ABSTRACT
The Origin Replication Complex subunit 4 (ORC4) is one in six subunits of the Origin Replication Complexes (ORCs) which is essential for initiating licensing at DNA replication origins and recruiting adaptor molecules necessary for various cellular processes. Previously, we reported that ORC4 also plays a vital role in polar body extrusion (PBE) during oogenesis in which half the chromosomes are extruded from the oocyte. We hypothesized that ORC4 might play a broader role in chromatin elimination. We tested its role in enucleation during the development of erythrocytes. Murine erythroleukemia (MEL) cells can be propagated in culture indefinitely and can be induced to enucleate their DNA by treatment with Vacuolin-1, thereby mimicking normal erythrocyte enucleation. We found that ORC4 appeared around the nuclei of the MEL cells with Vacuolin-1 treatment, gradually increasing in thickness before enucleation. We then tested whether ORC4 was required for MEL enucleation by down regulating ORC4 with siRNA-ORC4 during Vacuolin-1 treatment and found that this prevented MEL enucleation. These data are consistent with the model that ORC4 is required for erythroblast enucleation just as it is for oocyte PBE. They suggest a new model in which ORC4 expression is a marker for the initiation to the enucleation pathway.
Introduction
The Origin Replication Complex (ORC), which is composed of six proteins, ORC1-6, is essential for initiating licensing at DNA replication origins and recruit adaptor molecules necessary for various cellular processes (Takeda et al. Citation2005). Previously, we reported that ORC4 plays a vital role in polar body extrusion (PBE) in addition to DNA licensing (Nguyen et al. Citation2015). During oogenesis, developing oocytes go through two round of asymmetric meiotic division in which half of the chromosomes are extruded into polar bodies that are much smaller than the remaining oocyte (Kloc et al. Citation2012; Wang et al. Citation2017). We found that ORC4 polymerizes to form a cage that surrounds the chromatin that will expelled from the oocyte during both meiotic divisions, and disrupting this cage by the injection of peptides that block the polymerization also prevented PBE (Nguyen et al. Citation2017). The newly described function of the ORC4 protein surrounding expelled chromatin suggested that ORC4 could play a similar role in other asymmetric cell divisions.
Three types of cells in the body exist without nuclei and must expel their DNA during differentiation, lens epithelium, keritnocytes and erythrocytes. The enucleation of the latter has been studied extensively and bears some similarity to oocyte PBE. In vivo, erythroblasts arrest their cell cycle and bind to macrophages initiating a complex series of cytoskeletal and membrane compositional changes that stimulate enucleation (Gromley et al. Citation2005; Yeo et al. Citation2019). During enucleation, the erythroblast nucleus condenses, and is then extruded as a pyrenocyte. The resulting reticulocyte is released into the blood where it matures, developing its characteristic biconcave shape. The reticulocyte is already capable of absorbing oxygen, but the fully mature erythrocyte, the red blood cell, is more efficient. The mechanism of erythroblast enucleation includes the involvement of Rho GTPases which regulate actin cytoskeleton, cell polarity and motility, and cell cycle (Kalfa and Zheng Citation2014). One interesting convergence between oocyte PBE and erythroblast enucleation is the dependence on vesicle transport for both processes. Crispino and colleagues (Keerthivasan et al. Citation2010; Citation2011) demonstrated that vesicle trafficking is required for erythroblast enucleation, both by using inhibitors and by knocking down clathrin or survivin, two proteins involved in this process. Schuh and colleagues (Schuh and Ellenberg Citation2007; Citation2008; Holubcova et al. Citation2013) demonstrated that in primary oocytes, the mitotic spindle that is located in center of the large cell migrates to the oolemma through a network of actin filaments that nucleates around vesicles that move to the spindle. This cytoskeletal reorganization is dependent on RhoGTPase and Rab 11. Thus, while there are striking differences between oocyte PBE and erythroblast enucleation – the oocyte is a large cell that ejects only half its chromosomes from a mitotic spindle while the erythroblast is a much smaller cell that ejects its entire nucleus from a G0 state, there are also similarities. Both cells eject chromatin that will be degraded. Both processes produce asymmetric daughter cells, one of which is destined to die, and both negate vesicle transport in the mechanisms that expel the DNA. We felt it was reasonable to test whether ORC4 cage formation was involved in erythrocyte enucleation.
