Abstract
For decades, the endometrium was considered to be a sterile environment. However, now this concept is disputed, and there is growing evidence that microbiota composition might affect endometrial receptivity. Routine clinical management of infertility is still limited to a microbiological assessment of the lower reproductive tract. The purpose of this study was to compare the abundance of various bacterial, fungal, and viral species, qualitatively and quantitatively, in vaginal, cervical, and endometrial biomaterial of infertile patients. A total of 300 samples from 100 infertile patients of a private assisted reproduction clinic were analyzed. A broad real-time polymerase chain reaction panel was used to identify 28 relevant microbial taxa as well as three members of the Herpesviridae family. All patients underwent endometrial biopsy for further histopathological evaluation. Analysis of the microbial diversity (within the boundaries of the detection panel) revealed that Shannon indexes in the cervix and vagina were similar (1.4 × 10−2 (1.6 × 10−3 – 6.5 × 10−1) vs 1.9 × 10−2 (2.3 × 10−3 – 5.3 × 10−1), respectively, p = 0.502), whereas endometrial indexes differed significantly from both regions (0 (0 – 1.4 × 10−1), p < 0.0001). Surprisingly, 17 microbial and viral taxa were detected in at least one sample. Endometrium exhibited a quite distinct microbiological profile, being different at the detection rates of 14 taxa (p < 0.05). Remarkably, 4% and 2% of endometrial samples were positive for Cytomegalovirus and Candida spp., respectively, while these were undetectable in corresponding cervical and vaginal samples. Prevalence of the Gardnerella vaginalis + Prevotella bivia + Porphyromonas spp. group in endometrium was associated with a low abundance of Lactobacillus spp. (p = 0.039). No noteworthy associations were identified between various microbiota characteristics and clinical parameters, such as chronic endometritis, uterine polyps and adhesions, endometriosis, and a history of sexually transmitted infections. These findings indicate that the microbiological profile of the endometrium is unique, and the analysis of the lower reproductive tract should supplement, rather than be a substitute for it.
Introduction
Human cells make up only approximately half of the body’s total cell count; the other half microorganisms. According to recent calculations, in a 70 kg ‘reference human’ the total number of bacteria is estimated to be 3.8 × 1013, and their total mass is about 0.2 kg (Sender et al. Citation2016). The gastrointestinal tract is the most microbiota-rich part of the human body (∼80% of total microorganisms) with the second most rich being the genitourinary tract (∼9%) (Peterson et al. Citation2009).
The co-evolution of Homo sapiens, as a biological species, with various microorganisms and viruses resulted in non-sterile organs being dependent on a proper microbial environment to maintain their functions. For example, it is well known that the gut microbiota plays an essential role in host nutrient, xenobiotic, and drug metabolism, as well as in protection against pathogens (Jandhyala et al. Citation2015). Furthermore, gut microbiota analysis is a routine procedure in the management of various gastrointestinal tract diseases, as stated in clinical guidelines (World Gastroenterology Organisation).
Intuitively, a similar principle should apply to endometrial microbiota and receptiveness of the endometrium, yet routine clinical procedures of infertility diagnosis and treatment do not involve the assessment of uterine microorganisms. Usually, microbiological testing is limited to the analysis of either vaginal or cervical swabs. There are several quite specific reasons for endometrial microbiota assessment being mostly ignored in clinical practice. Firstly, for decades the endometrium was considered to be a sterile environment, as the cervix was thought to be an impenetrable barrier for pathogens (Tissier Citation1900; Butler Citation1958). Secondly, any collection of endometrial biomaterial is a rather invasive procedure, especially compared to sampling the lower reproductive tract. Thirdly, even though there is growing evidence that the uterus might not be sterile (Romero et al. Citation2004; Mitchell et al. Citation2015; Moreno et al. Citation2016), the topic is still debated and the results of recent studies regarding the association of certain microbiological profile with impaired endometrial receptivity are contradictory (Franasiak et al. Citation2016; Kyono et al. Citation2019; Diaz-Martínez et al. Citation2021; Moreno et al. Citation2022), and meta-analyses of these data have not been reported. Moreover, there is even no consensus on the microbiological profile of such a common condition as chronic endometritis (CE), since there is a discrepancy in the reported microbiota composition in patients with this disease (Tsypurdeeva et al. Citation2018; Liu et al. Citation2019; Chen et al. Citation2021; Lozano et al. Citation2021). Lastly, why even consider analyzing the endometrium since microorganisms are transferred upstream from the lower parts of the reproductive tract? In other words, the vaginal or cervical microbiota could potentially reflect the endometrial microbiota.
