ABSTRACT
The analysis of products of conception (POC) is clinically important to establish the cause of early pregnancy loss. Data from such analyses can lead to specific interventions in subsequent natural or assisted conceptions. The techniques available to examine the chromosomal composition of POC have limitations and can give misleading results when maternal cell contamination (MCC) is overlooked. The aim of this study was to develop a protocol for MCC assessment and to formulate POC material handling, testing, and reporting recommendations. Using array comparative genomic hybridization, we tested 86 POC samples, of which 47 sample pairs (DNA extracted from the POC sample and maternal DNA) were assessed for the presence of MCC. MCC was evaluated using an approach we developed, which exploited the genotyping of 14 STR, AMEL, and SRY loci. POC samples showing the clear presence of villi (63.9%) did not contain any signs of the maternal genome and can therefore be reliably tested using conventional methods. The proportion of 46,XX karyotype in the unselected sample batch was 0.39, which fell to 0.23 in visually good samples and was 0.27 in samples having no signs of contamination upon MCC testing. MCC assessment can rescue visually poor samples from being discarded or wrongly genotyped. We demonstrate here that classification based on visual POC material evaluation and MCC testing leads to predictable and reliable POC genetic testing outcomes. Our formulated recommendations covering POC material collection, transportation, primary and secondary processing, as well as the array of pertinent considerations discussed here, can be implemented by laboratories to improve their POC genetic testing practices. We anticipate our protocol for MCC assessment and recommendations will help reduce the misconception regarding the etiology of miscarried fetuses and foster informed decision-making by clinicians and patients dealing with early pregnancy loss.
Introduction
Miscarriage is a traumatizing experience for the patient and places a burden on the practice of obstetrics and gynecology. The magnitude of miscarriage appears to be increasing, particularly in developed countries where the population is aging fairly rapidly and consequently the age of mothers at childbirth is rising (Heazell et al. Citation2018). Fetal chromosomal aberrations play the biggest role in the etiology of miscarriage. The majority of embryos with an aberrant karyotype decease during the first weeks of pregnancy (Romero et al. Citation2015; Davis et al. Citation2017). Specifically, 40–75% of the fetuses aborted between 6 and 7 weeks of gestation have an abnormal karyotype, 20–25% aborted between 12 and 17 weeks, and only 2–7% aborted between 17 and 28 weeks (Agarkova Citation2010).
The analysis of products of conception (POC) is clinically important to establish the cause of early pregnancy loss and choose specific interventions in subsequent natural or assisted conceptions. Various techniques are currently used to detect chromosomal aneuploidies and structural rearrangements in POC, including but not limited to karyotyping, quantitative fluorescent polymerase chain reaction, multiple ligation probe amplification, array comparative genomic hybridization (aCGH), single-nucleotide polymorphism/variation (SNP) arrays, and next-generation sequencing.
G-banding of chorionic villi metaphases is considered to be the gold standard in routine POC testing. However, its application is limited due to the quality and viability of chorion cells in the primary biological material. Molecular testing has overcome some of the disadvantages inherent to conventional cytogenetic techniques and can be used as a rescue molecular karyotyping (Lomax et al. Citation2000) if culture failure occurs or POC material is degraded.
All of the techniques examining POC chromosomal composition have drawbacks and especially can give misleading results when maternal DNA contamination (commonly referred to as maternal cell contamination (MCC)) is overlooked. POC degradation is not unusual given the fact that a missed abortion can go unnoticed for several days during which cell maceration has already started (De La Rochebrochard and Thonneau Citation2002).
A bias toward an increased number of normal female karyotype reports in comparison to normal male karyotype reports has been noted (Bell et al. Citation1999; Jarrett et al. Citation2001; Lathi et al. Citation2014). However, not all laboratories fully address this important issue and its etiology (Nikitina et al. Citation2005). There are cases of possible misconception of a higher incidence of normal female karyotype results with the 69,XXX karyotype (Shen et al. Citation2016) when maternal DNA contamination was not considered.
While there are several factors possibly influencing the differing sex ratio in spontaneous abortions (Jarrett et al. Citation2001) like unrecognized 46,XX molar samples, maternal age at gestation, X-linked lethal mutations acting in utero, and sex chromosome-specific failure of chromosome preparation (Hassold et al. Citation1983; Eiben et al. Citation1990), studies have demonstrated that up to 59% of the normal female karyotypes reported in POC testing are in fact cases of MCC, when contamination completely obscures the fetal material. The overall MCC rates of POC samples across different laboratories vary, but can occur in as high as in 89.7% of the cases (Jarrett et al. Citation2001; Lathi et al. Citation2014; Romero et al. Citation2015), thus indicating different sample management and demonstrating that the general awareness of MCC in this context is limited and needs to be improved.
The recorded bias toward a higher number of 46,XX karyotype reports together with the existing problem of MCC in POC testing points to a limited awareness of the technical limitations and critical aspects of methodologies used for POC analysis. Crucially, this failing needs to be acknowledged by laboratory specialists and consulting physicians. Therefore, the aim of this study was to develop a protocol for MCC assessment and to formulate POC material handling, testing, and reporting recommendations.
