Abstract
This study investigated the expression and clinical significance of long intergenic noncoding RNA 00665 (LINC00665) in ovarian cancer (OC), as well as its effect on the malignant biological behavior of OC cells. The expression of LINC00665, miR-148b-3p, and Krüppel-like factor 5 (KLF5) in OC tissues and cells were determined by RT-qPCR. Western blot was used to detect the protein expression of KLF5. The expression patterns of LINC00665 in nuclear and cytoplasm fractions were undertaken using RT-qPCR. In addition, CCK-8 assay, clone formation assay, transwell, scratch test, and flow cytometry were respectively used to detect the cell activity, proliferation, invasiveness, healing of cells, and apoptosis rate of OC cells. Furthermore, the interactions between LINC00665 and miR-148b-3p and between miR-148b-3p and KLF5 were verified by the luciferase reporter assay, and the correlations among these three genes were analyzed. LINC00665 expression was upregulated both in OC cell lines and tissues. Si-LINC00665 inhibited cell proliferation, invasion, and migration and induced apoptosis to a certain extent. The subcellular fraction assay revealed LINC00665 to be located mainly in the cytoplasm. miR-148b-3p was a target of LINC00665, and KLF5 was directly targeted by miR-148b-3p. Si-LINC00665 inhibited KLF5 expression, miR-148b-3p inhibitor promoted KLF5 expression, and si-KLF5 inhibited LINC00665 expression. Interestingly, the expression of LINC00665 was reversely associated with miR-148b-3p expression but positively correlated with KLF5. Furthermore, miR-148b-3p expression was negatively correlated with KLF5. In addition, si-KLF5 inhibited the malignant biological behavior of OC cells, whereas miR-148b-3p inhibitor had the opposite effect. Most importantly, the si-LINC00665 could reverse the promotion effect of the miR-148b-3p inhibitor on the malignant biological behavior of OC cells. LINC00665 can be used as an effective prognostic indicator of OC, which has the potential to be a new therapeutic target.
Introduction
Ovarian cancer (OC), one of the three malignancies of the female reproductive system, is a leading cause of death in women (Mezzanzanica Citation2015; Zhou Z et al. Citation2021). As the ovary is located deeper in the pelvic cavity, early detection of ovarian cancer is difficult, and often many females go to clinic with symptoms at an advanced stage. In addition, due to metastasis and its wide spread of cancer, the five-year survival rate remains in the range of 30–40%, and the mortality rate among the gynecological malignant tumors is highest (Webb and Jordan Citation2017). Therefore, exploring the pathogenesis of OC is of great significance.
It has been reported that long-non-coding RNAs (lncRNAs) can present with different expression patterns in cancer. The lncRNAs exert their activities through a variety of complex molecular mechanisms (Peng et al. Citation2017), such as affecting the epigenetic activity of tumor cells in the nucleus and gene transcription and post-transcriptional modification of mRNA (Jathar et al. Citation2017). It has been reported that long intergenic noncoding RNA 00665 (LINC00665) is upregulated in OC (Gao L et al. Citation2020). A report on the potential mechanism of lncRNAs in lymphocyte infiltration showed that LINC00665 positively correlates with lymphocyte infiltration in OC (Wu et al. Citation2020). Importantly, LINC00665 has been documented to be a key regulator of many malignancies. LINC00665 has been shown to promote breast cancer cell metastasis by triggering EMT (Zhou JL et al. Citation2020). Dysfunction of LINC00665 plays a critical role in the progression of non-small cell lung carcinoma, and its down-regulation can reduce cell proliferation and invasion (Wang et al. Citation2020). Furthermore, LINC00665 is up-regulated in osteosarcoma, and its high expression is associated with a poor prognosis of osteosarcoma (Zhang et al. Citation2020). Although LINC00665 plays an important role in cancer, its regulatory mechanism in the occurrence and development of OC remains unclear.
Numerous studies have shown that lncRNAs carry some miRNA “seed sequences” and bind microRNAs (miRNAs) like sponges, thus preventing miRNAs from binding to their target mRNA in some cancer cells and specific tissues (Qi et al. Citation2015). A recent study revealed some of the most predictive miRNAs markers in cancers, and described three to four biomarkers in each cancer type that showed highest correlations compared with the healthy controls. Among these, miR-29c-3p, miR-92a-3p, miR-101-3p, and miR-148b-3p had the highest correlation in OC (Tomeva et al. Citation2022). Furthermore, through bioinformatics, we found a potential binding relationship between LINC00665 and miR-148b-3p, a miRNA that plays a key role in disease progression. Earlier studies have shown miR-148b-3p to have an anticancer role in human cancers including, gastric carcinoma (Li X et al. Citation2018), pituitary adenoma (He et al. Citation2017), and breast carcinoma (Chen et al. Citation2018). Thus, we hypothesized that LINC00665 may participate in OC by binding to miR-148b-3p; however, this needs to be verified further. MiRNAs are known to bind to the 3′ untranslated region (3′ UTR) of target genes to control gene expression (Du and Zhang Citation2015). Using bioinformatics, we found that Krüppel-like factor 5 (KLF5) is a potential target gene of miR-148b-3p. A previous study showed that a regulatory crosstalk exists between miR-148b-3p and KLF5 in atherosclerosis (Jiang et al. Citation2021). KLF5, a member of the KLF family, is known to be highly expressed in a variety of cancers (Dong et al. Citation2013; Ma et al. Citation2018), and is considered a potential therapeutic target as well as a prognostic marker for epithelial OC (Siraj et al. Citation2020). Therefore, the LINC00665/miR-148b-3p/KLF5 axis may exist in OC that plays an important regulatory role in the development of OC.
This study illustrates the interaction of the LINC00665/miR-148b-3p/KLF5 axis in OC. In addition, we elucidated that the effects of LINC00665 on the biological function of OC cells are through regulating miR-148b-3p/KLF5. Hence, LINC00665 can possibly serve as a new biomarker toward molecular therapy of OC.
Results
LINC00665 was upregulated in OC tissues and cells
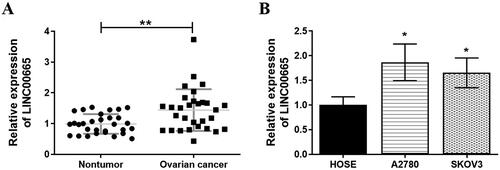
To clarify the expression of LINC00665 in OC, 30 pairs of OC tissue samples and normal para-tumor tissue samples were collected and analyzed. The RT-qPCR results showed that the expression of LINC00665 was significantly increased in OC tissues as compared with the normal para-tumor tissues (; p < 0.01). In addition, the analysis also showed that in comparison to the normal human ovarian surface epithelial (HOSE) cells, LINC00665 expression was significantly increased in OC cell lines (SKOV3 and A2780) (; p < 0.05).
