotides, glucose metabolites and amino acids (Gilchrist et al. Citation2008; Coticchio et al. Citation2015). In the process of this reciprocal interaction, oocytes appear to tightly control their neighboring somatic cells, directing them to perform functions required to provide the optimal micro-environment for oocyte development (Matzuk et al. Citation2002). Gene expression levels in CCs should reflect oocyte developmental potential.
Growth differentiation factor 9 (GDF9) and bone morphogenetic protein 15 (BMP15) are both important factors secreted by oocytes that regulate proliferation and differentiation, metabolism, and expansion in CCs (Chang et al. Citation2016). For example, prostaglandin synthase 2 (PTGS2), acetyl hyaluronic acid synthase 2 (HAS2), versican (VCAN) and gremlin 1 (GREM1) expressed in the CCs during expansion all act as GDF9 downstream targets. Studies demonstrated that expression of PTGS2, VCAN, HAS2 and GREM1 genes in cumulus cells correlates with embryo morphological characteristics of a higher quality embryo (McKenzie et al. Citation2004; Zhang et al. Citation2005; Cillo et al. Citation2007; Adriaenssens et al. Citation2010; Wathlet et al. Citation2011; Uyar et al. Citation2013).
In addition, oocytes rely on CCs to provide energy substrates such as pyruvate and lactate to meet their energy needs. Oocyte-derived BMP15 and fibroblast growth factors also cooperate to enhance glycolytic activity and expression of glycolytic enzymes in CCs from large antral follicles (Sugiura et al. Citation2007; Sutton-McDowall et al. Citation2010). These enzymes include phosphofructokinase (PFK), pyruvate kinase (PKM2) and lactic dehydrogenase (LDHA). This suggests that gene expression related to glycolysis and cumulus expansion could be associated with oocyte development and maturation.
Recent data have indicated that the expression levels of some genes in CCs could be used as indicators to predict oocyte developmental competence (McKenzie et al. Citation2004; Zhang et al. Citation2005; Cillo et al. Citation2007; Adriaenssens et al. Citation2010; Wathlet et al. Citation2011; Uyar et al. Citation2013). However, the results varied among laboratories, which could have resulted from different methods and patients (Fragouli et al. Citation2014; Burnik Papler et al. Citation2015a, Citation2015b; Kordus and LaVoie Citation2017). Bio-markers of CC gene expression require validation.
In this study, we chose to examine key genes related to glucose metabolism (PFKP, PKM2, LDHA and glucosaminefructo-6-phosphate transaminase (GFPT)) and cumulus expansion (HAS2, VCAN, PTGS2, human tumor necrosis factor 6 protein (TNFAIP6) and penetrin 3 (PTX3) and syndecan-4 (SDC4)) to verify the association between the level of CC gene expression from the individual oocyte and the corresponding oocyte developmental competence. We further investigated the effect of age and body mass index (BMI) on the level of CC gene expression.
Results
A total of 354 CC samples associated with individual cumulus-oocyte complex from 48 ICSI patients were collected, and they were comprised of 26 from immature oocytes, and 328 from mature oocytes. Maturation rate, fertilization rate, cleavage rate, high-quality embryo rate and implantation rate were 92.66%, 75.91%, 97.99%, 66.39% and 41.86%, respectively.
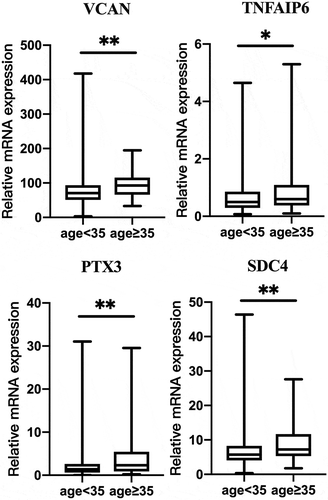
From the 10 candidate genes selected, three (PFKP, PKM2, VCAN) had a statistical difference between mature and immature oocyte group expression (P < 0.05; ). The level of VCAN expression differed significantly between high-quality and poor-quality embryos (P < 0.05). Additionally, expression levels of the PKM2 and VCAN genes in the implanted embryo group were lower than that in non-implantation group (P < 0.05). In addition, we analyzed that the effect of patient age and BMI on the 10 candidate CC genes. They were divided into two groups, less than 35 years old and 35 years old or more. Interestingly, the levels of VCAN, TNFAIP6, PTX3 and SDC4 were higher (P < 0.05) in patients of age ≥35 years old than that in age <35 years old (P < 0.05; ).