The aim of the present study was to compare the abundance of various bacterial, protozoan, fungal, and viral species qualitatively and quantitatively in vaginal, cervical, and endometrial biomaterial of infertile patients. As most studies dedicated to the analysis of microbiota in endometrium were focused on prokaryotic 16S ribosomal RNA (rRNA) sequencing (Chen et al. Citation2017; Riganelli et al. Citation2020; Ichiyama et al. Citation2021; Diaz-Martínez et al. Citation2021), it was decided to use real-time polymerase chain reaction (PCR) to analyze not only the key microbial taxa that are expected to occur in endometrial samples but also some other pathogens that are known to cause endometritis (Kleinman et al. Citation1984; Austin & de León Citation1987; Hollier et al. Citation1997).
Results
All analyzed samples passed the sampling quality control and exhibited the presence of human DNA (except one cervical sample which was excluded from further analysis). Negative control samples (as well as those obtained at the DNA isolation step) showed no contamination by human, protozoan, bacterial, fungal, or viral (at least regarding the studied species) DNA. As the total bacterial loads were unequal, being the highest in vaginal samples, followed by cervical and then endometrial samples (7.9 × 107(2.5 × 107 – 2.5 × 108) vs 1.3 × 107(2.7 × 106 – 4.0 × 107) vs 6.3 × 103(1.6 × 103 – 4.0 × 104) copies of bacterial genomes, respectively, p < 0.0001 for all comparisons), quantitative data of microbiological analysis was normalized by the total bacterial load in each corresponding sample and subjected for further analysis as ‘abundance’ (%).
While all vaginal and cervical specimens were positive for bacterial DNA, it was absent in 16% of the endometrial samples. It is important to mention that even though these samples had overall lower human DNA values than other endometrial samples (1.8 × 104(3.5 × 103 – 2.4 × 105) vs 1.6 × 105(2.1 × 105 – 1.0 × 106) copies, respectively, p = 0.03), no significant association between human DNA concentration and bacterial DNA detection was found (p = 0.128), yet there was a negligible correlation for quantitative values of these parameters (rS=0.288, p = 0.004). Thus, it is unclear whether the absence of bacterial DNA in these samples could be explained by low amounts of collected biomaterial or by true sterility.
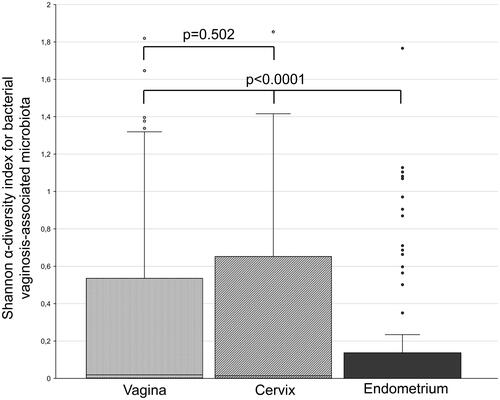
Analysis of the α-diversity of the microbiologic communities revealed that Shannon indexes in the cervix and vagina were comparable (1.4 × 10−2 (1.6 × 10−3 – 6.5 × 10−1) vs 1.9 × 10−2 (2.3 × 10−3 – 5.3 × 10−1), respectively, p = 0.502), whereas endometrium differed significantly compared to both above-mentioned environments (0(0–1.4 × 10−1), p < 0.0001) (). Results of the microbial and viral analysis of each individual sample are visualized in a form of a heatmap (Supplemental material 1), whereas numerical values are available in Supplemental material 2. Comparative microbiological analysis of the three studied sites of the reproductive tract was separated into two parts: qualitative and quantitative.
Figure 1. Diversity of microbial communities in vaginal, cervical, and endometrial samples of infertile patients. Vaginal, cervical, and endometrial samples were obtained from each study participant (n = 100). The abundances of vaginosis-associated bacteria and fungi were analyzed using a real-time PCR kit ‘Femoflor 16’ (cat# R1-P801-S3/6) (DNA-Technology, Moscow, Russia)), which includes reagents for the detection of 24 relevant bacterial and fungal taxa. The line inside the box represents the median value; the borders of the box – are the first and third quartiles; whiskers – the maximum value before the upper limit (third quartile + 1.5 × interquartile range); circles above whiskers are outliers.