Figure 1. LINC00665 was upregulated in ovarian cancer tissues and cells. (A) A total of 30 histologically verified ovarian cancer tissue and normal para-tumor tissue samples were collected. The RT-qPCR assay results showed that LINC00665 was significantly highly expressed in OC tissues. **p < 0.01 (paired t-tests), compared with the normal para-tumor tissue. (B) The RT-qPCR assay also showed that LINC00665 was significantly highly expressed in ovarian cancer cell lines (SKOV3 and A2780). *p < 0.05, **p < 0.01 (ANOVA with Dunnett's post hoc test) compared with the normal human ovarian surface epithelial cells (HOSE) group.
Si-LINC00665 inhibited the proliferation, invasion, migration and promoted the apoptosis of ovarian cancer cells
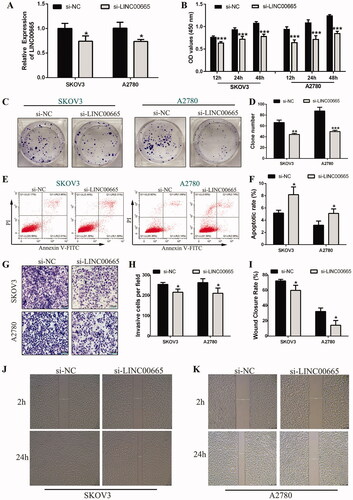
To investigate the biological roles of LINC00665 in OC cells, small interfering RNA (siRNA) was used to intervene LINC00665 in SKOV3 and A2780 cells for subsequent cell functional tests. As shown in , the expression of LINC00665 was significantly decreased after si-LINC00665 was transfected into SKOV3 and A2780 cells (p < 0.05). The effects of si-LINC00665 on the function of SKOV3 and A2780 cells were further analyzed. The CCK-8 assay showed that the proliferative abilities of SKOV3 and A2780 cells in the si-LINC00665 group were both significantly reduced at 12, 24, and 48 h compared with the siRNA negative control (si-NC) group (; p < 0.001). The colony formation assay also showed that the number of clones of SKOV3 and A2780 cells in the si-LINC00665 group was significantly reduced (; p < 0.01). Flow cytometry analysis revealed that the apoptotic rate of SKOV3 and A2780 cells in the si-LINC00665 group was significantly increased as compared with the si-NC group (; p < 0.05). The transwell invasion assay illustrated that in comparison to the si-NC group, the invasive ability of SKOV3 and A2780 cells in the si-LINC00665 group was significantly inhibited (; p < 0.05). Furthermore, the wound-healing assay revealed that the migration ability of SKOV3 and A2780 cells in the si-LINC00665 group was also significantly inhibited (; p < 0.05). Thus, si-LINC00665 could inhibit the proliferation, invasion, and migration of OC cells and promoted the apoptosis of OC cells.
Figure 2. Si-LINC00665 inhibited the proliferation, invasion, migration, and promoted the apoptosis of ovarian cancer cells. (A) The gene expression of LINC00665 was inhibited by LINC00665 small interfering RNA (siRNA) in SKOV3 and A2780 cells, and the interference effect was evaluated by RT-qPCR analysis. (B) Cell counting kit-8 assay revealed that the proliferative abilities of SKOV3 and A2780 cells transfected with si-LINC00665 were significantly decreased at 12, 24, and 48 h. (C,D) Colony formation assay also revealed that the colony numbers of SKOV3 and A2780 cells transfected with si-LINC00665 were significantly reduced. (E,F) Flow cytometry analysis revealed apoptotic rates in the si-LINC00665 transfected SKOV3 and A2780 cells were significantly increased. (G,H) Transwell invasive assay illustrated that the invasive abilities in the si-LINC00665 transfected SKOV3 and A2780 cells were significantly decreased. (I,K) Wound-healing assay showed the wound areas at 24 h after wounding in the si-LINC00665 transfected SKOV3 and A2780 cells were significantly decreased. *p < 0.05, **p < 0.01, ***p < 0.001 (unpaired t‑test) compared with the si-NC group.
miR-148b-3p as a target miRNA of LINC00665
The subcellular localization of lncRNA is one of the main determinants of its molecular function, and cytoplasmic lncRNA can serve as competing endogenous RNAs (ceRNA) (Carlevaro-Fita and Johnson Citation2019). To study the regulatory mechanism of LINC00665 in OC, the subcellular location of LINC00665 in SKOV3 and A2780 cells was detected by subcellular RNA fractions analysis. The results showed that LINC00665 was mainly distributed in the cytoplasm of SKOV3 and A2780 cells (), implying that LINC00665 could possibly serve as a “sponge” for miRNAs. Using the online software StarBase (http://starbase.sysu.edu.cn), we searched for miRNAs with base pairs complementary to the LINC00665 sequence. It was noted that LINC00665 can potentially have a binding relationship with miR-148b-3p, and the presumed binding site between LINC00665 and miR-148b-3p is shown in . To further clarify the interaction between LINC00665 and miR-148b-3p, luciferase gene reporter assay was performed which confirmed that the luciferase activity decreased when the LINC00665 wild sequence (LINC00665-WT) was co-transfected with miR-148b-3p mimic (; p < 0.001). However, there was no change in luciferase activity when LINC00665 mutant (LINC00665-MUT) was co-transfected with miR-148b-3p mimic (; p > 0.05). This suggested that miR-148b-3p can directly interact with LINC00665 through the putative binding site. Further analysis of the miR-148b-3p expression in OC tissues and cells showed that miR-148b-3p in OC tissues was significantly decreased compared with non-tumor tissues (; p < 0.05), and that the expression was also significantly decreased compared with the HOSE cells (; p < 0.05). In addition, Pearson correlation analysis showed that the level of miR-148b-3p was negatively correlated with LINC00665 (; p < 0.01), and si-LINC00665 significantly increased the expression of miR-148b-3p in SKOV3 and A2780 cells (; p < 0.05). In addition, the RT-qPCR results revealed that the LINC00665 level in the miR-148b-3p mimic group was significantly downregulated compared with the si-NC group (p < 0.05), whereas in the miR-148b-3p inhibitor group the level was significantly upregulated compared with the NC-inhibitor group (; p < 0.05). These results indicated that LINC00665 contains a binding site of miR-148b-3p, which could possibly inhibit the expression of miR-148b-3p, and that miR-148b-3p could also inhibit the expression of LINC00665 through a feedback loop. Thus, LINC00665 could act as a “sponge” for miR-148b-3p.