Results and discussion
Visual POC inspection and MCC genetic testing
POC specimens are considered samples not containing any identifiable material from the fetus proper (e.g., cord, amnion), but rather consisting of villi, membranous material (Jarrett et al. Citation2001) and other tissues of unspecified origin. Visual inspection of the primary POC material (n = 86) resulted in the following observations: 55 were good quality samples, 19 compromised quality POCs with signs of tissue maceration, and 12 samples where no tissue with typical villous morphology could be detected – marked ‘no chorion.’ Four FFPE samples were marked as compromised and one had no signs of villi.
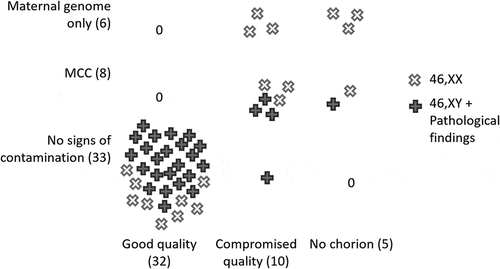
Forty-seven sample pairs (POC and maternal genomic DNA) subjected to polymorphic microsatellite (short tandem repeat, STR) loci genotyping based MCC detection protocol developed by us revealed that in 33 (70.2%) of the POC samples maternal genome was not detected; of those, one was marked as compromised quality, while the remainder demonstrated good quality chorions. Eight samples (17.0%) showed the presence of MCC; of those, six were classified as compromised quality chorions and two showed no villi upon visual inspection; one sample with MCC was positive for the SRY region. Six POC samples (12.8%) showed only maternal genome; three were of poor quality and three showed no visual presence of villi.
Data from 41 sample pairs (excluding samples showing only maternal genome) was used to calculate allelic frequencies (AF) of the STR loci included in the assay and is presented in S1. Each STR locus revealed 9.7 ± 1.5 alleles (average±standard deviation (SD)). Out of 14 loci, 8.5 ± 2.0 (average±SD) were informative in distinguishing fetal and maternal genotypes. Theoretical probability of the assay being not-informative, i.e., giving false perception that fetal sample contains only maternal genome, was calculated to be 1.9E-08. Thus, it can be assumed that MCC detection system provides reliable results and can be used with high confidence.
Chromosomal microarray analysis
All POC samples (n = 86) were subjected to aCGH analysis irrespective of biological material quality and MCC testing. In total, 34 samples corresponded to normal female karyotype and 16 to normal male karyotype (sex ratio 2.1:1). The remaining 36 (41.9%) samples exhibited some kind of chromosomal abnormality, out of those 12 contained an XX sex chromosome set, 11 contained XY (sex ratio 1:0.9), and 13 were associated with sex chromosome copy number variations (). The majority of chromosomal imbalances were autosomal trisomies, followed by pure monosomy X (four cases). Of seven cases showing some kind of sex chromosome discrepancy, four indicated a mosaic form of X monosomy – arrmos(X)x1, while three cases were unable to be resolved using aCGH analysis alone. Lastly, following structural aberrations were detected: loss of 8p23.2p11.21, gain of 22q13.2q13.33, and combined gain of 11p15.5p15.2 and 15q26.1q26.3 in one sample.
Table 1. Chromosomal pathology distribution across positive samples (n= 36).
Analysis of POC with high risk of MCC
A result indicative of the 46,XX karyotype should be treated with caution since it might arise from the analysis of maternal cells, especially in samples of unsatisfactory visual quality. As seen from the , the poorer the quality of the samples included in the analysis (visually inspected), the higher the proportion of 46,XX samples and the lower the fraction of 46,XY and chromosomally abnormal samples. This was also true for samples tested for MCC if samples with partial MCC were included in the calculations. A significant difference (p-value 0.02) in the observed genotypes distribution was seen between the group having no signs of contamination upon MCC testing and all samples group. 46,XY samples were completely absent in the groups ‘Compromised quality + No chorion’ and ‘No chorion.’ The ‘Only maternal genome’ group was not included since it contained solely 46,XX results upon aCGH testing as expected.
Figure 1. Distribution of karyotype results across different POC evaluation groups. Samples were grouped based on the MCC testing results (no signs of contamination; MCC; maternal genome only – not included in the figure since those contains only 46,XX results as expected) and visual sample evaluation (good quality chorion; compromised quality; no chorion). * Significant difference (p-value 0.02) in the observed genotypes distribution was seen between the groups ‘No signs of contamination’ and ‘All samples’. MCC – maternal cell contamination
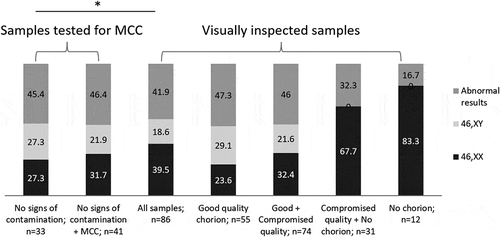
The origin of cells/tissue having the 46,XY karyotype or any chromosomal abnormality (n = 28) is indubitable (i.e., fetal). visually represents that vast majority of these cases (82.1%) concentrated among samples showing good visual quality and no signs of contamination upon MCC investigation. Nevertheless, four cases (14.3%) were found amongst compromised quality POC, three of them having a certain amount of MCC. One case indicative of a sex chromosome discrepancy upon aCGH analysis was localized in the ‘no chorion’ group and also displayed MCC but was positive for the SRY region. Based on standard criteria samples of compromised visual quality would have been discarded (Romero et al. Citation2015), because having viable cells or POC with identifiable villi was crucial for cytogenetic and molecular cytogenetic techniques, e.g. FFPE samples typically required pathologist conclusion on fetal cells presence prior to DNA extraction. Here we demonstrate that simple and quick step of MCC evaluation can rescue some percentage of poor primary biological samples and increase number of correct diagnoses.