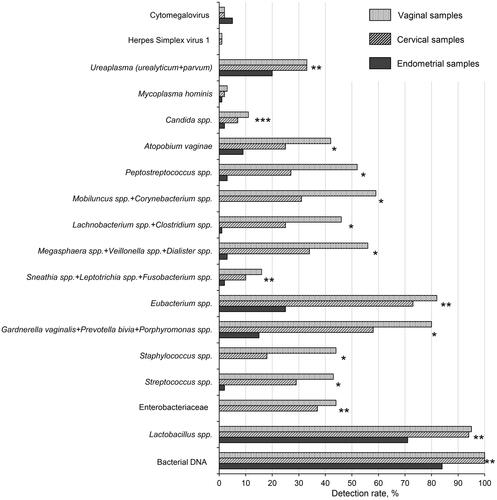
A comparison of the detection rates of each analyzed group or species of microorganisms and viruses is presented in . Such pathogenic agents as Mycoplasma genitalium, Trichomonas vaginalis, Neisseria gonorrhoeae, Chlamydia trachomatis, and Herpes simplex virus 2 were completely absent in all samples. Moreover, among 31 analyzed taxa 14 were detected in none of the studied samples. Therefore, only the remaining 17 taxa were subjected to further analysis. Paired cervical and vaginal samples differed significantly in the detection rates of 8/17 microorganisms (p < 0.05). At the same time, endometrium exhibited a quite distinct microbiological profile, being different (negatively) in the detection rates of 14/17 taxa (p < 0.05), except Candida spp. in which case significant differences were observed only compared to vaginal samples. Due to the generally lower microbiological diversity of the uterus, the detection of a certain taxon in the endometrium, but not in the lower parts of the reproductive tract, is of particular interest. In total, there were six cases fitting the above description: Cytomegalovirus (CMV) – 3 patients; Candida spp. – 1 patient; CMV + Candida spp. – 1 patient, Atopobium vaginae – 1 patient.
Figure 2. Comparison of detection rates of microorganisms and viruses in vaginal, cervical, and endometrial samples of infertile patients. Vaginal, cervical, and endometrial samples were obtained from each study participant (n = 100). Samples were analyzed using real-time PCR kits ‘Femoflor 16’, ‘TNC Complex’, ‘Herpes Multiplex’ (cat# R1-P801-S3/6, R1-P111-S3/9, R1-P210-S3/9, respectively, (DNA-Technology, Moscow, Russia)). The recommended manufacturer cycle threshold value for qualitative analysis was 24, whereas during quantitative analysis DNA levels of less than 103 copies were considered negative. The detection of bacterial DNA was based on the amplification of a conservative procaryotic sequence. Some closely related taxa were analyzed collectively due to limitations of the applied real-time PCR assay. In such cases, the names of the taxa are listed together and joined by a “+” sign. P-values were calculated using Cochran’s Q test. *significant difference for all types of samples (p < 0.05). **significant difference for endometrial samples vs other types of samples. ***significant difference for endometrial samples vs vaginal samples.
No significant association between CD56+ (natural killers, which may be indicative of CMV reactivation process (Jang et al. Citation2019; Basílio-Queirós et al. Citation2022)) cell count in the biomaterial and CMV-positive endometrium was found (p = 0.229). The findings of the histopathological analysis of these samples were not indicative of any cell alterations characteristic of acute inflammation caused by this virus.
The quantitative comparison revealed the absence of any differences between the cervix and vagina for microorganisms which were positive in both environments (). On the other hand, the quantitative structure of the endometrial microbiota differed significantly compared to the vagina in 3 out of 15 taxa (CMV and Herpes Simplex virus 1 were excluded as their analysis was only qualitative) and to the cervix in 2 out of 15 (p < 0.05). Thus, an abundance of microorganisms simultaneously detectable in all three environments was relatively equal, highlighting the balance along the microbiological continuum of the reproductive tract. Among all the studied taxa, only the abundance of the Gardnerella vaginalis + Prevotella bivia + Porphyromonas spp. group had a significant impact on the abundance of Lactobacillus spp. in the endometrium (p = 0.039).
Table 1. Results of comparative quantitative microbiota analysis in vaginal, cervical, and endometrial samples.