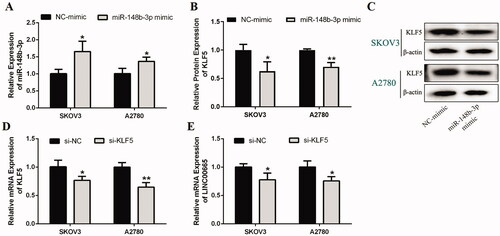
Figure 3. miR-148b-3p was a target miRNA of LINC00665. (A,B) The subcellular of RNA fractions analysis revealed the location of LINC00665 in the cytoplasm of SKOV3 or A2780 cells. (C) The putative binding site between LINC00665 and miR-148b-3p. (D) Luciferase gene reporter assay showed the activity of luciferase reporter gene decreased when co-transfected with the LINC00665 wild sequences and miR-148b-3p mimic. ***p < 0.001 (unpaired t‑test), compared with the NC-mimic group. (E) A total of 30 histologically verified ovarian cancer tissue and normal para-tumor tissue samples were collected. The RT-qPCR assay revealed the low expression of miR-148b-3p in ovarian cancer tissues. *p < 0.05 (paired t-tests), compared with the normal para-tumor tissue. (F) The RT-qPCR assay also showed low expression of miR-148b-3p in ovarian cancer cell lines (SKOV3 and A2780). *p < 0.05 (ANOVA with Dunnett's post hoc test), compared with the normal human ovarian surface epithelial cells (HOSE) group. (G) Pearson correlation analysis showed that the miR-148b-3p level was significantly negatively correlated with the LINC00665 level. (H) The RT-qPCR assay revealed that si-LINC00665 significantly increased the expression of miR-148b-3p in SKOV3 and A2780 cells. *p < 0.05 (unpaired t‑test), compared with the si-NC group. (I) The RT-qPCR assay displayed the mRNA levels of LINC00665 under miR-148b-3p mimic or inhibitor treatment in SKOV3 and A2780 cells. *p < 0.05, **p < 0.01 (unpaired t‑test), compared with the NC-mimic group; #p < 0.05 (unpaired t‑test), compared with the NC-inhibitor group. (J) The RT-qPCR assay revealed that si-LINC00665 significantly decrease the expression of KLF5 in SKOV3 and A2780 cells. *p < 0.05 (unpaired t‑test), compared with the si-NC group. (K,L) Western blot analyses displayed the protein levels of KLF5 under si-LINC00665 and miR-148b-3p inhibitor treated SKOV3 and A2780 cells. **p < 0.01, ***p < 0.001 (unpaired t‑test), compared with the si-NC group; #p < 0.05 (unpaired t-test), compared with the si-LINC00665 group. KLF5: Krüppel-like factor 5.
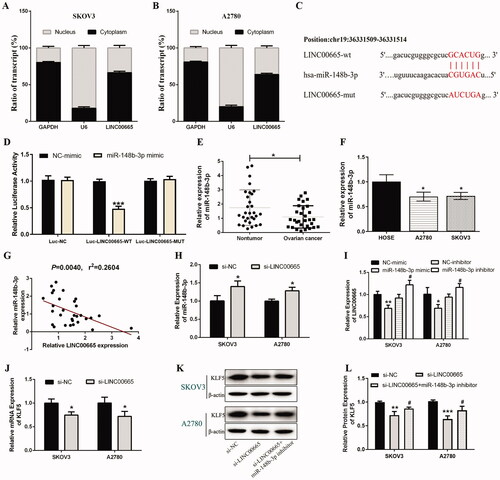
Furthermore, the mRNA expression level of KLF5 was significantly reduced in the si-LINC00665 transfected SKOV3 and A2780 cells compared with the si-NC group (; p < 0.05). Next, the effect of miR-148b-3p on KLF5 expression in SKOV3 and A2780 cells was analyzed, and the results revealed that the protein expression of KLF5 in the si-LINC00665 group was significantly lower than that in the si-NC group (p < 0.05), and the decreased KLF5 expression was significantly upregulated after si-LINC00665 and miR-148b-3p inhibitor co-transfection (; p < 0.05). These results suggested that KLF5 in OC can be regulated by LINC00665 and miR-148b-3p, which may be a downstream target gene of miR-148b-3p.
KLF5 was a direct target of miR-148b-3p
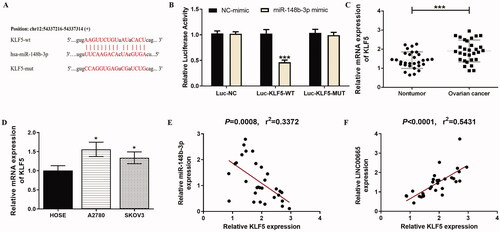
As non-coding regulatory small molecules, miRNAs exert their effects by inhibiting the expression of target genes. The relationship between miR-148b-3p and KLF5 was predicted by TargetScan (http://www.targetscan.org/vert_71/), and it was found that there was a binding site between the 3′‑UTR of KLF5 and miR-148b-3p due to the complementary base pairing (). Furthermore, the luciferase reporter assay showed that the relative luciferase activity of the group co-transfected with KLF5 wild sequence (KLF5-WT) and miR-148b-3p mimic was significantly lower than that of the group co-transfected with KLF5-WT and NC-mimic (; p < 0.001). However, when the KLF5 mutant sequence (KLF5-MUT) was co-transfected with miR-148b-3p mimic, the luciferase activity remained unaltered compared with the group co-transfected with KLF5-MUT and NC-mimic (; p > 0.05). Analysis of the expression of KLF5 in OC tissues and cells showed that the expression of KLF5 in OC tissues was significantly higher than non-tumor tissues (; p < 0.001), and the expression of KLF5 in all OC cell lines was also significantly increased compared with the HOSE cells (; p < 0.05). Besides, the KLF5 level was found to be negatively correlated with the miR-148b-3p level (; p < 0.001), and be positively correlated with the LINC00665 level (; p < 0.001). Therefore, these results suggested that KLF5 was a direct target of miR-148b-3p.