Figure 2. Distribution of MCC-high- and -low-risk karyotypes across different product of conception evaluation groups. Y-axis depicts sample evaluation based on MCC genetic testing using STR genotyping. X-axis depicts visual examination of primary biological sample. MCC-high risk are samples corresponding to ‘46,XX’ karyotype (depicted as white X’s) can arose from analysis of fetal cells or maternal cells thus masking any genuine fetal karyotype. MCC-low risk samples are the ones showing 46,XY karyotype or any chromosomal pathology (depicted as gray crosses). MCC – maternal cell contamination
Recommendations to reduce MCC in POC testing
The identification of a chromosomal abnormality in POC material not only provides an explanation for the miscarriage but also removes the need for further investigations. Current American Society for Reproductive Medicine (ASRM) recommendations state that chromosome testing of miscarriage specimens may be of psychological benefit to the patient and may aid treatment decisions in the setting of treatment of recurrent pregnancy loss (‘Evaluation and treatment of recurrent pregnancy loss: A committee opinion,’ Citation2012).
The main reasons for unsuccessful POC testing are loss of cell viability (applies to cytogenetic techniques) and MCC (applies to a majority of methodologies) (). Therefore, the ASRM recommends that in the case of a 46,XX result, maternal blood should be obtained for the differentiation of maternal from fetal source of the euploid result by the means of microsatellite analysis (‘Evaluation and treatment of recurrent pregnancy loss: A committee opinion,’ Citation2012). Solutions to MCC assessment are well documented for cases of prenatal genetic testing when the material for testing is obtained by the means of amniocentesis or chorionic villi sampling of ongoing pregnancy. Authorities in the field of medical genetics (American College of Medical Genetics, Association for Molecular Pathology) agree and emphasize that the presence of MCC that may interfere with the interpretation of fetal results must be excluded, using STR testing or exploiting technology capable of distinguishing MCC, e.g., SNP-arrays (Nagan et al. Citation2011; American College of Medical Genetics and Genomics Citation2018; Monaghan et al. Citation2020).
Table 2. Comparison of different methodologies used for POC analysis
Assessment of STR loci is widely used in human genetic identification (e.g., CODIS core loci (Karantzali et al. Citation2019)), because STRs are highly variable and evenly distributed throughout the genome; and while there are various ways to assess MCC in POC (Hassold et al. Citation1983; Bell et al. Citation1999), STR genotyping using commercially available or in-house tests is an easy and reliable way to perform MCC testing (Jarrett et al. Citation2001; Amos et al. Citation2005; Schrijver et al. Citation2007).
Although MCC testing of prenatal samples is recommended in guidelines, only 60% of surveyed laboratories across the United States performed it without exception (Schrijver et al. Citation2007). Unfortunately, the exact data for the POC testing is unknown. However, as can be seen from the issues raised in literature the numbers might be similar. While the problem of MCC is universal to POC and prenatal samples, there are major differences in processing both types of samples. Therefore, due to lack of comprehensive practice guidelines, we aimed to formulate recommendations addressing the entire workflow of POC samples handling from preanalytical, through the analytical stages ().
Figure 3. POC handling and testing workflow. Recommendations addressing the entire workflow of POC samples handling from preanalytical, through the analytical stages suitable for molecular/molecular cytogenetic techniques. POC – product of conception. FFPE – formalin-fixed paraffin-embedded samples; PBS – phosphate buffer saline; STR – short tandem repeats; aCGH – array comparative genomic hybridization; NGS – next-generation sequencing, fPCR – fluorescent polymerase chain reaction
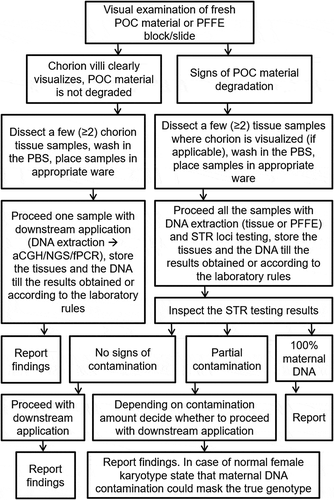
As a starting point, clinicians carrying out the collection of primary material after curettage of a missed abortion need to be trained to visually inspect the tissues and collect the correct ones. It is not unusual that the transportation medium is unsuitable, e.g., distilled water or formalin. To reduce number of rejected samples, it is the responsibility of the laboratory to provide clinicians with unequivocal sampling guidelines and collection containers with the appropriate medium.
In comparison to curettage medically induced abortion is considered safer procedure for the female (Kovavisarach and Jamnansiri Citation2005; Niinimäki et al. Citation2006; Behnamfar et al. Citation2013). This allows for the patient to collect the miscarried material herself in a stationary or home setting – the same also applies to the situations when the spontaneous abortion is clinically expected. While the laboratory cannot control the quality of the collected material, it can provide these patients with the instructions and a suitable container with the proper medium. Alternatively, if access to the collection medium is not possible, transportation without medium is possible. In all cases, the POC sample has to be transferred to the laboratory as quickly as possible, preferably within 24 hours, in order to preserve the viability of cells and ensure appropriate sample evaluation and processing prior to genetic testing. Sampling of maternal material (blood/buccal swab) concurrently with POC sampling/sample delivery to the laboratory is a beneficial practice to implement to avoid repeated visits to the clinic.