Lastly, logistic regression analysis was performed to determine associations between characteristics of endometrial microbiota and clinical parameters, such as CE, uterine polyps and adhesions, endometriosis, and history of sexually transmitted infections. The only significant association was observed for the presence of Megasphaera spp. + Veillonella spp. + Dialister spp. and CE (odds ratio (OR) 19.3; confidence interval: 1.6 – 243.7; p = 0.02), although the detection rate of this microbial group was so low (3%) that these results must be interpreted with caution.
Discussion
Even though in recent years the concept of sterile endometrium is being actively disputed, the true origin of microbial genetic material in samples obtained from a uterine cavity is still uncertain (Romero et al. Citation2004; Mitchell et al. Citation2015; Moreno et al. Citation2016). The major concern is the possibility of contamination from the lower parts of the reproductive tract, as transcervical sampling is the only available option for most patients of an assisted reproduction clinic. Any approach aimed to reduce the chances of contamination by cervical microbiota, such as the introduction of an additional external catheter which prevents the contact of the main sampling device with cervical mucus, as was utilized in the present study, cannot guarantee truly sterile biomaterial collection. However, Winters et al. (Citation2019) demonstrated that in samples collected at hysterectomy, bacterial DNA was detected in 15/25 cases, whereas Lactobacillus spp. was present in 3/13 cases, with a mean abundance of 0.006%. Similar results regarding the low frequency of Lactobacillus spp. in hysterectomy samples were obtained in two small pilot studies (n = 10 in both) (Walther–António et al. Citation2016; Miles et al. Citation2017). On the other hand, in a larger cohort (n = 58) Mitchell et al. (Citation2015) demonstrated that the upper genital tract (endometrium + upper endocervix) was dominated by Lactobacillus iners (45%) and Lactobacillus crispatus (33%) in swabs collected in sterile conditions. Moreover, in patients undergoing laparoscopy or laparotomy (n = 95) Chen et al. (Citation2017) reported similar microbiome profiles between endometrial samples obtained transcervically vs surgical access through the abdomen, with Lactobacillus spp. being the most dominant genus, although its abundance (30.6%) appeared to be almost three times less than in our cohort. These contradictory results indicate that the issue of transcervical contamination at the sampling stage remains unresolved, but its impact may not be as crucial as it had been thought (Baker et al. Citation2018).
Undetectable total bacterial DNA in a proportion of endometrial samples and in six unique cases where the analyzed agent was present in the endometrium, but not in the lower parts of the reproductive tract, could be regarded as favouring a low probability of contamination in the present study. However, it is worth noting that in vitro, readily available, commercial diagnostic real-time PCR kits, as was used here, have been developed for clinical tasks. The manufacturers set cycle threshold values in the automated analysis software relatively high to ensure the absence of false-positive results. Thus, the analytical system is developed with specificity rather than sensitivity in mind, in contrast to prokaryotic 16S rRNA sequencing which is often used in exploratory studies of endometrial microbiota (Chen et al. Citation2017; Riganelli et al. Citation2020; Ichiyama et al. Citation2021; Diaz-Martínez et al. Citation2021). For example, Reschini et al. Citation2022 reported that a commercially available real-time PCR kit for the detection of bacterial vaginosis detected Lactobacillus spp. in only 53% of endometrial samples, whereas simultaneous Next Generation Sequencing analysis of 16S rRNA detected this group in 83% of the samples. However, examination of these contradictory cases (n = 15) reveals that the abundance of this microorganism was exceptionally low [5 (1–25) %] and, moreover, it was significantly lower than in other positive samples (p < 0.05). Therefore, the real-time PCR approach implemented in the present study is, indeed, less susceptible to contamination, but less sensitive. The absence of total bacterial DNA in a proportion of samples should be interpreted accordingly.
In concordance with previously published data, it was demonstrated that the endometrium, compared to the vagina, has a clearly distinct qualitative composition of the microbiota (Riganelli et al. Citation2020; Ichiyama et al. Citation2021). The cervical profile is slightly closer, but still far from being capable of reflecting the microbiological community of the uterus (Chen et al. Citation2017). Anyway, these results should not be immediately interpreted as clinically relevant. The role of uterine microbial composition in the reduction of endometrial receptivity is yet to be uncovered. Severe methodological heterogeneity of the available studies on this topic makes it challenging to compare already contradictory results, while to date no proper meta-analyses have been conducted. There is no consensus, neither for the prevalence of uterine Lactobacillus spp. in patients with receptive endometrium (Kyono et al. Citation2019; Ichiyama et al. Citation2021; Moreno et al. Citation2022), nor for the increased α-diversity index (Franasiak et al. Citation2016; Diaz-Martínez et al. Citation2021).