Figure 4. KLF5 was directly targeted by miR-148b-3p. (A) The putative binding site between KLF5 3′-UTR and miR-148b-3p. (B) Luciferase gene reporter assay showed that the luciferase activity increased when co-transfected with KLF5 wild sequences and miR-148b-3p mimic. ***p < 0.001 (unpaired t‑test), compared with the NC-mimic group. (C) A total of 30 histologically verified ovarian cancer tissue and normal para-tumor tissue samples were collected. The RT-qPCR assay showed that KLF5 was highly expressed in OC tissues. ***p < 0.001 (paired t-tests), compared with the normal para-tumor tissue. (D) The RT-qPCR assay also showed high expression of KLF5 in ovarian cancer cell lines (SKOV3 and A2780). *p < 0.05 (ANOVA with Dunnett's post hoc test), compared with the normal human ovarian surface epithelial cells (HOSE) group. (E) Pearson correlation analysis showed that the miR-148b-3p level was significantly negatively correlated with KLF5. (F) Pearson correlation analysis showed that the LINC00665 level was significantly positively correlated with KLF5. KLF5: Krüppel-like factor 5.
Regulatory effects of miR-148b-3p on KLF5, and KLF5 on LINC00665
To further confirm the regulatory relationship between miR-148b-3p and KLF5 in OC, miR-148b-3p mimic was used to upregulate the expression of miR-148b-3p in OC cells. As shown in , the expression of miR-148b-3p in SKOV3 and A2780 cells was significantly increased after transfection with miR-148b-3p mimic (p < 0.05), indicating that the transfection was successful. Protein expression analysis of KLF5 showed that the expression of KLF5 in both SKOV3 and A2780 cells was significantly decreased after miR-148b-3p mimic transfection (; p < 0.05), suggesting that miR-148b-3p directly targets KLF5, and then negatively regulates the expression of KLF5. In addition, KLF5 expression was significantly downregulated when si-KLF5 was transfected into SKOV3 and A2780 cells (; p < 0.05), and the LINC00665 expression was also significantly inhibited by si-KLF5 (; p < 0.05). The collective results confirmed that miR-148b-3p negatively regulated KLF5, whereas KLF5 could upregulate LINC00665 in a kind of feedback loop, and that there was a LINC00665/miR-148b-3p/KLF5 axis in OC.
Figure 5. Regulatory effects of miR-148b-3p on KLF5 and KLF5 on LINC00665. (A) The expression of miR-148b-3p was upregulated by miR-148b-3p mimic in SKOV3 and A2780 cells as detected by RT-qPCR. (B,C) Western blot assay revealed that miR-148b-3p mimic significantly decreased the expression of KLF5 in SKOV3 and A2780 cells. *p < 0.05, **p < 0.01 (unpaired t‑test), compared with the NC mimic group. (D) The gene expression of KLF5 was inhibited by si-KLF5 in SKOV3 and A2780 cells, and the interference effect was evaluated by RT-qPCR analysis. *p < 0.05, **p < 0.01 (unpaired t‑test), compared with the si-NC group. (E) The RT-qPCR assay revealed that si-KLF5 significantly decreased the expression of LINC00665 in SKOV3 and A2780 cells. *p < 0.05 (unpaired t‑test), compared with the si-NC group. KLF5: Krüppel-like factor 5.
LINC00665/miR-148b-3p/KLF5 axis regulated the malignant biological behavior of OC cells
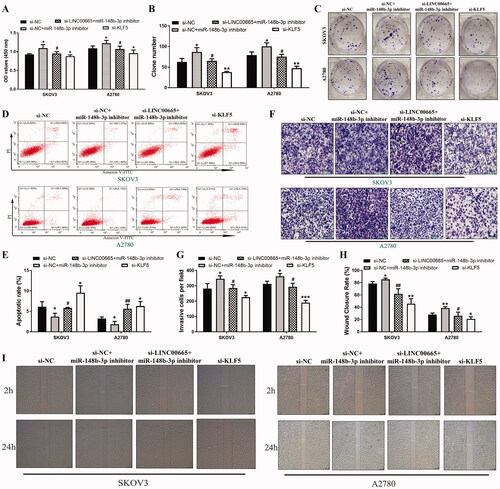
To determine the regulating effect of LINC00665/miR-148b-3p/KLF5 axis on malignant biological behavior of OC cells, miR-148b-3p inhibitor, si-LINC00665 + miR-148b-3p inhibitor, or si-KLF5 were transfected into SKOV3 and A2780 cell lines. In both the SKOV3 and A2780 cells, the CCK-8 () and colony formation assays () revealed that as compared with the si-NC group, the proliferative ability of OC cells in the si-NC + miR-148b-3p inhibitor group was significantly increased, whereas the proliferation in the si-KLF5 group was significantly decreased (p < 0.05). Furthermore, the proliferative ability of OC cells in the si-LINC00665 + miR-148b-3p inhibitor group was significantly reduced compared with the si-NC + miR-148b-3p inhibitor group (p < 0.05). Flow cytometry analysis () showed that compared with the si-NC group, the apoptotic rate of cells in the si-NC + miR-148b-3p inhibitor group was significantly decreased, and that in the si-KLF5 group, the rate was significantly increased (p < 0.05). Compared with the si-NC + miR-148b-3p inhibitor group, the apoptotic rate in the si-LINC00665 + miR-148b-3p inhibitor group was significantly increased (p < 0.05). In addition, the transwell invasive assay () and the scratch test () illustrated that as compared with the si-NC group, the invasive and migration ability of cells in the si-NC + miR-148b-3p inhibitor group was significantly increased, and that in the si-KLF5 group, the activity was significantly decreased (p < 0.05). In addition, the invasive and migration ability of cells in the si-LINC00665 + miR-148b-3p inhibitor group was significantly reduced compared with the si-NC + miR-148b-3p inhibitor group (p < 0.05). Taken together, the results confirmed that LINC00665/miR-148b-3p/KLF5 axis regulates OC cell proliferation, apoptosis, migration, and invasion.
Figure 6. Regulating effect of LINC00665/miR-148b-3p/KLF5 axis on the malignant biological behavior of ovarian cancer cells. Si-NC, si-NC + miR-148b-3p inhibitor, si-LINC00665 + miR-148b-3p inhibitor, or si-KLF5 were transfected into SKOV3 and A2780 cell lines, respectively. (A) The proliferative abilities of SKOV3 and A2780 cells were detected by cell counting kit-8 assay. (B,C) The colony numbers of SKOV3 and A2780 cells were detected by colony formation assay. (D,E) The apoptotic rates of SKOV3 and A2780 cells were detected by flow cytometry. (F,G) The invasive abilities of SKOV3 and A2780 cells were detected by transwell invasive assay. (H,I) The wound-healing assay showed the wound areas at 2 and 24 h after wounding in SKOV3 and A2780 cells. *p < 0.05, **p < 0.01, ***p < 0.001 (ANOVA with Dunnett's post hoc test), compared with the si-NC group; #p < 0.05, ##p < 0.01 (ANOVA with Dunnett's post hoc test), compared with the si-NC + miR-148b-3p inhibitor group. KLF5: krüppel-like factor 5.