The next important step is the visual inspection of primary biological material by trained laboratory personnel. We did not find it necessary to perform POC dissection under a microscope since typical villous morphology can be clearly visualized with the naked eye. Chorion tissues need to be carefully separated and thoroughly washed with an appropriate buffer (saline, PBS) to remove any maternal decidua, blood, mucus. From our investigations, we advise to avoid selection of poor specimens, i.e., grossly necrotic tissues with no signs of typical villous morphology, if no MCC testing is performed, as they rarely contain fetal genome. It was suggested before that POC samples where only villi or a combination of villi and membranous material was identified and which resulted in normal female karyotype should be viewed with suspicion (Rodgers et al. Citation1996). In contrast, our experience shows that DNA extracted from samples displaying clear villous morphology does not have any signs of maternal genome and consequently is able to be processed without MCC testing. Sampling of multiple (two or three) pieces and written witnessing of the sampled tissue quality are beneficial for downstream application results interpretation and proper reporting.
Depending on the POC testing methodology used, the impact of an arbitrary amount of maternal genome on the result can range from absolutely no effect to complete obscuration of the genuine fetal genome. Therefore, it is recommended that MCC systems capable of quantifying the amount of MCC is developed and also that the extents to which certain methodologies can tolerate the presence of MCC are validated. We exploited our MCC detection protocol solely for the qualitative evaluation of MCC. However, based on the peak height observed in our fluorescent PCR assay, it was possible to detect as low as approximately 5% of the maternal genome in the POC sample, although such an amount is unlikely to obscure results obtained by molecular cytogenetic techniques and can be considered negligible.
It is the responsibility of the laboratory to evaluate the practicability and validity of any particular methodology before its application in clinical practice. Reporting should follow specific diagnostic reporting practices and all reports must be complemented with adequate counseling.
Further considerations to improve POC testing practice
In the era of genetic testing diversity, it is essential to choose the most precise, detailed, and reliable techniques adding to cost-efficiency and overall utility for POC analysis, e.g., aCGH which has demonstrated certain advantages over classic cytogenetic methodologies (Lomax et al. Citation2000; Hyde and Schust Citation2015). A great tool for MCC confirmation without the need for additional MCC testing is SNP microarrays which also yields higher diagnostic return (Lathi et al. Citation2014; Levy et al. Citation2014). Additionally, more sophisticated primary biological material sampling practices (if applicable) could be adopted, e.g., precise chorion or embryo proper sampling in utero using hysteroscopic embryoscopy (Robberecht et al. Citation2012).
If, for any reason, MCC testing is unavailable, it is essential to estimate the probability of obtaining an unreliable 46,XX result. To demonstrate how biased POC reporting could be if a laboratory adopted a procedure of testing all samples without visual evaluation and MCC testing, we applied a suitable mathematical model developed by Nikitina and colleagues (Nikitina et al. Citation2005) to our results. The proposed model exploits samples having XX or XY sex chromosomes set excluding all cases with sex chromosome discrepancies (e.g., XO and XX/XY) and is largely based on the initially obtained MCC coefficient (k). Since we have performed MCC testing, we were able to calculate actual k (). The model demonstrated that the percentage of samples obscured by MCC could be as high as 21.24% (16 samples in our scenario), which significantly (p-value 0.0006) changes the expected distribution of the genotypes across the samples.
Table 3. Observed and expected genotype distribution when corrected for maternal cell contamination based on the approach described in Nikitina et al. Citation2005
Samples with the 46,XX karyotype would be expected to fall from 0.47 to 0.25 when corrected for MCC, which is similar to our value of 0.27 obtained from samples having no signs of contamination upon MCC testing. Despite the theoretical nature of the model, it may be useful for estimating the MCC value in an established culture regimen in any laboratory, thus serving as a valuable tool for quality control (Nikitina et al. Citation2005).
We have demonstrated how different approaches of visual POC material evaluation complemented with MCC testing lead to predictable and reliable POC genetic testing outcomes. While our formulated recommendations covering all steps of POC material processing cannot influence the quality of primary biological samples, those once implemented by laboratories can actually improve their POC genetic testing practices, acknowledge, and diminish problem of MCC. Ultimately, through a combination of different techniques and the development of an up-to-date approaches to POC handling and testing, from the perspective of reproductive counseling, a significant advancement could be achieved with the elucidation of the cause of miscarriage for most cases.
Materials and methods
Patients
Patients experiencing miscarriage or spontaneous abortion before the 13th week of gestation were recruited to the study. POC material was either obtained by uterine curettage performed by a gynecologist or collected by patients themselves and transferred in 0.9% sodium phosphate buffer or without medium to the laboratory within one to 2 days after collection. Cases of fetus proper (n = 2) were excluded. Five samples were FFPE tissues. In total, 86 POC samples were included in the study. The gestational age at the time of early pregnancy loss was 8.6 ± 2.2 (average±SD) weeks and the patient age was 32.7 ± 5.5 (average±SD) years. Peripheral blood samples were obtained only from 47 women for genetic analysis of MCC, unfortunately we could not obtain blood specimens from the rest of the patients.