Figure 1. The relationship between PFKP, PKM2 and VCAN gene expression levels in cumulus cells and the corresponding oocyte development potential. Data were present as the median values (MD) and the interquartile range (IQR) and analyzed using the Wilcoxon rank sum test. *P < 0.05. **P < 0.01.
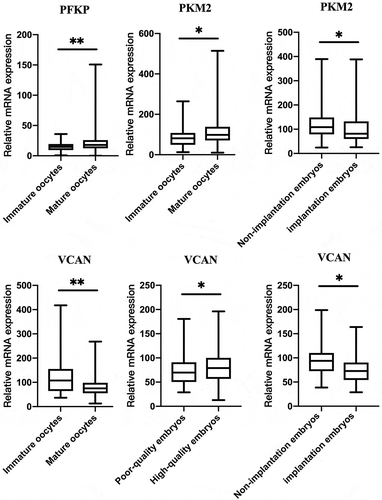
Figure 2. Effect of patient age on the gene expression in the cumulus cells. Age ≥ 35 (n = 103); age < 35 (n = 253). Data were present as the median values (MD) and the interquartile range (IQR) and analyzed using the Wilcoxon rank sum test. *P < 0.05. **P < 0.01.
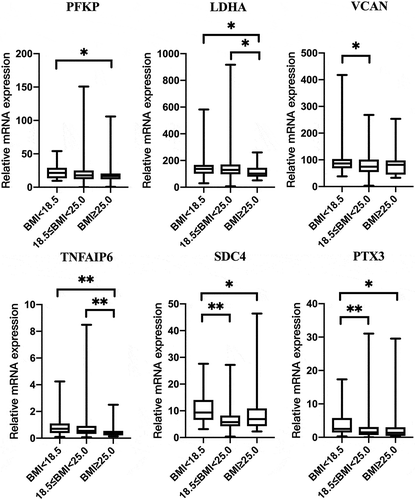
Patients were divided into three groups according to baseline BMI: underweight (BMI < 18.5), normal weight (18.5 ≤ BMI < 25.0) and overweight (BMI ≥ 25.0), in line with WHO guidelines (Body mass index – BMI Citation2011). PFKP, LDHA, VCAN, TNFAIP6, PTX3 and SDC4 expression levels in CCs differed among three groups (P < 0.05; ).
Figure 3. Effect of BMI of patients on the gene expression of the cumulus cells. Underweight (n = 45); normal weight (n = 279); overweight (n = 32). Data were present as the median values (MD) and the interquartile range (IQR) and analyzed using the Kruskal–Wallis test. *P < 0.05. **P < 0.01.
Multivariate logistic regression was performed to examine the association of the candidate genes with oocyte developmental competence (). PFKP expression was positively correlated with oocyte maturity (P = 0.014), and TNFAIP6 was negatively correlated to fertilization (P = 0.044). VCAN was positively correlated to Day 3 embryo grade (P = 0.024). VCAN expression level was not related to implantation competence after adjusted for age and baseline BMI (P = 0.065). Other genes were analyzed including LDHA, PTGS2, HAS2, PTX3, SDC4 and GFPT. Their expression levels were not associated with oocyte maturity, fertilization, embryo grade and implantation P > 0.05.
Table 1. The relationship between the gene expression levels in cumulus cells and the embryo development outcomes.
Discussion
The present study showed that expression of glycolytic genes PFKP, PKM2, LDHA and GFPT in CCs is associated with mature oocytes to a greater degree than with immature oocytes, although LDHA and GFPT gene expression levels did not differ between both groups. Moreover, PFKP expression was positively correlated with oocyte maturity after adjusting for age and BMI. The results suggest that the CC glycolytic level of glucose increases during oocyte maturation to meet the oocyte demand for energy. Most of the glucose is metabolized via glycolysis by granulosa cells yielding small molecule energy metabolites, such as pyruvic acid, lactic acid, that are then are transferred to the oocyte (Sutton et al. Citation2003). However, they were not associated with embryo grade, in agreement with the others (Gebhardt et al. Citation2011).