We obtained MEL (Murine Erythroleukemia) cells from Dr. John Crispino. MEL cells can be propagated in culture indefinitely and exhibit many characteristics of erythroblasts (Singer et al. Citation1974; Antoniou Citation1991), including enucleation (Volloch and Housman Citation1982; Patel and Lodish Citation1987). In this study, we investigated the role of the licensing ORC4 protein in erythroblast enucleation by using MEL cells and demonstrated that ORC4 is required MEL cell enucleation.
Results
Vacuolin-1 induces enucleation in MEL cells
Before attempting to test the role that ORC4 plays in erythrocyte enucleation we established the cell culture system for MEL cells in our lab. We chose the MEL model to study the role of ORC4 in erythrocyte enucleation because the process could be controlled and cells could be easily counted in our hands. As the MEL cells were cultured and passaged, they appeared relatively homogeneous (). Vacuolin-1 can induce primary erythrocytes to enucleate to form mature erythrocytes (Keerthivasan et al. Citation2012), and MEL can also be induced to enucleate by vacuolin-1 (John Crispino, unpublished data). We tested whether vacuolin-1 could induce MEL cells to enucleate, in vitro, in our hands. MEL cells growing in culture were treated with vacuolin-1 for 48 h, and many of the cells enucleated (). These enucleation events were more obvious in our immunocytochemical staining experiments when the chromatin that was being expelled was clearly stained with DAPI (, below). These results suggested that MEL cells provide a reasonable model for erythrocyte enucleation in culture that would be susceptible to experimental manipulation.
Figure 1. Induction of enucleation in MEL cells with Vacuolin-1 stimulates MEL cells to enucleate. (A) MEL cells were grown continuously in culture without any treatment. The cells are relatively uniform, with no spontaneous enucleation. (B) MEL cells were treated with Vacuolin-1 for 48 hrs to induce enucleation. Arrows point to enucleating cells. The larger, homogenous looking cells will become reticulocytes, and the smaller cells that appear to be budding off are the pyrenocytes that contain the extruded nuclei. Arrows, enucleating cells. Bar = 10 µm
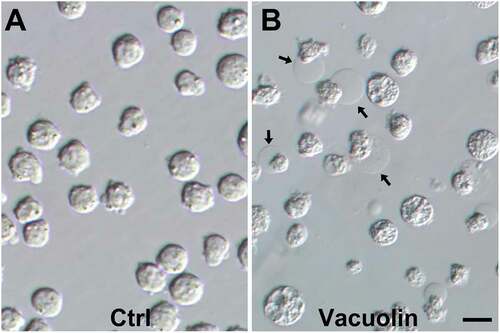
Figure 2. Four types of MEL cells are detected by ORC4 staining after Vacuolin-1 treatment that suggest a progressive model towards enucleation. MEL cells were treated with Vacuolin-1 for 48 hrs and stained with antibodies to ORC4. Four different types of cells were identified. (A–D) Type A cells had no ORC4 staining. (E–F) Type B cells had a small amount of ORC4 staining surrounding the nuclei of the cells. (I–L) Type C cells had more intense staining of ORC4 around slightly condensed nuclei. (M–P) Type D cell were enucleating MEL cells which had more intense ORC4 staining throughout both the pyrenocyte and reticulocyte. Cells were stained for DAPI (A, E, I, M) and ORC4 (B, F, J, N). Merged fluorescence (C, G, K, O), and phase (D, H, L, P) images are shown. Bar = 5 µm
ORC4 staining identifies three types of MEL cells before enucleation begins
We next began to test our hypothesis that ORC4 plays a role in erythrocyte enucleation similar to the role it plays in polar body extrusion in developing oocytes (Nguyen et al. Citation2015; Citation2017). We examined MEL cells by immunocytochemistry (ICC) using antibodies to ORC4. When MEL cells were growing exponentially without treatment to induce enucleation, we noticed two types of cells. Type A had no ORC4 staining and relatively large nuclei (). Type B cells had a small amount of visible ORC4 surrounding the nucleus and just under the plasma membrane (). When MEL cells were induced to enucleate with vacuolin-1 treatment, we noticed two additional types of cells. Type C had much stronger expression of ORC4 surrounding the nucleus, and the nucleus, itself, was slightly condensed (). Type D cells were in the process of enucleation. ORC4 surrounded the chromatin that was being expelled ().