Primary biological material handling
Upon receipt, each POC sample was visually inspected by trained laboratory personnel. Visual appearance of the sample was recorded as follows: ‘good quality chorion’ – if presenting typical villous morphology; ‘poor quality chorion’ – if presenting tissue maceration and only a few villi could be dissected; or ‘no chorion visualized’ – if no tissues with typical villous morphology could be localized. Even in the absence of typical villous morphology, tissue sampling was performed for all the samples. Tissues were washed in 1x PBS [Thermo Fisher Scientific, UK] and three pieces of each sample were placed in separate tubes for subsequent nucleic acid extraction. DNA was extracted from fresh POC, FFPE tissues, and peripheral blood following the manufacturer’s standard protocol [Qiagen, Germany].
POC chromosome analysis
Chromosome analysis was performed by aCGH for all the POC samples following the manufacturer’s protocol [24sure; Illumina, USA]. Microchip slides were scanned with an InnoScan scanner [Innopsys, France]. Images in Tiff format were imported into BlueFuse Multi v.4.0 [Illumina] and the resulting copy number karyotypes were assessed. The given methodology detects unbalanced chromosomal material changes >5Mb and can detect polyploidy if sex chromosomes are represented by at least one X and Y chromosomes.
MCC testing system design
To test for MCC signs in samples where maternal DNA was available (n = 47), we designed a detection system employing fluorescent PCR with visualization by capillary electrophoresis. The system encompassed 14 microsatellite (STR) loci, the AMEL region giving different amplicon lengths on X and Y, and the SRY region for the more precise genotyping of chromosome Y. In order to detect MCC, maternal DNA, and DNA extracted from the POC sample need to be tested in parallel.
All loci of interest were located through the University of California Santa Cruz’s genome browser (https://genome-preview.ucsc.edu/index.html). For all genomic regions, fluorescently tagged (5′ end 6-FAM or HEX) primers for fluorescent PCR were located manually and specificity was confirmed using IDT’s primer tool (https://eu.idtdna.com/calc/analyzer) and Primer-BLAST (Ye et al. Citation2012). Primer pairs were multiplexed into four mixes, each containing four loci (S2).
The PCR mixture for each multiplex was as follows: 3.6 μl of ddH2O, 7.5 μl of Type-it Master Mix [Qiagen], 0.32 μl of each primer (0.2 μM, eight primers for one multiplex), and 1.5 μl of DNA (~50 ng/μl) in a total reaction volume of 15.16 μl. The cycling conditions were: initial denaturation for 5 min at 95°C, followed by 28 cycles of 30 s at 95°C, 1 min 30 s at 60°C, and 30 s at 72°C; final extension for 10 min at 72°C. For the initial PCR product visualization as a quality control step, the amplified products were run on 2% agarose gels. Amplicon separation and sizing were carried out using capillary electrophoresis [ABI Prism 3500 DNA Analyzer; Applied Biosystems, USA] and GeneMapper v.4.0 software [Applied Biosystems], respectively.
STR marker is considered not-informative when the paternal allele transmitted to the conceptus being the same size as the non-inherited maternal allele – such a situation can result in the false consistency of MCC. The probability of this event decreases with the usage of a reasonable number of highly polymorphic STR markers, increasing overall informativeness of the assay (Nagan et al. Citation2011). A proposed minimum number of STR markers to be analyzed is four to six (Jarrett et al. Citation2001). Fourteen STR markers exploited here have previously been used for linkage analysis in preimplantation genetic testing and therefore are known to be biallelic and polymorphic, and the individual primer pair performance has already been checked (Volozonoka et al. Citation2018). Since population AF of the STR markers used is unavailable, those were calculated from the genotyping data of POC and maternal genomic DNA sample pairs (n = 47); allele shared between mother and fetus was excluded from AF counting. The highest allelic frequencies of each locus were multiplied, thus estimating the probability of the whole assay being not informative.
MCC testing results were classified as follows: ‘MCC’ – in case of informative STR marker characterized by three alleles visible on electropherogram, two of which match the alleles of the mother (or two alleles, if mother was homozygous); ‘maternal genome only’ – characterized by the complete allelic match of the two samples across all loci; ‘no signs of contamination’ – characterized by the second allele in a fetus distinguishable from the mothers’ alleles across informative markers. The developed STR testing system only allows for the qualitative not the quantitative evaluation of MCC based on STR loci differences between the genomes being compared.
Statistical analysis
Data were described with average values and SD if appropriate. Genotypes across different sample categories were compared using the Chi-square test. Statistical significance was assumed at p < 0.05. Statistical analyses used the Microsoft Excel 2019, Version 16.0.
Ethical considerations
The study was conducted in accordance with the Declaration of Helsinki’s ethical principles and its protocol was approved by the Central Medical Ethics Committee (14.04.2016.Nr.1/16-04-14). Patients considered for genetic testing were counseled and the testing principles were clearly explained. All the recruited patients signed an informed consent. Patients acknowledged that they could not be identified from the article as personal data were fully anonymized.