In the present study, among the analyzed members of the Herpesviridae family, only CMV was detectable in the endometrium (5%), and only in 1% of cases was it detectable in the lower parts of the reproductive tract of the same patient. Recently, a slightly higher detection rate of this virus in endometrial mucosa of patients with reproductive problems (3 out of 24) was reported by Blazheva (Citation2022). It is well known that CMV is the most common agent responsible for congenital infection, which leads to neurological complications (Walker et al. Citation2013). The role of CMV, detected either immunologically in blood, or genetically in cervical swabs, in infertility is still debated, and universal screening for it is not recommended (Eggert–Kruse et al. Citation2009). However, the relationship of endometrial CMV to infertility and infection of the fetus during the following pregnancy has not yet been studied. Our findings suggest that this topic would be worth exploring.
It is also remarkable that only 2% of the Candida spp. positive endometrium samples had a paired positive sample from the lower parts of the reproductive tract (overall cervical and vaginal detection rates were 7 and 11%, respectively). The data regarding the pathological role of fungi in the endometrium is scarce and is mostly limited to small studies and individual case reports (Rodriguez et al. Citation1972; Austin & de León Citation1987; Gazvani et al. Citation2013). In a recent study, Liu et al. (Citation2022) demonstrated certain cervical and vaginal mycobiome structural alterations, particularly for some members of the Candida genera, in patients with uterine adhesions compared to healthy controls. They also demonstrated the ability of Candida parapsilosis to modulate inflammatory activity, fibrosis, and bacterial microbiota in a rat model of uterine adhesions. The complete discrepancy of the Candida spp. detection in the upper and lower reproductive tract observed in our study allows speculation that the analysis of the endometrial mycobiome could be even more informative than cervical or vaginal comparisons with regard to various uterine conditions related to infertility.
The absence of any reliable associations of the tested microbiological parameters of the endometrium with clinical data (CE, uterine polyps and adhesions, endometriosis, history of sexually transmitted infections) could be explained by the low incidence of these conditions in our cohort. Some studies refer to a threshold value of CD138+ cells for CE diagnosis as low as a single cell in 10 high power fields (HPF) (Hirata et al. Citation2021). Selection of this criterion instead of 5 cells in 30 HPF in our cohort resulted in a dramatic shift of CE incidence from 11% to 83%, with Gardnerella vaginalis + Prevotella bivia + Porphyromonas spp. group as well as Atopobium vaginae being unexpectedly prevalent in non-CE patients (p = 0.002 and p = 0.026, respectively), which contradicts the results of other studies (Cicinelli et al. Citation2014; Tsypurdeeva et al. Citation2018; Liu et al. Citation2019; Lozano et al. Citation2021). Overall, there is no consensus on the microbiota composition associated with CE, mainly due to variations of the selected criteria for its diagnosis in the above-cited publications. Anyway, CE remains to be an important clinical condition, as it was shown in a recent meta-analysis (n = 4145) that patients with CE had lower ongoing pregnancy or live-birth and clinical-pregnancy rates (OR 1.97, p = 0.02 and OR 2.28, p = 0.002, respectively) (Vitagliano et al. Citation2022). Moreover, the same meta-analysis demonstrated that confirmed CE resolution after antibiotic therapy may improve endometrial receptivity leading to similar in vitro fertilization outcomes as compared to unaffected patients. Thus, further investigation, more methodologically uninformed, of CE aetiology and pathogenesis is needed.
In conclusion, it was demonstrated that the endometrium of infertile patients has a quite distinct microbial profile as well as viral status, at least for certain members of the Herpesviridae family, compared to the profile of lower parts of the reproductive tract. Various immunological and biochemical interactions between the members of the microbial community and the endometrium may play a pivotal role in the regulation of its receptivity, and this role is yet to be uncovered.