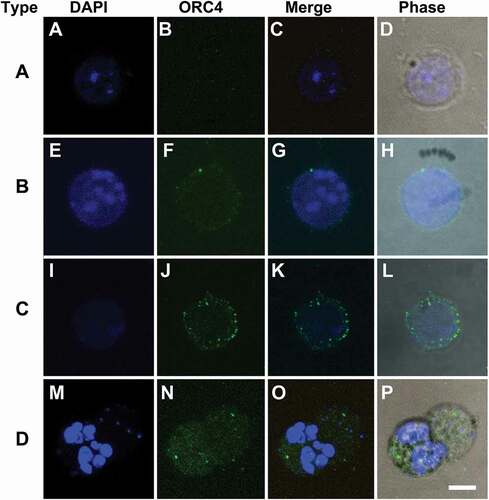
We quantitated the numbers of each type of cell in MEL cells that were growing in culture without treatment (control) and 24 and 48 h after the addition of vacuolin-1 (). We found that when cells were growing in culture without treatment, the cell types as determined by ORC4 staining were asymmetrically divided into a smaller number with no ORC4 staining (Type A) and a larger proportion with low levels of ORC4 near the plasma membrane. After treatment with vacuolin-1 for 24 h, a greater percentage of the cells were Type B. After 48 h of vacuolin-1 treatment, cells with thicker layers of ORC4 around the nucleus (Type C) appeared, and a significant portion of the cells were visibly enucleating (Type D). A low level of punctate ORC4 was apparent in the reticulocyte. The best estimate for the number of cells that progress to full enucleation is the percentage of cells that progressed to Type D, which was 30.9% ()
Figure 3. MEL cells treated with Vacuolin-1 progress to the enucleation stage in 48 hrs. MEL cells were treated with Vacuolin-1 for 0, 24 or 48 hrs and analyzed by immunocytochemistry with antibodies to ORC4 combined with DAPI staining to identify the cell types. The four cell types that were examined were: Type A cells which had no ORC4 staining; Type B cells that had a small amount of ORC4 staining surrounding the nuclei of the cells; Type C cells which had more intense staining of ORC4 around slightly condensed nuclei; and Type D cells which were enucleating MEL cells which had more intense ORC4 staining throughout both the pyrenocyte and reticulocyte. Three experiments were performed analyzing a total of 730 cells with at least 234 cells in each category. The different cell types were counted
Only Type A cells are actively replicating DNA
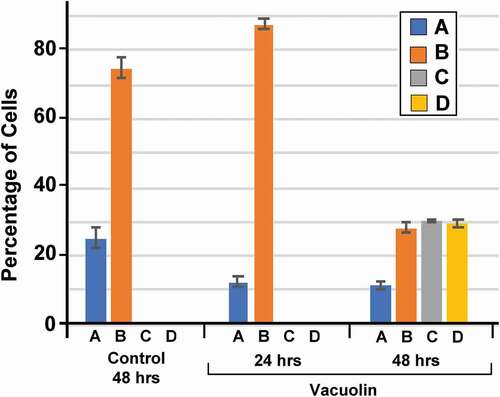
The graph in suggest a model of the progression of enucleation in MEL cells in which cycling cells have no detectable cytoplasmic ORC4 (Type A) and gradually progress to cells that have clear enucleating chromatin (Type D). If this were true, one might expect Type A cells to be actively replicating DNA in order to maintain the MEL population, while enucleating cells should not be replicating DNA. We tested this by pulsing a population of MEL cells that had been treated with vacuolin-1 for 48 h with EdU for one hour. The cells were then fixed and double stained for ORC4, to recognize the four distinct cell types and for the presence of EdU, a thymidine analogue that indicates DNA synthesis. We found that Type A cells () had much stronger incorporation of EdU than either Type B () or Type C () and Type D cells had no detectable EdU incorporation. These results are consistent with a model for MEL cell progression in which Type A cells maintain the population and Types B, C and D are progressively accumulating ORC4 while transitioning to enucleation.