Our results showed that VCAN and CC gene expression associated with mature oocyte (M Ⅱ) was lower than that within the immature oocyte (GV and M Ⅰ) though VCAN expression was not correlated with oocyte maturity after logistic regression. This result is consistent in several independent studies, that also indicated that cumulus cells around mature oocytes had a lower VCAN expression as compared with that from immature oocytes (McKenzie et al. Citation2004; Cillo et al. Citation2007; Ekart et al. Citation2013). Versican expression is regulated by FSH, luteinizing hormone (LH) and human chorionic gonadotropin (HCG) (Richards et al. Citation2005; Dunning et al. Citation2007). RT-PCR and western blotting (Dunning et al. Citation2007) previously showed that the expression of versican mRNA in CCs and mural granulosa cells peaked at 6 h, began to decline at 12 h, and decreased significantly at 16 h after mice were injected with HCG. This revealed that the transcription and translation of versican gene in follicles in vivo were significantly down-regulated after the oocyte matured. Thus, CCs associated with mature oocytes exhibit a low expression of VCAN.
On one hand, our study suggests that VCAN CC gene expression is positively correlated with Day 3 embryo grade, which is in agreement with the study of Adriaenssens et al. (Adriaenssens et al. Citation2010). However, we found that levels of VCAN expression in implantation embryos were lower than that in non-implantation embryos. Analysis showed that MD of the relative level of VCAN expression in high-quality D3 embryos was 79.38, which was similar level with implantation embryos of 72.80.
On the other hand, other researchers have reported that VCAN expression was higher in good-quality blastocysts or the live birth group (Gebhardt et al. Citation2011; Ekart et al. Citation2013). Wathlet’s study also revealed that CCs in the pregnancy group had a higher level of VCAN expression than that in a non-pregnancy group (Wathlet et al. Citation2011). However, it is difficult to reconcile that the level of VCAN expression in CCs was negatively correlated to the embryo or blastocyst morphology in Wathlet’s et al. (Wathlet et al. Citation2011). Versican has EGF-like effects on gene expression in oocyte-cumulus complexes, and it thus significantly increased during cumulus expansion and exemplified by the cumulus-specific genes PTGS2, TNFAIP6 and HAS2 when versican protein was supplied into media for in vitro mouse oocyte maturation (Dunning et al. Citation2015). This supports the view that versican is important for the final maturation of oocytes. In view of the change in VCAN CC expression during follicular maturation, we inferred that there can be an appropriate range of VCAN gene expression in cumulus cells corresponding to high-quality oocytes, and variance from this level can have negative effects.
Other factors may also explain contradictory results for relationship between VCAN CC gene expression and oocyte developmental competence. Firstly, the gene expression level within CCs varies widely among individuals. Wathlet et al. pointed out that both between- and within-patient variation was large for almost all genes (Wathlet et al. Citation2011). Secondly that the number of patients studied is small (McKenzie et al. Citation2004; Zhang et al. Citation2005; Cillo et al. Citation2007; Adriaenssens et al. Citation2010; Gebhardt et al. Citation2011; Wathlet et al. Citation2011; Ekart et al. Citation2013), thus sample bias could lead to different results. In the present study, 354 cumulus cell masses were collected; however, they were just from 48 women, which may reflect a bias of age or BMI. Thirdly, materials and methods in different studies were also different. Some pooled CCs, and some collected CCs from an individual oocyte.
In our study, patient age and BMI affected some gene expression in cumulus cells. Other studies also revealed that the expression of some genes in CCs was affected by patient clinical characteristics (Adriaenssens et al. Citation2010; Wathlet et al. Citation2011; Fragouli et al. Citation2014; Kordus and LaVoie Citation2017). Indeed, the transcriptome and proteome of granulosa cells differs considerably between young and older women (McReynolds et al. Citation2012; Al-Edani et al. Citation2014). Perhaps this variance in results reflects the impact of their focus on different groups of people. Nevertheless, the current study shows that VCAN CC gene expression may be associated with the corresponding embryo developmental potential. Perhaps the gene expression level of VCAN would complement the morphological evaluation as a useful tool to select oocytes with better developmental competence.