Author contributions
Conceptualization: LV, DP, LG, LK, AM; data curation: LV, LG, LK, AM; methodology: LV, DP, LG, LK, AM; software: LV, DP; visualization: LV; writing - original draft: LV; writing – review and editing: LV, DP, LK, IK, VF, AM; funding acquisition: LG, IK, VF; project administration: LG, IK, VF, AM; resources: LG, IK, VF, AM; supervision: LG, IK, AM.
Supplemental Material
Download Zip (25.1 KB)Disclosure statement
The authors report no conflict of interest.
Supplemental data
Supplemental data for this article can be accessed here.
References
- Agarkova I. 2010. Non-developing pregnancy: assessment of risk factors and prognosis (review) (Неразвивающаяся беременность: оценка факторов риска и прогнозирование (обзор)). Медицинский Альманах. 4:82–88.
- American College of Medical Genetics and Genomics. 2018. Standards and guidelines for clinical genetics laboratories. 2018 ed, p. 906. Revised January 2018. Available at: https://www.acmg.net/PDFLibrary/Standards-Guidelines-Cytogenetics.pdf.
- Amos J, Feldman GL, Grody WW. 2005. Technical standards and guidelines for CFTR mutation testing. Am Coll Med Genet. doi:https://doi.org/10.1097/01.GIM.0000031065.72493.BC.
- Ari E, Ozdemir O, Djurovic J, Silan F. 2018. Prenatal diagnosis of aneuploidies and microdelation/duplication in amniotic fluid and fetal aborted material by QF-PCR and MLPA analysis. Biomed Genet Genomics. 3(1). doi:https://doi.org/10.15761/bgg.1000136.
- Ballif BC, Kashork CD, Saleki R, Rorem E, Sundin K, Bejjani BA, Shaffer LG. 2006. Detecting sex chromosome anomalies and common triploidies in products of conception by array-based comparative genomic hybridization. Prenat Diagn. 26(4):333–339. doi:https://doi.org/10.1002/pd.1411.
- Behnamfar F, Mahdian M, Rahimi F, Samimi M. 2013. Misoprostol abortion: ultrasonography versus beta-hCG testing for verification of effectiveness. Pak J Med Sci. 29(6). doi:https://doi.org/10.12669/pjms.296.3361.
- Bell KA, Van Deerlin PG, Haddad BR, Feinberg RF. 1999. Cytogenetic diagnosis of “normal 46,XX” karyotypes in spontaneous abortions frequently may be misleading. Fertil Steril. 71(2):334–341. doi:https://doi.org/10.1016/S0015-0282(98)00445-2.
- Berezowsky J, Zbieranowski I, Demers J, Murray D. 1995. DNA ploidy of hydatidiform moles and nonmolar conceptuses: A study using flow and tissue section image cytometry. Mod Pathol: an official journal of the United States and Canadian Academy of Pathology, Inc. 8(7):775-781.
- Bin LS, Xie YJ, Chen Z, Zhou Y, Wu JZ, Zhang ZQ, … Fang Q. 2015. Improved assay performance of single nucleotide polymorphism array over conventional karyotyping in analyzing products of conception. J Chin Med Assoc. doi:https://doi.org/10.1016/j.jcma.2015.03.010.
- Bruno DL, Burgess T, Ren H, Nouri S, Pertile MD, Francis DI, Slater HR. 2006. High-throughput analysis of chromosome abnormality in spontaneous miscarriage using an MLPA subtelomere assay with an ancillary FISH test for polyploidy. Am J Med Genet A. 140A(24):2786–2793. doi:https://doi.org/10.1002/ajmg.a.31552.
- Davis AR, Horvath SK, Castaño PM. 2017. Trends in gestational age at time of surgical abortion for fetal aneuploidy and structural abnormalities. Am J Obstet Gynecol. 216(3):278.e1–278.e5. doi:https://doi.org/10.1016/j.ajog.2016.10.031.
- De La Rochebrochard E, Thonneau P. 2002. Paternal age and maternal age are risk factors for miscarriage; results of a multicentre European study. Hum Reprod. 17(6):1649–1656. doi:https://doi.org/10.1093/humrep/17.6.1649.
- Donaghue C, Mann K, Docherty Z, Mazzaschi R, Fear C, Ogilvie C. 2010. Combined QF-PCR and MLPA molecular analysis of miscarriage products: an efficient and robust alternative to karyotype analysis. Prenat Diagn. 30(2):133–137. doi:https://doi.org/10.1002/pd.2424.
- Dong Y, Yi Y, Yao H, Yang Z, Hu H, Liu J, Liang Z. 2016. Targeted next-generation sequencing identification of mutations in patients with disorders of sex development. BMC Med Genet. 17(1). doi:https://doi.org/10.1186/s12881-016-0286-2.
- Dória S, Lima V, Carvalho B, Moreira ML, Sousa M, Barros A, Carvalho F. 2010. Application of touch FISH in the study of mosaic tetraploidy and maternal cell contamination in pregnancy losses. J Assist Reprod Genet. 27(11):657–662. doi:https://doi.org/10.1007/s10815-010-9460-1.
- Eiben B, Bartels I, Bahr-Porsch S, Borgmann S, Gatz G, Gellert G, … Hansmann I. 1990. Cytogenetic analysis of 750 spontaneous abortions with the direct-preparation method of chorionic villi and its implications for studying genetic causes of pregnancy wastage. Am J Hum Genet. 47(4):656.