Figure 4. Only Type A cells have measureable levels of DNA replication. MEL cells were treated with Vacuolin-1 for 48 hrs, then incubated with EdU for 30 min and fixed for immunocytochemistry. Cells were stained for DAPI (blue, A, E, I, M) and ORC4 (note in this panel ORC4 staining is red, B, F, J, N), and EdU (green, C, G, K, O). Merged fluorescent and phase (D, H, L, P) images are shown. Cells were then scored for (A-D) Type A cells which had no ORC4 staining; (E-H) Type B (cells that had a small amount of ORC4 staining surrounding the nuclei of the cells; (I-L) Type C cells which had more intense staining of ORC4 around slightly condensed nuclei; and (M-P) Type D cells which were enucleating MEL cells which had more intense ORC4 staining throughout both the pyrenocyte and reticulocyte. Three experiments were performed analyzing a total of 732 cells with at least 236 cells in each category Bar = 5 µm
ORC4 is required for MEL enucleation
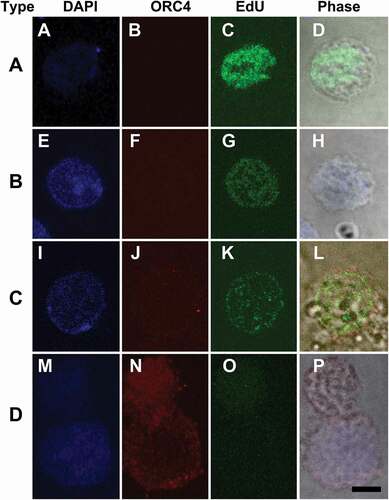
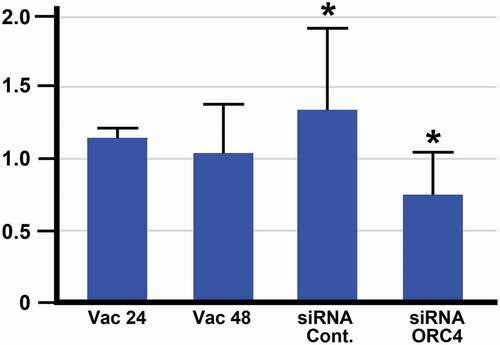
We were previously able to demonstrate that in murine oogenesis, the formation of the ORC4 cage was required for polar body extrusion. We tested whether ORC4 synthesis was required for the progression of MEL cells from exponentially growing to enucleating cells. MEL cells were transfected with plasmids containing expression vectors for either control or ORC4 siRNA, and then induced to progress to enucleation with vacuolin-1. Cells were stained for ORC4, and the four types of cells were counted after 48 hrs. Control MEL cells that were not treated with vacuolin-1 maintained an approximately 1 to 2.5 ratio of Type A to Type B cells (). MEL cells that were treated with control siRNA that contained a scrambled sequence progressed to enucleated cells just as cells with no siRNA with vacuolin-1 did (). However, when cells were transfected with siRNA directed against Orc4 mRNA and then treated with vacuolin-1 for 48 h, no enucleated cells could be detected. Moreover, the ratio of Type A to Type B was closer to 1 to 1, suggesting that even the progression from Type A, with no cytoplasmic ORC4 to Type B, with a low level of cytoplasmic ORC4, was decreased. Quantitative RT-PCR of the MEL cells treated with control versus Orc4-directed siRNA showed a significant decrease in Orc4 mRNA, but not a complete inhibition of Orc4 transcription ().