Materials and methods
Ethical approval
This project was approved by the Ethics Committee of Tongji Medical College of Huazhong University of Science and Technology and all experiments were performed in accordance with approved guidelines of Huazhong University of Science and Technology (IORG No. IORG0003571), and informed consent was obtained from all participants.
Patients and treatment
The study involved 48 intracytoplasmic sperm injection (ICSI) patients, and all of them underwent a standard institutional stimulation protocol of ovarian down-regulation with gonadotropin-releasing hormone (GnRH) agonist followed by controlled ovarian stimulation with recombinant FSH (Gonal-F, Merck-Serono, Switzerland) and highly purified urinary gonadotropin (hMG, Livzon, China) at the reproductive medicine center of Tongji Hospital of Tongji Medical College in Huazhong University of Science and Technology. All patients needed ICSI due to male factor.
The cumulus cell masses (n = 354) associated with individual oocyte from 48 female patients were collected and used to extract RNA. Patient ages ranged from 22 to 42 years (mean±SD: 31.85 ± 5.17) and their body mass indexes (BMI) were between 17.2 and 37.1 kg/m2 (mean±SD: 21.61 ± 3.23). Basal FSH ranged from 3.46 to 13.62 IU/L (mean±SD: 7.15 ± 1.91). Infertility during 2–20 years (5.85 ± 4.77 years) and 24 (56%) of couples were considered primary infertility and others were secondary infertility. In the current study, the main cause of infertility was male factor (asthenozoospermia or oligozoospermia), and excluded endometriosis and abnormal chromosome.
Cumulus cell collection
Individual oocyte-corona-cumulus complex was treated by hyaluronidase (Sigma-Aldrich Co., St. Louis, MO, USA), and then the naked oocytes were placed in a marked drops of culture medium. Injections were given one by one when ICSI. Thus, there is a one-to-one correspondence between cumulus cell mass and the oocyte, and it is easier to track oocyte development.
Assessment of oocytes and embryos
The oocyte and embryo morphological assessment was recorded from the day of collection of the cumulus–oocyte complexes (Day 0) until Day 6 of embryo culture. Oocytes were classified by the meiotic stage of germinal vesicle (GV), into metaphase I (M I) or M II. M II oocytes were subjected to ICSI at 39–40 h post-hCG treatment, and cultured individually in 25–30 μl micro-drops of G-1 Plus media (G5 Series, Vitrolife, Sweden) under paraffin oil (Vitrolife) in a 37°C incubator (MINC; Cook, Brisbane, Australia) under 5% O2 and 6% CO2. The embryo transfer was carried out in G-2 Plus media (Vitrolife) using standard protocols.
Embryos were assessed at 18, 42 and 66 h after ICSI using a common grading system (Gardner and Sakkas Citation2003). The high-quality (grades 1 and 2) were embryos with more than six blastomeres of regular size and less than 25% fragmentation. One or two high-quality embryos were transferred or frozen on Day 3, and the rest were cultured to the blastocyst stage to be transferred or frozen. Clinical pregnancy status was identified by the presence of a fetal heartbeat detected with an ultrasound scan at 5 weeks after embryo transfer. Implantation message was confirmed after the ultrasound scan and either all the embryos implanted or none did so. Thus, patients with a gestational sac after two embryos were transferred were excluded.
RNA extraction and complementary DNA synthesis
The total RNA was extracted with TRIzol Reagent (life technologies, Invitrogen, America) in line with the manufacturer’s instructions. The cells were homogenized in 500 μl of TRIzol Reagent and extracted using 100 μl of chloroform. The organic and inorganic phases were separated, then the mixture was placed at room temperature for 3 min before 250 μl isopropanol was added. The total RNA was precipitated in isopropanol and then washed in 75% (v/v) ethanol. The supernatant was discarded and the precipitate left at room temperature for 5 min. The RNA pellet was subsequently eluted with 20 μl nuclease-free water. The RNA was quantified by measuring its absorbance at 260 nm with a nucleic acid protein meter (Biophotometer plus, Eppendorf, Germany). The RNA was reverse-transcribed into complementary DNA (cDNA) using PrimeScript™ RT Master Mix (Perfect Real Time) (RR036A, Takara, Dalian, China) and frozen at −80°C.