Discussion
Targeted therapy is a new approach for the treatment of malignant tumors that can inhibit the proliferation of cancer cells through specific factors or even directly kill the cancer cells. Therefore, it is essential to understand the molecular mechanisms underlying the occurrence and development of OC for the molecular targeted therapy (Rehmani et al. Citation2020). As a new regulatory factor, lncRNA is currently a an emerging research topic in the field of oncology, and its biological functions are mainly known to be exhibited through the regulation of gene activation or expression inhibition (Geisler and Coller Citation2013; Wahlestedt Citation2013). In this study, we found that LINC00665 expression was upregulated in OC cell lines and tissues. Elevated expression of LINC00665 in OC cells indicates that LINC00665 may trigger a series of pathophysiological processes as an oncogene. Our results showed that the knockdown of LINC00665 inhibited the proliferation, invasion, and migration of ovarian cancer cells, and promoted the apoptosis of ovarian cancer cells to a certain extent.
Previously, multiple lncRNAs have been reported to regulate various biological processes in ovarian cancer cells (Li T et al. Citation2018; Wang J et al. Citation2018; Sun et al. Citation2019). For instance, lncRNA MEG3 regulates the proliferation, invasion, and migration of ovarian cancer cells via regulating PTEN (Wang J et al. Citation2018). Similarly, LncRNA TP73-AS1 promotes proliferation and metastasis of ovarian cancer cells by regulating the expression of MMP2 and MMP9 (Wang X et al. Citation2018). As a poorly researched lncRNA, LINC00665 has been identified as an oncogene associated with human diseases in a variety of cancer cells, such as breast (Zhou JL et al. Citation2020), non-small cell lung (Wang et al. Citation2020), osteosarcoma (Zhang et al. Citation2020), and prostate cancer (Chen et al. Citation2020). The findings of our study are consistent with the previous reports.
Regulatory networks of non-coding RNAs are involved in the development of many types of tumors (Yuan et al. Citation2018). MiRNAs, as a kind of non-coding RNAs, regulate gene expression following transcription. Numerous experimental studies have determined that miRNAs dysregulation has a profound impact on the development and progression of human cancers (Peng and Croce Citation2016). They control a wide range of biological processes, including oncogenic effects activated by transcription of oncogenes (Esquela-Kerscher and Slack Citation2006). Multiple biological processes such as cell differentiation, proliferation, and apoptosis can also be regulated through its feedback mechanisms by acting on multiple target RNAs (Hill and Tran Citation2021). In addition, studies have shown that lncRNAs and other transcriptional substances can bind to miRNAs through miRNA recognition elements and play the role of ceRNA (Karreth and Pandolfi Citation2013). For instance, studies have reported that LncRNA HOTAIR is a ceRNA in gastric cancer cells through modulating miR-331-3p (Liu et al. Citation2014). As another example, it has been documented that LncRNA EBLN3P may act as a ceRNA to modulate DOCK4 expression by competitively sponging miR-144-3p (Li et al. Citation2020). These findings stimulated the concept that LINC00665 might function as the ‘sponge’ of miRNAs. Therefore, we analyzed the potential targets of LINC00665 through bioinformatics and found that LINC00665 had binding sites with mir-148b-3p. The luciferase gene reporter assay confirmed that miR-148b-3p could directly bind to LINC00665 through its own miRNA binding site. In addition, the expression of mir-148b-3p was negatively correlated with LINC00665. This is because LINC00665 could inhibit the expression of mir-148b-3p, and likewise mir-148b-3p could negatively regulate LINC00665. This confirmed our hypothesis that LINC00665 could act as the "sponge" of miRNA. Further studies showed that LINC00665 acted as a ceRNA to drive proliferation, invasion, migration, and apoptosis of ovarian cancer cells via the spongization of miR-148b-3p. Moreover, we identified the down-expression level of miR-148b-3p in OC. It is worth mentioning that mir-148b-3p has been previously reported to inhibit the development of a variety of cancers (He et al. Citation2017; Chen et al. Citation2018; Li X et al. Citation2018). Thus, our results are in line with these findings.
In this study, we found that LINC00665 was mainly localized in the cytoplasm, suggesting its post-transcriptional regulation. Subsequently, we further verified that KLF5 mRNA and the resulting proteins were targets of miR-148b-3p, which was consistent with a previous study (Yu L et al. Citation2018). We then constructed the LINC00665/miR-148b-3p/KLF5 axis. KLF5 is one of the most widely studied member of the KLFs family. It belongs to the family of DNA-binding transcriptional regulators and has various biological functions. In recent years, studies have found that KLF5 is closely related to the occurrence and development of tumors and that it is abnormally expressed in different types of tumors wherein it plays an important role (Yu L et al. Citation2018; Jia et al. Citation2019). In this study, we also found that a significant up-regulation of KLF5 was shown in OC. Interestingly, a negative correlation between LINC00665 and miR-148b-3p expression, a positive correlation between LINC00665 and KLF5 expression, and a negative correlation between miR-148b-3p and KLF5 expression were found in ovarian cancer tissues. Thus, we further confirmed the existence of the LINC00665/miR-148b-3p/KLF5 axis. KLF5 expression is often abnormal in human cancers, and there is growing evidence that KLF5 represents a novel therapeutic target for cancer treatment (Gao Y et al. Citation2015). Our findings are further supported by the fact that KLF5 is an important transcription factor involved in the survival, migration, invasion, and apoptosis of osteosarcoma cells (Zhang et al. Citation2019). Importantly, we found that down-regulation of LINC00665 inhibited the expression of KLF5 by promoting miR-148b-3p in OC cells, thus inhibiting the malignant biological behavior of OC.
As discussed above, our data suggested that LINC00665, by sponging miR-148b-3p/KLF5 axis, affected cell proliferation, invasion, and migration and induced apoptosis to a certain extent, suggesting an important regulation of LINC00665 on the OC tumorigenesis. LINC00665 can be used as an effective prognostic indicator of OC. By elucidating the varied mechanisms of LINC00665 in the malignant biological behavior of OC, we demonstrated that LINC00665 has the potential to be a new therapeutic target.
Materials and methods
Patients and tissue samples
Thirty pairs of OC tissue samples and normal para-tumor tissue samples were obtained from patients diagnosed with OC in the Fifth People's Hospital of Qinghai Province. The diagnosis, staging, and risk status of OC were determined according to the regulations of the International Federation of Obstetrics and Gynecology. We excluded patients with other malignancies and those who had received preoperative chemoradiotherapy. After surgical removal, tissues were collected and stored in liquid nitrogen for further investigations. This study was approved by the Ethics Committee of The Fifth People's Hospital of Qinghai Province.