Figure 5. Treatment with siRNA against Orc4 mRNA inhibits the progression of MEL cells to enucleation. MEL cells were incubated for 48 hrs with no treatment (Control); or transfected with plasmids containing siRNA with a scrambled sequence (siRNA-Ctrl) or siRNA directed against Orc4 mRNA (siRNA-ORC4) for 48 hrs with Vacuolin-1. Cells were then fixed, stained for ORC4, analyzed for Type A-D, and counted. Type A cells which had no ORC4 staining; Type B cells that had a small amount of ORC4 staining surrounding the nuclei of the cells; Type C cells which had more intense staining of ORC4 around slightly condensed nuclei; and Type D cells which were enucleating MEL cells which had more intense ORC4 staining throughout both the pyrenocyte and reticulocyte. Three experiments were performed analyzing a total of 732 cells with at least 236 cells in each category
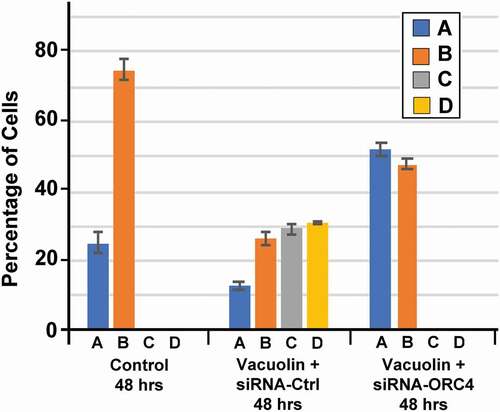
Figure 6. RT-PCR analysis shows a significant decrease in Orc4 mRNA when MEL cells are treated with siRNA directed against Orc4. MEL cells were treated with plasmids containing genes for siRNA with scrambledOrc4 sequence (siRNA Cont.) or siRNA directed against Orc4. Cells were then treated with Vacuolin-1 for 48 hrs and then analyzed by RT-PCR for the presence of Orc4 mRNA. Control cells were not transfected and treated with Vacuolin-1 for 24 (Vac 24) or 48 (Vac 48) hours. There was a significant decrease in Orc4 mRNA with siRNA ORC4 as compared with the control siRNA
Discussion
MEL cells are derived from erythroid progenitor cells infected with the Friend virus (Antoniou Citation1991). These cells exhibit many of the characteristics of erythroid differentiation. Based on previously unpublished data (John Crispino, personal communication) that MEL cells could be induced to enucleate with vacuolin-1, we have now provided evidence to support this. The enucleation differs somewhat from in vivo enucleation primarily because the expelled nucleus is larger than expected. However, we observed several such events and consider this model to be representative of the major components of enucleation.
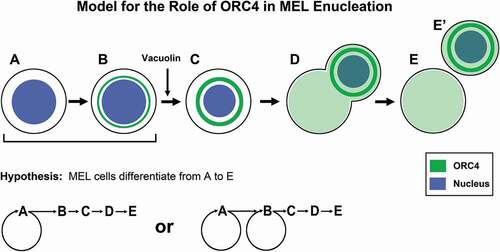
We propose a model for the progression of MEL cells to enucleation when induced by vacuolin-1 based on ORC4 staining and cell morphology, summarized in . Based on the fact that Type A were the cells that incorporated the highest amount of EdU in pulsed experiments, and that they are always present under any condition, we suggest that these are the cells that maintain the MEL cell population. It is also possible that Type B cells have some replicative potential, but we were not able to detect that. Normal erythroblasts also arrest their cell cycle when they initiate terminal differentiation to erythrocytes (Keerthivasan et al. Citation2011; Ovchynnikova et al. Citation2018). Only when MEL cells are activated with vacuolin-1 do they continue this progression to Type C and D cells. Type C MEL cells have condensed nuclei, again mimicking the in vivo progression of erythroblasts to enucleated erythrocytes (Jayapal et al. Citation2010; Ji et al. Citation2011; Yeo et al. Citation2019). While this condensation was not as dramatic as occurs in vivo, in which erythroblast nuclei condense up to seven fold in volume before enucleation (Sasaki et al. Citation1982), the condensation was enough to identify Type C cells as being morphologically distinct from Type B. The novel feature that we observed was a more intense staining for ORC4 that surrounds the nucleus, suggesting that as the MEL cells progress towards enucleation, they accumulate perinuclear ORC4. Finally, we identified cells that were clearly enucleating that had strong ORC4 staining signals ( and ). We were never able to observe, convincingly, the separated pyrenocyte-like cells or the reticulocyte-like cells because we could not be sure of identifying these cells. We noticed that in this model, the chromatin appeared to begin to degrade when it was being expelled, perhaps a result of the lack of condensation. One important aspect of our model for the progression of MEL cells through enucleation is that ORC4 accumulates in the cells as they progress. When ORC4 was knocked down by siRNA expression, progression to Type C cells and to enucleation (Type D cells) was arrested (). Interestingly, the percentage of cells that were Type A increased significantly in cultures treated with siRNA directed against Orc4 mRNA, suggesting that even the progression from Type A to Type B may be dependent on ORC4 production.