Quantitative real-time PCR
The primers used in this study were designed and synthesized by Sangon Biotechnology Ltd (Shanghai, China). The primer sequences and the sizes of the amplified products were shown in .
Table 2. Oligonucleotide primers used for RT-PCR assays.
The reverse transcriptase polymerase chain reaction mixture contained 2 µl of cDNA template, 10 µl SYBR® Premix Ex Taq™ II (Tli RNaseH Plus, Code No. RR820A, Takara), 0.5 µl of each the forward and reverse primer and nuclease-free water to 20 μl. Each sample was run in triplicate, and negative controls were included in each response. For predegeneration, the samples were heated to 95°C for 30 s (RocheLight Cycler 96 System). The amplification process used 40 cycles of 95°C for 5 s (denaturation), and 63°C for 30 s (annealing and extension). After the reaction, the melting curve was checked and the PCR amplification products were subjected to electrophoresis on 1.5% agarose gel. The 2(-Delta-CT) (2−ΔCT) method was used to transform the CT values into relative normalized expression levels. ΔCT was counted by CT target gene-CT housekeeping gene and actin was used as the housekeeping gene. Owing to low expression of some gene in CCs compared with β-actin, all gene relative levels were treated as 1000 × 2−ΔCT, and used in subsequent statistical analysis.
Statistical analysis
Data were described with median values and the interquartile range (IQR), or mean values and standard deviation (SD), if appropriate. Gene levels in two groups were compared using the Wilcoxon rank sum test. Furthermore, multivariate logistic regression was performed between the expression levels of 10 candidate genes and oocyte development outcomes. Kruskal–Wallis test was used for comparison of body mass index (BMI) groups. Statistical analyses used the Statistical Package for Social Sciences program, Version 12.0 (SPSS Inc., Chicago, IL, USA). Statistical significance was assumed at P < 0.05.
Author contributions
Processed data, interpreted the data and wrote the manuscript: QS; Carry out experimental work and helped with the analysis of the data: MC; Took part in experimental work and analyzed data: XZ; Took part in experimental work and revised the manuscript: YL; Selected suitable patients, scored the embryos and collected clinical data: XR; Designed experiments, analyzed data, and revised the manuscript: LZ.
Acknowledgments
We thank Prof Yuan Liang and Ms Xiaoyu Wang for their valuable advice during data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, Coucke W, Devroey P, Smitz J. 2010. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod. 25(5):1259–1270.
- Al-Edani T, Assou S, Ferrieres A, Bringer Deutsch S, Gala A, Lecellier CH, Ait-Ahmed O, Hamamah S. 2014. Female aging alters expression of human cumulus cells genes that are essential for oocyte quality. Biomed Res Int. 2014:964614.
- Body mass index – BMI. 2011. http://www.euro.who.int/en/health-topics/disease-prevention/nutrition/a-healthy-lifestyle/body-mass-index-bmi.
- Burnik Papler T, Vrtacnik Bokal E, Lovrecic L, Kopitar AN, Maver A. 2015a. No specific gene expression signature in human granulosa and cumulus cells for prediction of oocyte fertilisation and embryo implantation. PLoS One. 10(3):e0115865.
- Burnik Papler T, Vrtacnik Bokal E, Maver A, Lovrecic L. 2015b. Specific gene expression differences in cumulus cells as potential biomarkers of pregnancy. Reprod Biomed Online. 30(4):426–433.
- Chang HM, Qiao J, Leung PC. 2016. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum Reprod Update. 23(1):1–18.
- Cillo F, Brevini TA, Antonini S, Paffoni A, Ragni G, Gandolfi F. 2007. Association between human oocyte developmental competence and expression levels of some cumulus genes. Reproduction. 134(5):645–650.
- Coticchio G, Dal Canto M, Mignini Renzini M, Guglielmo MC, Brambillasca F, Turchi D, Novara PV, Fadini R. 2015. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum Reprod Update. 21(4):427–454.