Materials and methods
Study design
The study included 100 infertile patients at a private, assisted reproduction clinic. The criterion for infertility was an inability to conceive for more than one year of unprotected intercourse. Patient recruitment took place between January 2021 and June 2022. All enrolled participants were scheduled for hysteroscopy with endometrial biopsy as part of their treatment on days 5–12 of the menstrual cycle. Histological and immunohistochemical (CD138 and CD56 immunostaining) analysis of endometrial biomaterial was outsourced to a private laboratory. The diagnostic criterion for chronic endometritis was ≥5 CD138+ cells in at least 1 out of 30 HPF (Li et al. Citation2021). All patients confirmed that they had not taken any antibiotics for at least three months prior to study inclusion, and they were not diagnosed with any sexually transmitted disease for the same period. The clinical and demographic characteristics of the subjects are provided in .
Table 2. Clinical and demographic characteristics of the participants.
Sample collection
Prior to any direct hysteroscopy-related interventions into the uterine cavity, the external genitalia was cleaned with an antiseptic wipe. Vaginal and cervical samples were collected using cytobrushes. Overall, the order of sampling was ascending to reduce the chances of microbiota contamination from the upper parts of the reproductive tract. Endometrial samples were obtained using a Pipelle catheter that was introduced into the uterine cavity through an external 3.5 mm diameter catheter to prevent any possible contact with cervical mucus.
Vaginal and cervical samples were immediately transferred into sterile tubes containing 0.5 ml of preservation solution. The endometrial aspirate was transferred into empty sterile tubes. All samples were stored at –80 °C before processing.
Microbiological analysis
After defrosting, all samples were thoroughly mixed by pulse vortexing. Endometrial samples were diluted with sterile 0.9% saline solution to a total volume of 0.5 ml and mixed again. DNA was isolated using “PREP-NA-PLUS” kit (cat# P-002/2, DNA-Technology, Russia). To ensure the absence of any systematic contamination at this stage, DNA isolation was also performed from negative control samples, according to the manufacturer’s recommendations.
The microbiological profiles were analyzed using the following commercial kits on a DT-Prime real-time PCR instrument (DNA-Technology, Moscow, Russia) according to the manufacturers’ protocols: ‘Femoflor 16’, ‘TNC Complex’, ‘Herpes Multiplex’ (cat# R1-P801-S3/6, R1-P111-S3/9, R1-P210-S3/9, respectively, (DNA-Technology, Moscow, Russia)). These reagents allowed quantitative analysis of the total bacterial load (based on the detection of conservative procaryotic sequences), Lactobacillus spp., Enterobacteriaceae, Streptococcus spp., Staphylococcus spp., Gardnerella vaginalis, Prevotella bivia, Porphyromonas spp., Eubacterium spp., Sneathia spp., Leptotrichia spp., Fusobacterium spp., Megasphaera spp., Veillonella spp., Dialister spp., Lachnobacterium spp., Clostridium spp., Mobiluncus spp., Corynebacterium spp., Peptostreptococcus spp., Atopobium vaginae, Candida spp., Mycoplasma hominis, Ureaplasma urealyticum, Ureaplasma parvum, Mycoplasma genitalium as well as qualitative analysis of Trichomonas vaginalis, Neisseria gonorrhoeae, Chlamydia trachomatis, Herpes simplex viruses 1 & 2, Cytomegalovirus. The number of Homo sapiens DNA was also measured in every sample to control the biomaterial collection quality (>103 copies per reaction mixture). Real-time PCR data was analyzed automatically in the RealTime_PCR software (DNA-Technology, Russia) developed for the above-mentioned PCR kits. The recommended manufacturer cycle threshold value for qualitative analysis was 24, whereas during quantitative analysis DNA levels of less than 103 copies were considered negative. Some closely related taxa were analyzed collectively due to limitations of the applied real-time PCR assay. In such cases, the names of the taxa are listed together and joined by a “+” sign.
The absence of contamination of the analytical system was ensured by testing the negative control samples provided by the manufacturer in addition to the previously mentioned negative control samples from the DNA isolation stage.