- Evaluation and treatment of recurrent pregnancy loss: A committee opinion. 2012. Fertility and sterility. 98(5):1103-1111. doi:https://doi.org/10.1016/j.fertnstert.2012.06.048.
- Fejgin MD, Pomeranz M, Liberman M, Fishman A, Amiel A. 2005. Fluorescent in situ hybridization: an effective and less costly technique for genetic evaluation of products of conception in pregnancy losses. Acta Obstet Gynecol Scand. 84(9):860–863. doi:https://doi.org/10.1111/j.0001-6349.2005.00757.x.
- Hassold T, Quillen SD, Yamane JA. 1983. Sex ratio in spontaneous abortions. Ann Hum Genet. 47(1):39–47. doi:https://doi.org/10.1111/j.1469-1809.1983.tb00968.x.
- Heazell AEP, Newman L, Lean SC, Jones RL. 2018. Pregnancy outcome in mothers over the age of 35. Curr Opin Obstet Gynecol. 30(6):337–343. doi:https://doi.org/10.1097/GCO.0000000000000494.
- Hedley D, Friedlander M, Taylor I, Rugg C, Musgrave E. 1983. Method for analysis of cellular DNA content of paraffin-embedded pathological material using flow cytometry. J Histochem Cytochem. 31(11):1333–1335. doi:https://doi.org/10.1177/31.11.6619538.
- Hyde KJ, Schust DJ. 2015. Genetic considerations in recurrent pregnancy loss. Cold Spring Harb Perspect Med. 5(3):a023119–a023119. doi:https://doi.org/10.1101/cshperspect.a023119.
- Jarrett KL, Michaelis RC, Phelan MC, Vincent VA, Best RG. 2001. Microsatellite analysis reveals a high incidence of maternal cell contamination in 46,XX products of conception consisting of villi or a combination of villi and membranous material. Am J Obstet Gynecol. 185(1):198–203. doi:https://doi.org/10.1067/mob.2001.114692.
- Karantzali E, Rosmaraki P, Kotsakis A, Le Roux-Le Pajolec MG, Fitsialos G. 2019. The effect of FBI CODIS core STR loci expansion on familial DNA database searching. Forensic Sci Int Genet. 43:102129. doi:https://doi.org/10.1016/j.fsigen.2019.07.008.
- Kovavisarach E, Jamnansiri C. 2005. Intravaginal misoprostol 600 μg and 800 μg for the treatment of early pregnancy failure. Int J Gynecology Obstetrics. 90(3):208–212. doi:https://doi.org/10.1016/j.ijgo.2005.04.016.
- Lathi RB, Gustin SLF, Keller J, Maisenbacher MK, Sigurjonsson S, Tao R, Demko Z. 2014. Reliability of 46,XX results on miscarriage specimens: A review of 1,222 first-trimester miscarriage specimens. Fertil Steril. 101(1):178–182. doi:https://doi.org/10.1016/j.fertnstert.2013.09.031.
- Lathi RB, Loring M, Massie JAM, Demko ZP, Johnson D, Sigurjonsson S, Rabinowitz M. 2012. Informatics enhanced SNP microarray analysis of 30 miscarriage samples compared to routine cytogenetics. PLoS ONE. 7(8). doi:https://doi.org/10.1371/journal.pone.0031282.
- Levy B, Sigurjonsson S, Pettersen B, Maisenbacher MK, Hall MP, Demko Z, Rabinowitz M. 2014. Genomic imbalance in products of conception: single-nucleotide polymorphism chromosomal microarray analysis. Obstet Gynecol. 124(2, PART 1):202–209. doi:https://doi.org/10.1097/AOG.0000000000000325.
- Liu S, Song L, Cram DS, Xiong L, Wang K, Wu R, Yang F. 2015. Traditional karyotyping vs copy number variation sequencing for detection of chromosomal abnormalities associated with spontaneous miscarriage. Ultrasound Obstetrics Gynecology. 46(4):472–477. doi:https://doi.org/10.1002/uog.14849.
- Lomax B, Tang S, Separovic E, Phillips D, Hillard E, Thomson T, Kalousek DK. 2000. Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet. 66(5):1516–1521. doi:https://doi.org/10.1086/302878.
- Monaghan KG, Leach NT, Pekarek D, Prasad P, Rose NC. 2020. The use of fetal exome sequencing in prenatal diagnosis: a points to consider document of the American College of Medical Genetics and Genomics (ACMG). Genet Med. doi:https://doi.org/10.1038/s41436-019-0731-7.
- Nagan N, Faulkner NE, Curtis C, Schrijver I (2011). Laboratory guidelines for detection, interpretation, and reporting of maternal cell contamination in prenatal analyses: A report of the association for molecular pathology. J Mol Diagn. doi:https://doi.org/10.1016/j.jmoldx.2010.11.013
- Niinimäki M, Jouppila P, Martikainen H, Talvensaari-Mattila A. 2006. A randomized study comparing efficacy and patient satisfaction in medical or surgical treatment of miscarriage. Fertil Steril. 86(2):367–372. doi:https://doi.org/10.1016/j.fertnstert.2005.12.072.
- Nikitina TV, Lebedev IN, Sukhanova NN, Sazhenova EA, Nazarenko SA. 2005. A mathematical model for evaluation of maternal cell contamination in cultured cells from spontaneous abortions: significance for cytogenetic analysis of prenatal selection factors. Fertil Steril. 83(4):964–972. doi:https://doi.org/10.1016/j.fertnstert.2004.12.009.