Cell culture
SKOV3 is a cell line with epithelial morphology that was isolated from the ovary of a 64-year-old, white, female with ovarian adenocarcinoma, and A2780 ovarian cancer cell line was established from an ovarian endometroid adenocarcinoma tumor in an untreated patient. The present study was conducted using these two OC cell lines. The human OC cell lines SKOV3 (BNCC310551), A2780 (BNCC351906), and normal human ovarian surface epithelial cells (HOSE, BNCC340096) were purchased from BeNa Culture Collection (Beijing, China). The cell lines were cultured in RPMI-1640 (HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS, HyClone) and incubated at 37°C in a 5% CO2 atmosphere.
Transfection of ovarian cancer cells
Small interfering RNA (siRNA) targeting LINC00665 (si-LINC00665) and siRNA targeting KLF5 (si-KLF5) were synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). The siRNA sequences against LINC00665 and KLF5 used in the study are as follows: si-LINC00665: 5′-UCCUCAGUCUUGGGCUAUU-3′; si-KLF5: 5′-AAGCUCACCUGAGGACUCA-3′. The miR-148b-3p mimic (cat no. miR10000759-1-5), NC mimic (cat no. miR1N0000001-1-5), miR-148b-3p inhibitor (cat no. miR20000759-1-5), and NC inhibitor (cat no. miR2N0000001-1-5) were purchased from RiboBio Co., Ltd (Guangzhou, China). OC cell lines SKOV3 and A2780 were transfected with si-LINC00665, si-KLF5, si-NC, miR-148b-3p mimic, NC-mimic, miR-148b-3p inhibitor, and NC inhibitor, respectively, using Lipofectamine™2000 (Invitrogen, USA) as per the manufacturer’s instructions. First, the cells were seeded into a 6-well plate with a density of 1 × 105 cells per well and cultured at 37°C overnight. Then, 100 pmol siRNA, miRNA mimic, or miRNA inhibitor were transfected into the cells with Lipofectamine 2000 Reagent in 2 mL medium free of penicillin-streptomycin and serum for 6 h. The transfection medium was then replaced by a complete RPMI-1640 medium and incubated for another 42 h. After transfection for 48 h, cells were harvested for subsequent experiments.
RT-qPCR analysis
Total RNA was extracted by conventional RNA extraction according to the TRIzol Reagent (cat no. 15596026, Invitrogen), and cDNA was obtained using the HiScript® III RT SuperMix for qPCR (+gDNA wiper) (cat no. R323-01, Vazyme, China) by incubating at 42°C for 2 min, next at 37°C for 15 min and then at 85°C for 5 s. RT-qPCR was carried out on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad; USA) through the HiScript® II One Step RT-PCR Kit (cat no. P611-01, Vazyme) at a final volume of 20 µL using the standard protocol, which was as follows: 50°C for 30 min, 94°C for 3 min; 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min; followed by 72°C for 5 min. For the quantification of miRNA level, RT-PCR was executed by the Bulge‑Loop™ miRNA RT‑qPCR Primer and Bulge‑Loop™ miRNA RT‑qPCR Starter kit (cat. no. C10211‑1; Guangzhou RiboBio Co., Ltd). The gene expression was analyzed at 95°C for 10 min; 40 cycles of 95°C for 2 s, 60°C for 20 s, and 70°C for 10 s. Data analysis was made up of the 2−ΔΔCt method with GAPDH or U6 small nuclear RNA genes as the endogenous controls. The primer sequences used in this study were as follows: LINC00665 Forward: 5′-TAAGCAGAGGGCTACAGAA-3′, Reverse: 5′-CAGGGAGATCAGGAGGAC-3′; miR-148b-3p Forward: 5′-TCTCTCCAGTCTACTCAGTGCATC-3′; Reverse: 5′-TATGGTTTTGACGACTGTGTGAT-3′; KLF5 Forward: 5′-ACACCAGACCGCAGCTCCA-3′, Reverse: 5′-TCCATTGCTGCTGTCTGATTTGTAG-3′; GAPDH Forward: 5′-TTGCCATCAATGACCCCTTCA-3′, Reverse: 5′-CGCCCCACTTGATTTTGGA-3′; U6 Forward: 5′-GCTTCGGCAGCACATATACTAAAAT-3′, Reverse: 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
CCK-8 assay
The CCK-8 assay was performed to detect the proliferation of SKOV3 and A2780 cells. The specific determination method was as follows: Cells with different treatment methods were seeded into a 96-well plate with a density of 5 × 103 per well for 24 h, and then 10 µl CCK8 reagent (cat no. CA1210, Beijing Solarbio Science & Technology Co., Ltd, China) was added to each well and the cells were further incubated for 1 h at 37°C. In total, six replicates were repeated for each treatment group, and the absorbance value at 450 nm was determined using a microplate microscope (Bio-Rad).
Colony formation assay
Following transfection of SKOV3 and A2780 cells for 48 h, cells of each group were seeded in a 60 mm cell culture dish with 1 × 103 cells. After 14 days, the cells were fixed with 4% paraformaldehyde (Sigma-Aldrich; USA), stained with 0.1% crystal violet solution (Sigma-Aldrich), and colonies containing ≥30 cells were counted. Representative colonies were photographed and counted with Olympus inverted fluorescence microscope (Tokyo, Japan).
Flow cytometry assay
SKOV3 and A2780 cells with different treatments were washed with PBS (Sigma-Aldrich), centrifuged at 300×g for 5 min, and resuspended in 100 µL PBS. Subsequently, 5 µl Annexin V-FITC (cat. no. KGA107; Nanjing KeyGen Biotech Co., Ltd, China) and 5 µl Propidium lodide (PI, cat. no. KGA107; Nanjing KeyGen Biotech Co., Ltd, China) were added and incubated at room temperature in dark for 15 min. Following incubation, 400 µl binding buffer was added, and the mixture was placed on ice for 1 h for flow cytometry. The apoptotic cells were measured by BD Caliber flow cytometry (Becton Dickinson, Franklin lakes, USA).
Transwell invasion assay
For the transwell invasion assay, 50 µl matrix glue was added to the transwell chamber and allowed to solidify at 37°C for 3 h. And a total of 100 µL FBS-free medium was added to the upper chamber and the complete medium was added to the lower chamber. About 1 × 105 SKOV3 and A2780 cells from each group were seeded in the upper chamber of the transwell chamber and cultured for 48 h. Cells that did not cross the membrane were erased. To fix the residual cells, paraformaldehyde was added for 15 min, and then stained using 0.1% crystal violet (Sigma-Aldrich) for 5 min. The crystal violet that did not bind to the cells was washed off with PBS. After proper air drying, five random fields per well were selected under a microscope (Nikon Corporation, Tokyo, Japan) for observation and counting the number of cells. Assays were independently repeated three times.