Figure 7. The data suggest a model for MEL cell progression that identifies new cell types using ORC4 as a marker. Our data suggest that MEL cells exist primarily as cell types A and B, with Type A generating Type B through replication (though it is also possible that Type B replicate DNA, though our data to do not support a robust division of Type B cells). Type B cells have a small amount of cytoplasmic ORC4, and these gradually progress to Type C, with larger amounts of cytoplasmic ORC 4 but no DNA replication. As the nuclei condense, they start to be evicted from the cell, becoming Type D cells. ORC4 appears punctate throughout the cytoplasm of both the reticulocyte and the pyrenocyte (that contains the extruded nucleus). We never observed Type E cells that we could clearly distinquish because the pyrenocytes are programmed to degrade
When comparing the possible involvement of ORC4 in erythroblast enucleation and oocyte polar body exclusion, there are many similarities and some important differences. Both MEL cells that are progressing to enucleation and maturing and fertilized oocytes have arrested cell cycles and are no longer replicating DNA. In both cell types, ORC4 is located in the cytoplasm in the perinuclear area. When fertilized mouse oocytes re-enter the cell cycle and begin replicating DNA in the one cell embryo stage, ORC4 moves from the perinuclear cytoplasm into the nucleus where it participated in DNA synthesis once again (Nguyen et al. Citation2015). In both MEL cells () and maturing oocytes (Nguyen et al. Citation2017), ORC4 seems to be required for the expulsion of the chromatin.
There are, however, important differences between the two events and the methods used to study them. GV oocytes have a large store of ORC4 just below the membrane before progressing to anaphase I when the first polar body that excludes chromatin is formed. In this case, siRNA had no effect on polar body exclusion (unpublished data). However, since the oocytes are large enough to be injected by micromanipulation, small peptides that were predicted to inhibit the polymerization of ORC4 to form the cytoplasmic cage that surrounds the chromatin that is expelled in the pronucleus did inhibit polar body extrusion (Nguyen et al. Citation2017). MEL cells in log phase growth do not have a significant store of cytoplasmic ORC4 – Type A cells have no detectable cytoplasmic ORC4 and Type B cells have minor amounts. Furthermore, it is likely that Type B cells are derived from Type A cells and may require ORC4 synthesis. In this case, siRNA treatment would be expected to halt enucleation if ORC4 synthesis was required for this process, and this is supported by our data (). Another important difference is that MEL cells mimic the terminal differentiation of erythroblasts to reticulocytes, while the maturation of oocytes in vitro mimics the initiation of embryogenesis. In the latter, ORC4 appears to return to its usual role in DNA synthesis, while in the former it does not. Finally, the most important difference in this comparison is that in erythroblasts all the chromatin is extruded as a condensed nucleus, while in oocyte polar body extrusion half of the DNA is extruded as condensed mitotic chromosomes two times. In both polar bodies, the chromatin is quickly decondensed into a small nucleus that is capable of DNA replication (Nguyen et al. Citation2017). The fact that the nuclear DNA replication licensing protein ORC4 is involved in chromatin expulsion in two such diverse examples of chromatin expulsion during a normal differentiation process is a testament to both the body’s creative use of similar pathways for different processes that have similar requirements and to the importance of the role of ORC4 in chromatin extrusion.
We recognize that our study suggesting that ORC4 is required for chromatin expulsion in MEL cell vacuolin-1 induced enucleation is not a definitive assessment of its role in the in vivo enucleation of erythroblasts. We are currently designing experiments to directly address through targeted deletion of Orc4 in embryonic erythroblast precursors but this will take time. The current study does strongly suggest that ORC4 plays a role in chromatin expulsion during blood cell development, in a manner similar to its role in oocyte polar body exclusion.
Materials and methodologies
a. Materials
MEL cells were donated by Dr. Crispino’s lab. All cell culture media, supplements, and buffers were from Thermo Fisher (Waltham, MA). Vacuolin-1 was obtained from Millipore (Burlington, MA). ORC4 antibodies, siRNA-Control, and siRNA-ORC4 plasmids were purchased from Santa Cruz Biotechnology (Dallas, Texas). Edu staining kit and Lipofectamin-2000TM were purchased from Thermo Fisher (Waltham, MA).