- Robberecht C, Pexsters A, Deprest J, Fryns J-P, D’Hooghe T, Vermeesch JR. 2012. Cytogenetic and morphological analysis of early products of conception following hystero-embryoscopy from couples with recurrent pregnancy loss. Prenat Diagn. 32(10):933–942. doi:https://doi.org/10.1002/pd.3936.
- Rodgers CS, Creasy MR, Fitchett M, Maliszewska CT, Pratt NR, Waters JJ. 1996. Solid tissue culture for cytogenetic analysis: A collaborative survey for the association of clinical cytogeneticists. J Clin Pathol. 49(8):638–641. doi:https://doi.org/10.1136/jcp.49.8.638.
- Romero ST, Geiersbach KB, Paxton CN, Rose NC, Schisterman EF, Branch DW, Silver RM. 2015. Differentiation of genetic abnormalities in early pregnancy loss. Ultrasound Obstetrics Gynecology. doi:https://doi.org/10.1002/uog.14713.
- Russo R, Sessa AM, Fumo R, Gaeta S. 2016. Chromosomal anomalies in early spontaneous abortions: interphase FISH analysis on 855 FFPE first trimester abortions. Prenat Diagn. 36(2):186–191. doi:https://doi.org/10.1002/pd.4768.
- Sahoo T, Dzidic N, Strecker MN, Commander S, Travis MK, Doherty C, Hovanes K. 2017. Comprehensive genetic analysis of pregnancy loss by chromosomal microarrays: outcomes, benefits, and challenges. Genet Med. 19(1):83–89. doi:https://doi.org/10.1038/gim.2016.69.
- Saxena D, Agarwal M, Gupta D, Agrawal S, Das V, Phadke SR. 2016. Utility and limitations of multiplex ligation-dependent probe amplification technique in the detection of cytogenetic abnormalities in products of conception. J Postgrad Med. doi:https://doi.org/10.4103/0022-3859.192664.
- Schaeffer AJ, Chung J, Heretis K, Wong A, Ledbetter DH, Martin CL. 2004. Comparative genomic hybridization-array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am J Hum Genet. 74(6):1168–1174. doi:https://doi.org/10.1086/421250.
- Schlesinger C, Raabe G, Ngo T, Miller K. 1990. Discordant findings in chorionic villus direct preparation and long term culture—mosaicism in the fetus. Prenat Diagn. 10(9):609–612. doi:https://doi.org/10.1002/pd.1970100910.
- Schrijver I, Cherny SC, Zehnder JL. 2007. Testing for maternal cell contamination in prenatal samples: A comprehensive survey of current diagnostic practices in 35 molecular diagnostic laboratories. J Mol Diagn. doi:https://doi.org/10.2353/jmoldx.2007.070017.
- Scott SA, Cohen N, Brandt T, Toruner G, Desnick RJ, Edelmann L. 2010. Detection of low-level mosaicism and placental mosaicism by oligonucleotide array comparative genomic hybridization. Genet Med. 12(2):85–92. doi:https://doi.org/10.1097/GIM.0b013e3181cc75d0.
- Shearer BM, Thorland EC, Carlson AW, Jalal SM, Ketterling RP. 2011. Reflex fluorescent in situ hybridization testing for unsuccessful product of conception cultures: A retrospective analysis of 5555 samples attempted by conventional cytogenetics and fluorescent in situ hybridization. Genet Med. 13(6):545–552. doi:https://doi.org/10.1097/GIM.0b013e31820c685b.
- Shen J, Wu W, Gao C, Ochin H, Qu D, Xie J, … Liu J. 2016. Chromosomal copy number analysis on chorionic villus samples from early spontaneous miscarriages by high throughput genetic technology. Mol Cytogenet. doi:https://doi.org/10.1186/s13039-015-0210-z.
- Simoni G, Brambati B, Danesino C, Rossella F, Terzoli GL, Ferrari M, Fraccaro M. 1983. Efficient direct chromosome analyses and enzyme determinations from chorionic villi samples in the first trimester of pregnancy. Hum Genet. 63(4):349–357. doi:https://doi.org/10.1007/BF00274761.
- Volozonoka L, Perminov D, Korņejeva L, Alkšere B, Novikova N, Pīmane EJ, Fodina V. 2018. Performance comparison of two whole genome amplification techniques in frame of multifactor preimplantation genetic testing. J Assist Reprod Genet. 35(8):1457–1472. doi:https://doi.org/10.1007/s10815-018-1187-4.
- Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 13(1):134. doi:https://doi.org/10.1186/1471-2105-13-134.
- Zhang H, Liu W, Chen M, Li Z, Sun X, Wang C. 2016. Implementation of a high-resolution single-nucleotide polymorphism array in analyzing the products of conception. Genet Test Mol Biomarkers. 20(7):352–358. doi:https://doi.org/10.1089/gtmb.2016.0035.
- Zhang YX, Zhang YP, Gu Y, Guan FJ, Li SL, Xie JS, Zhong N. 2009. Genetic analysis of first-trimester miscarriages with a combination of cytogenetic karyotyping, microsatellite genotyping and arrayCGH. Clin Genet. 75(2):133–140. doi:https://doi.org/10.1111/j.1399-0004.2008.01131.x.