Wound-healing assay
In the migration experiment, a maker pen was first used to draw a line evenly behind a 6-well plate, with at least five lines passing through each well. SKOV3 and A2780 cells were inoculated into the 6-well plates at a density of 5 × 105 cells per well. When the cells covered 80% of the plates, they were scratched perpendicular to the back horizontal line with the medium lance. Thereafter the cells were washed with PBS, and cultured in a humidified incubator at 37°C and 5% CO2 for 2 h and 24 h, and the scratch healing was observed and photographed under a microscope (Nikon Corporation). Image J software (National Institutes of Health, Bethesda, MD, USA) was then used to measure the distance between the scratches and analyze the migration distance of cells.
Nuclear and cytoplasmic RNA fraction isolation
Adherent SKOV3 and A2780 cells were placed in an ice bath, and cell lysis buffer was added for 10 min. The entire experimental process was carried out according to the Ambion PARISTM kit (cat no. AM1921, Invitrogen) instructions provided by the manufacturer. The lysate was collected and centrifuged at 3000 r/min for 10 min (centrifugation radius was 10 cm). The supernatant was the cytoplasm and the precipitation part was mainly the nucleus. The supernatant was placed in the ice bath. The precipitate was further lysed and also placed in an ice bath. Next, RNA was extracted and RT-qPCR performed.
Luciferase reporter assay
LINC00665 (wild-type and mutant) and KLF5 3′-UTR (wild-type and mutant) were constructed into pGL3-basic Luciferase reporter vectors (Promega, Madison, WI, USA) to generate wildtype reporters (LINC00665-WT and KLF5-WT) or mutant reporters (LINC00665-MUT and KLF5-MUT) as the luciferase reporter plasmids. Cells were inoculated into 24‑well plates at a density of 5 × 104 cells per well and cultured at 37°C. On reaching about 70% confluency, cells were co‑transfected with luciferase reporter plasmids (LINC00665-WT/LINC00665-MUT or KLF5-WT/KLF5-MUT), miR-148b-3p mimic/NC mimic using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, USA) according to the operating instructions. Luciferase activity of transfected cells was assayed after 48 h of transfection using a dual-Luciferase reporter assay system (cat. no. E1910; Promega Corporation).
Western blot
SKOV3 and A2780 cells were collected as described and homogenized using RIPA lysis buffer (cat. no. P0013B; Biotime Biotechnology, China), and the released proteins quantified with BCA protein assay kit (cat. no. P0010; Beyotime Biotechnology, China). Using a sample amount of 30 µg per well, the proteins were separated by 10% SDS-PAGE and then transferred to the PVDF membrane (EMD Millipore, Boston, USA) by semi-dry transfer method and blocked with 5% milk in TBST at room temperature for 2 h. Then rabbit anti-KLF5 (ab137676; 1:1,000; Abcam, UK) was added and incubated overnight at 4°C. Anti-β-actin (ab8227; 1:4,000; Abcam) was used as an inner loading control. After being rinsed with TBST, the HRP‑conjugated second antibody (ab205718, 1: 10,000, Abcam) was added and incubated with the membrane for 2 h at room temperature. The protein expression bands on the membrane were detected by the ECL color developing solution (Bio-Rad) and a ChemiDoc™ MP imaging system (Bio-Rad).
Statistical analysis
All data were analyzed by SPSS19.0 statistical software (SPSS Inc. Chicago, IL, USA). Gene expression in OC tissue and normal para-tumor tissue samples was analyzed by paired t-test. The differences between the two groups were determined by unpaired Student's t‑test, and the differences among the multiple groups were analyzed using one‑way analysis of variance (ANOVA) with Dunnett's post hoc test. Pearson correlation analysis was used to analyze the correlation between the expressions of LINC00665, miR-148b-3p, and KLF5. Prism 6.0 software (GraphPad, Inc., La Jolla, CA, USA) was used for analysis. Data were expressed as mean ± standard deviation. p < 0.05 was considered statistically significant in the differences.
Ethics approval
This clinical trial study has been approved by the Ethics Committee of The Fifth People's Hospital of Qinghai Province and registered in China Clinical Trial Center (no. 2020427 A). All patients signed informed consent before treatment.
Authors’ contributions
Conceived and designed the experiments: SW, CL, YL, YX; performed the experiments: SW, CL, JQ, XC; analyzed the data and contributed to the acquisition of reagents and materials: JB, RL; wrote the manuscript: SW, YX.
| Abbreviations | ||
| OC | = | ovarian cancer |
| lncRNAs | = | long-non-coding RNAs |
| KLF5 | = | krüppel-like factor 5 |
| siRNA | = | small interfering RNA |
| HOSE | = | normal human ovarian surface epithelial cells |
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analyzed in this study are included in this published article. All data used or analyzed during the current study are available from the corresponding author on reasonable request. The datasets supporting the conclusions of this article are included in the article.
Additional information
Funding
Reference
- Carlevaro-Fita J, Johnson R. 2019. Global positioning system: understanding long noncoding RNAs through subcellular localization. Mol Cell. 73(5):869–883.
- Chen W, Yu Z, Huang W, Yang Y, Wang F, Huang H. 2020. LncRNA LINC00665 promotes prostate cancer progression via miR-1224-5p/SND1 Axis. Onco Targets Ther. 13:2527–2535.
- Chen X, Wang YW, Gao P. 2018. SPIN1, negatively regulated by miR-148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res. 37(1):100.
- Dong Z, Yang L, Lai D. 2013. KLF5 strengthens drug resistance of ovarian cancer stem-like cells by regulating survivin expression. Cell Prolif. 46(4):425–435.
- Du J, Zhang L. 2015. Integrated analysis of DNA methylation and microRNA regulation of the lung adenocarcinoma transcriptome. Oncol Rep. 34(2):585–594.
- Esquela-Kerscher A, Slack FJ. 2006. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer. 6(4):259–269.
- Gao L, Li X, Nie X, Guo Q, Liu Q, Qi Y, Liu J, Lin B. 2020. Construction of novel mRNA-miRNA-lncRNA regulatory networks associated with prognosis of ovarian cancer. J Cancer. 11(23):7057–7072.
- Gao Y, Ding Y, Chen H, Chen H, Zhou J. 2015. Targeting Krüppel-like factor 5 (KLF5) for cancer therapy. Curr Top Med Chem. 15(8):699–713.