B. Mel cells
MEL, stored at −80°C, were thawed quickly at 37°C. Then the cells were centrifuged to discard supernatant. Finally, the MEL cells were cultured in a 15 mL flask with RPMI 1640 media under 5% CO2 at 37°C.
C. Immunocytochemistry (ICC)
Immunocytochemistry for ORC4 and EdU incorporation and staining was performed as described in (Nguyen et al. Citation2015).
D. Enucleation of MEL cells using Vacuolin-1
MEL cells were incubated with 20 µM of Vacuolin-1 for 24 h or 48 h to induce them to enucleate. All cells were incubated at under 5% CO2 at 37°C. After each treatment, MEL cells were analyzed by immunocytochemistry or the EdU assay.
E. Transfection of MEL cells with siRNA
The transfection of MEL cells was performed by using Lipofectamin-2000TM and 200 nM of siRNA-control and siRNA-ORC4 supplement with 20 µM vacuolin-1, according to the manufacturer’s protocol. The concentration of MEL cells used for siRNA transfection was 1.5 − 2.0 x 106 cells in 15 mL flask with 6 mL of RPMI 1640 containing serum and antibiotics. One vial containing 100 µL Opti-MEM and 2 µL Lipofectamin-2000TM was mixed with one vial containing 100 µL Opti-MEM and 200 nM siRNA-ORC4. This transfection mixture was incubated for 25 minutes at RT. The MEL cells were centrifuged at 2,300 x g for 4 minutes to discard supernatant and the pellet of MEL cells were mixed with 2 mL Opti-MEM and the transfection mixture, then incubated for 6 h under 5% CO2 at 37°C. Then, MEL cells were again centrifuged to remove supernatant and incubated with RPMI 1640 supplemented with 20 µM vacuolin-1 for 24 h or 48 h. At the desired time points, MEL cells were either analyzed for DNA synthesis by the EdU assay or fixed and analyzed immunocytochemistry, as described above.
F. Quantitative RT-PCR
Total MEL cell RNA was extracted with Trizol and DNaseI treatment (Ambion, Austin, TX, USA), and purified using an RNeasy kit (Qiagen, Valencia, CA, USA). Reverse transcription of polyadenylated RNA was performed with Superscript Reverse Transcriptase III, according to the manufacturer’s guidelines (Invitrogen, Carlsbad, CA, USA). RT-qPCR was performed using SYBR Green PCR Master mix on an ABI QuantStudio 12 K Flex machine (Applied Biosystems, Carlsbad, CA, USA). PCR reactions were incubated at 95°C for 10 min followed by 40 PCR cycles (10 s at 95°C and 60 s at 60°C). Three experiments were analyzed, all reactions were carried out in quadruplicates per assay, and four different loading controls, using ubiquitously expressed actin as a control. DCt value for each individual sample was calculated by subtracting either the average Ct or geometric mean of loading control(s) from the average Ct of a tested gene. DDCt value was calculated by subtracting the DCt of each vacuolin-1 treated MEL flask from the untreated MEL controls. The data were expressed as a fold value of expression level.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Author’s contributions
Performed all the experiments and contributed to the design of the work: HN; contributed to most of the experiments: AU; designed the overall work and directed the experiments performed: WSW.
Acknowledgments
The authors would like to thank Dr. John Crispino for important discussions and advice during this work and for the gift of the MEL cells.
Disclosure statement
The authors report no conflicts of interest. This work was supported by NIH Grant Nos. R01 GM123048 and P30 GM131944 to WSW.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Antoniou M. 1991. Induction of erythroid-specific expression in Murine Erythroleukemia (MEL) cell lines. Methods Mol Biol. 7:421–434.
- Gromley A, Yeaman C, Rosa J, Redick S, Chen CT, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. 2005. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 123(1):75–87. doi:https://doi.org/10.1016/j.cell.2005.07.027.
- Holubcova Z, Howard G, Schuh M. 2013. Vesicles modulate an actin network for asymmetric spindle positioning. Nat Cell Biol. 15(8):937–947. eng. doi:https://doi.org/10.1038/ncb2802.
- Jayapal SR, Lee KL, Ji P, Kaldis P, Lim B, Lodish HF. 2010. Down-regulation of Myc is essential for terminal erythroid maturation. J Biol Chem. 285(51):40252–40265. doi:https://doi.org/10.1074/jbc.M110.181073.