- Dunning KR, Lane M, Brown HM, Yeo C, Robker RL, Russell DL. 2007. Altered composition of the cumulus-oocyte complex matrix during in vitro maturation of oocytes. Hum Reprod. 22(11):2842–2850.
- Dunning KR, Watson LN, Zhang VJ, Brown HM, Kaczmarek AK, Robker RL, Russell DL. 2015. Activation of mouse cumulus-oocyte complex maturation in vitro through EGF-like activity of versican. Biol Reprod. 92(5):116.
- Ekart J, McNatty K, Hutton J, Pitman J. 2013. Ranking and selection of MII oocytes in human ICSI cycles using gene expression levels from associated cumulus cells. Hum Reprod. 28(11):2930–2942.
- Fragouli E, Lalioti MD, Wells D. 2014. The transcriptome of follicular cells: biological insights and clinical implications for the treatment of infertility. Hum Reprod Update. 20(1):1–11.
- Gardner DK, Sakkas D. 2003. Assessment of embryo viability: the ability to select a single embryo for transfer–a review. Placenta. 24 Suppl B:S5–S12.
- Gebhardt KM, Feil DK, Dunning KR, Lane M, Russell DL. 2011. Human cumulus cell gene expression as a biomarker of pregnancy outcome after single embryo transfer. Fertil Steril. 96(1):47–52.e42.
- Gilchrist RB, Lane M, Thompson JG. 2008. Oocyte-secreted factors: regulators of cumulus cell function and oocyte quality. Hum Reprod Update. 14(2):159–177.
- Kordus RJ, LaVoie HA. 2017. Granulosa cell biomarkers to predict pregnancy in ART: pieces to solve the puzzle. Reproduction. 153(2):R69–R83.
- Matzuk MM, Burns KH, Viveiros MM, Eppig JJ. 2002. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science (New York, NY). 296(5576):2178–2180.
- McKenzie LJ, Pangas SA, Carson SA, Kovanci E, Cisneros P, Buster JE, Amato P, Matzuk MM. 2004. Human cumulus granulosa cell gene expression: a predictor of fertilization and embryo selection in women undergoing IVF. Hum Reprod. 19(12):2869–2874.
- McReynolds S, Dzieciatkowska M, McCallie BR, Mitchell SD, Stevens J, Hansen K, Schoolcraft WB, Katz-Jaffe MG. 2012. Impact of maternal aging on the molecular signature of human cumulus cells. Fertil Steril. 98(6):1574–1580.e1575.
- Richards JS, Hernandez-Gonzalez I, Gonzalez-Robayna I, Teuling E, Lo Y, Boerboom D, Falender AE, Doyle KH, LeBaron RG, Thompson V, et al. 2005. Regulated expression of ADAMTS family members in follicles and cumulus oocyte complexes: evidence for specific and redundant patterns during ovulation. Biol Reprod. 72(5):1241–1255.
- Rienzi L, Vajta G, Ubaldi F. 2011. Predictive value of oocyte morphology in human IVF: a systematic review of the literature. Hum Reprod Update. 17(1):34–45.
- Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O’Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ. 2007. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development. 134(14):2593–2603.
- Sutton ML, Gilchrist RB, Thompson JG. 2003. Effects of in-vivo and in-vitro environments on the metabolism of the cumulus-oocyte complex and its influence on oocyte developmental capacity. Hum Reprod Update. 9(1):35–48.
- Sutton-McDowall ML, Gilchrist RB, Thompson JG. 2010. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction. 139(4):685–695.
- Uyar A, Torrealday S, Seli E. 2013. Cumulus and granulosa cell markers of oocyte and embryo quality. Fertil Steril. 99(4):979–997.
- Wang Q, Sun QY. 2007. Evaluation of oocyte quality: morphological, cellular and molecular predictors. Reprod Fertil Dev. 19(1):1–12.
- Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke G, Ron El R, Devroey P, Smitz J. 2011. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod. 26(5):1035–1051.
- Zhang X, Jafari N, Barnes RB, Confino E, Milad M, Kazer RR. 2005. Studies of gene expression in human cumulus cells indicate pentraxin 3 as a possible marker for oocyte quality. Fertil Steril. 83 Suppl 1:1169–1179.