- Geisler S, Coller J. 2013. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 14(11):699–712.
- He W, Huang L, Li M, Yang Y, Chen Z, Shen X. 2017. MiR-148b, MiR-152/ALCAM axis regulates the proliferation and invasion of pituitary adenomas cells. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol. 792.
- Hill M, Tran N. 2021. miRNA interplay: mechanisms and consequences in cancer. Disease Models Mech. 14(4).
- Jathar S, Kumar V, Srivastava J, Tripathi V. 2017. Technological developments in lncRNA biology. Adv Exp Med Biol. 1008:283–323.
- Jia J, Zhang HB, Shi Q, Yang C, Ma JB, Jin B, Wang X, He D, Guo P. 2019. KLF5 downregulation desensitizes castration-resistant prostate cancer cells to docetaxel by increasing BECN1 expression and inducing cell autophagy. Theranostics. 9(19):5464–5477.
- Jiang X, Chen L, Wu H, Chen Y, Lu W, Lu K. 2021. Knockdown of circular ubiquitin-specific peptidase 9 X-linked alleviates oxidized low-density lipoprotein-induced injuries of human umbilical vein endothelial cells by mediating the miR-148b-3p/KLF5 signaling pathway. J Cardiovasc Pharmacol. 78(6):809–818.
- Karreth FA, Pandolfi PP. 2013. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 3(10):1113–1121.
- Li H, Wang M, Zhou H, Lu S, Zhang B. 2020. Long noncoding RNA EBLN3P promotes the progression of liver cancer via alteration of microRNA-144-3p/DOCK4 Signal. Cancer Manag Res. 12:9339–9349.
- Li T, Chen Y, Zhang J, Liu S. 2018. LncRNA TUG1 promotes cells proliferation and inhibits cells apoptosis through regulating AURKA in epithelial ovarian cancer cells. Medicine (Baltimore)). 97(36):e12131.
- Li X, Jiang M, Chen D, Xu B, Wang R, Chu Y, Wang W, Zhou L, Lei Z, Nie Y, et al. 2018. miR-148b-3p inhibits gastric cancer metastasis by inhibiting the Dock6/Rac1/Cdc42 axis. J Exp Clin Cancer Res. 37(1):71.
- Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, et al. 2014. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:92.
- Ma Y, Wang Q, Liu F, Ma X, Wu L, Guo F, Zhao S, Huang F, Qin G. 2018. KLF5 promotes the tumorigenesis and metastatic potential of thyroid cancer cells through the NF-κB signaling pathway. Oncol Rep. 40(5):2608–2618.
- Mezzanzanica D. 2015. Ovarian cancer: a molecularly insidious disease. Chin J Cancer. 34(1):1–3.
- Peng WX, Koirala P, Mo YY. 2017. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 36(41):5661–5667.
- Peng Y, Croce CM. 2016. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 1:15004.
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. 2015. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 52(10):710–718.
- Rehmani H, Li Y, Li T, Padia R, Calbay O, Jin L, Chen H, Huang S. 2020. Addiction to protein kinase Cɩ due to PRKCI gene amplification can be exploited for an aptamer-based targeted therapy in ovarian cancer. Signal Transduct Target Ther. 5(1):140.
- Siraj AK, Pratheeshkumar P, Divya SP, Parvathareddy SK, Alobaisi KA, Thangavel S, Siraj S, Al-Badawi IA, Al-Dayel F, Al-Kuraya KS. 2020. Krupple-like factor 5 is a potential therapeutic target and prognostic marker in epithelial ovarian cancer. Front Pharmacol. 11:598880.
- Sun Q, Li Q, Xie F. 2019. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco Targets Ther. 12:6297–6307.
- Tomeva E, Switzeny OJ, Heitzinger C, Hippe B, Haslberger AG. 2022. Comprehensive approach to distinguish patients with solid tumors from healthy controls by combining androgen receptor mutation p.H875Y with cell-free DNA methylation and circulating miRNAs. Cancers. 14(2):462.
- Wahlestedt C. 2013. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 12(6):433–446.
- Wang H, Wang L, Zhang S, Xu Z, Zhang G. 2020. Downregulation of LINC00665 confers decreased cell proliferation and invasion via the miR-138-5p/E2F3 signaling pathway in NSCLC. Biomed Pharmacother. 127:110214.
- Wang J, Xu W, He Y, Xia Q, Liu S. 2018. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm Res. 67(11-12):927–936.
- Wang X, Yang B, She Y, Ye Y. 2018. The lncRNA TP73-AS1 promotes ovarian cancer cell proliferation and metastasis via modulation of MMP2 and MMP9. J Cell Biochem. 119(9):7790–7799.
- Webb PM, Jordan SJ. 2017. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 41:3–14.
- Wu M, Shang X, Sun Y, Wu J, Liu G. 2020. Integrated analysis of lymphocyte infiltration-associated lncRNA for ovarian cancer via TCGA, GTEx and GEO datasets. PeerJ. 8:e8961.
- Yu L, Xu Q, Yu W, Duan J, Dai G. 2018. LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop. Biochem Biophys Res Commun. 506(4):819–825.
- Yu L, Xu Q, Yu W, Duan J, Dai G. 2018. LncRNA cancer susceptibility candidate 15 accelerates the breast cancer cells progression via miR-153-3p/KLF5 positive feedback loop. Biochem Biophys Res Commun. 506(4):819–825.
- Yuan Y, Jiaoming L, Xiang W, Yanhui L, Shu J, Maling G, Qing M. 2018. Analyzing the interactions of mRNAs, miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA networks in glioblastoma. J Neurooncol. 137(3):493–502.
- Zhang DW, Gu GQ, Chen XY, Zha GC, Yuan Z, Wu Y. 2020. LINC00665 facilitates the progression of osteosarcoma via sponging miR-3619-5p. Eur Rev Med Pharmacol Sci. 24(19):9852–9859.
- Zhang Z, Luo G, Yu C, Yu G, Jiang R, Shi X. 2019. MicroRNA-493-5p inhibits proliferation and metastasis of osteosarcoma cells by targeting Kruppel-like factor 5. J Cell Physiol. 234(8):13525–13533.
- Zhou JL, Zou L, Zhu T. 2020. Long non-coding RNA LINC00665 promotes metastasis of breast cancer cells by triggering EMT. Eur Rev Med Pharmacol Sci. 24(6):3097–3104.
- Zhou Z, Wang X, Ren X, Zhou L, Wang N, Kang H. 2021. Disease burden and attributable risk factors of ovarian cancer from 1990 to 2017: findings from the Global Burden of Disease Study 2017. Front Public Health. 9:619581.