- Ji P, Murata-Hori M, Lodish HF. 2011. Formation of mammalian erythrocytes: chromatin condensation and enucleation. Trends Cell Biol. 21(7):409–415. doi:https://doi.org/10.1016/j.tcb.2011.04.003.
- Kalfa TA, Zheng Y. 2014. Rho GTPases in erythroid maturation. Curr Opin Hematol. 21(3):165–171. doi:https://doi.org/10.1097/MOH.0000000000000032.
- Keerthivasan G, Liu H, Gump JM, Dowdy SF, Wickrema A, Crispino JD. 2012. A novel role for survivin in erythroblast enucleation. Haematologica. 97(10):1471–1479. doi:https://doi.org/10.3324/haematol.2011.061093.
- Keerthivasan G, Small S, Liu H, Wickrema A, Crispino JD. 2010. Vesicle trafficking plays a novel role in erythroblast enucleation. Blood. 116(17):3331–3340. doi:https://doi.org/10.1182/blood-2010-03-277426.
- Keerthivasan G, Wickrema A, Crispino JD. 2011. Erythroblast enucleation. Stem Cells Int. 2011:139851. doi:https://doi.org/10.4061/2011/139851.
- Kloc M, Ghobrial RM, Borsuk E, Kubiak JZ. 2012. Polarity and asymmetry during mouse oogenesis and oocyte maturation. Results Probl Cell Differ. 55:23–44.
- Nguyen H, James NG, Nguyen L, Nguyen TP, Vuong C, Ortega MA, Jameson DM, Ward WS. 2017. Higher order oligomerization of the licensing ORC4 protein is required for polar body extrusion in murine meiosis. J Cell Biochem. 118(9):2941–2949. doi:https://doi.org/10.1002/jcb.25949.
- Nguyen H, Ortega MA, Ko M, Marh J, Ward WS. 2015. ORC4 surrounds extruded chromatin in female meiosis. J Cell Biochem. 116:778–786. Eng. doi:https://doi.org/10.1002/jcb.25033.
- Ovchynnikova E, Aglialoro F, von Lindern M, van den Akker E. 2018. The shape shifting story of reticulocyte maturation. Front Physiol. 9:829. doi:https://doi.org/10.3389/fphys.2018.00829.
- Patel VP, Lodish HF. 1987. A fibronectin matrix is required for differentiation of murine erythroleukemia cells into reticulocytes. J Cell Biol. 105(6 Pt 2):3105–3118. doi:https://doi.org/10.1083/jcb.105.6.3105.
- Sasaki K, Matsumura G, Ito T. 1982. Morphometric analysis of postnatal erythropoiesis in the spleen and bone marrow of the mouse. Arch Histol Jpn. 45(3):247–255. doi:https://doi.org/10.1679/aohc.45.247.
- Schuh M, Ellenberg J. 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 130(3):484–498. eng.
- Schuh M, Ellenberg J. 2008. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 18(24):1986–1992. eng. doi:https://doi.org/10.1016/j.cub.2008.11.022.
- Singer D, Cooper M, Maniatis GM, Marks PA, Rifkind RA. 1974. Erythropoietic differentiation in colonies of cells transformed by friend virus. Proc Natl Acad Sci U S A. 71(7):2668–2670. doi:https://doi.org/10.1073/pnas.71.7.2668.
- Takeda DY, Shibata Y, Parvin JD, Dutta A. 2005. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 19(23):2827–2836. eng. doi:https://doi.org/10.1101/gad.1369805.
- Volloch V, Housman D. 1982. Terminal differentiation of murine erythroleukemia cells: physical stabilization of end-stage cells. J Cell Biol. 93(2):390–394. doi:https://doi.org/10.1083/jcb.93.2.390.
- Wang JJ, Ge W, Liu JC, Klinger FG, Dyce PW, De Felici M, Shen W. 2017. Complete in vitro oogenesis: retrospects and prospects. Cell Death Differ. 24(11):1845–1852.
- Yeo JH, Lam YW, Fraser ST. 2019. Cellular dynamics of mammalian red blood cell production in the erythroblastic island niche. Biophys Rev. 11(6):873–894.