Statistical analysis
The statistical analyses were performed with ‘IBM SPSS Statistics 26.0’ software (IBM, Armonk, USA). The distribution normality of the data was assessed using Shapiro–Wilk tests. As the absence of normal distribution was verified for all the relevant data, variables were compared with non-parametric tests. A comparison of dependent qualitative data in multiple groups was performed with Cochran’s Q test. Dependent quantitative data in multiple groups were compared with Friedman’s test, whereas in cases where quantitative data was available only for two groups, the Wilcoxon signed-rank test was used. Due to the low detection rate of most taxa, quantitative comparisons of the microbiota in different sites of the reproductive tract were carried out only for samples positive for the compared microorganism. The exclusion of negative samples avoided duplication of the qualitative assessment results and defined the position of the tested taxon in the structure of the microbiota community where it is already present. Bonferroni correction and other similar methods were not used to adjust the results of multiple comparisons due to the low number of simultaneous comparisons and the exploratory design of the study (Armstrong Citation2014). Mann–Whitney U tests were used to compare independent quantitative data. Associations between variables were evaluated with logistic regression. The correlation of quantitative variables was assessed via the Spearman coefficient (rS), which was interpreted using the Chaddock scale. Shannon’s indexes were calculated to describe the diversity of the microbiological communities. Note that due to limitations of the selected real-time PCR microbiological panel in the present study, Shannon’s index will only reflect the diversity within bacterial vaginosis-associated microbiota boundaries and should not be directly compared to other α-diversity data obtained in 16S rRNA sequencing studies. A p-Value <0.05 was considered statistically significant. Data are presented as [median (interquartile range)] unless stated otherwise.
Ethics approval
The study was approved by the Local ethics committee of the Medical Research and Educational Center of Lomonosov Moscow State University (protocol no. 12/20, dated: 21.12.2020), and it was conducted in accordance with the principles of the Declaration of Helsinki. All patients signed written informed consent to take part in the study.
Author's contributions
Conceptualization: MJ, OP; Methodology: KK, MJ; Software: DA, AS; Validation: LS, OP; Experiments: KK; Formal analysis: LS; Investigation: EM, MJ; Resources: DA, LS; Data curation: DA, AS; Writing-original draft preparation: MJ; Writing-review and editing: LS, OP; Visualization: AD, MJ; Supervision: LS; Project administration: EM, LS; Funding acquisition: LS, OP. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download MS Excel (51.9 KB)Supplemental Material
Download MS Word (1.1 MB)Disclosure statement
The authors report no conflicts of interest.
Data availability statement
Results of microbiological analysis and clinical data of study participants are presented in Supplemental material 2.
Additional information
Funding
References
- Armstrong RA. 2014. When to use the Bonferroni correction. Ophthalmic Physiol Opt. 34(5):502–508.
- Austin KL, de León R. 1987. Candida albicans endometritis as a cause of reproductive dysfunction. Report of a case. Rev Med Panama. 12(2):79–85.
- Baker JM, Chase DM, Herbst-Kralovetz MM. 2018. Uterine microbiota: residents, tourists, or invaders? Front Immunol. 9:208.
- Basílio-Queirós D, Venturini L, Luther-Wolf S, Dammann E, Ganser A, Stadler M, Falk CS, Weissinger EM. 2022. Adaptive NK cells undergo a dynamic modulation in response to human cytomegalovirus and recruit T cells in in vitro migration assays. Bone Marrow Transplant. 57(5):712–720.
- Blazheva S. 2022. P-415 study of the incidence of herpes viruses in the endometrium of women with evidence of chronic endometritis and reproductive problems. Hum Reprod. 37(Supplement_1): deac107.392.
- Butler B. 1958. Value of endometrial cultures in sterility investigation. Fertil Steril. 9(3):269–273.
- Chen C, Song X, Wei W, Zhong H, Dai J, Lan Z, Li F, Yu X, Feng Q, Wang Z, et al. 2017. The microbiota continuum along the female reproductive tract and its relation to uterine-related diseases. Nat Commun. 8(1):875.
- Chen P, Chen P, Guo Y, Fang C, Li T. 2021. Interaction between chronic endometritis caused endometrial microbiota disorder and endometrial immune environment change in recurrent implantation failure. Front Immunol. 12:748447.
- Cicinelli E, Matteo M, Tinelli R, Pinto V, Marinaccio M, Indraccolo U, De Ziegler D, Resta L. 2014. Chronic endometritis due to common bacteria is prevalent in women with recurrent miscarriage as confirmed by improved pregnancy outcome after antibiotic treatment. Reprod Sci 21(5):640–647. http://www.ncbi.nlm.nih.gov/pubmed/24177713.
- Diaz-Martínez MDC, Bernabeu A, Lledó B, Carratalá-Munuera C, Quesada JA, Lozano FM, Ruiz V, Morales R, Llácer J, Ten J, et al. 2021. Impact of the vaginal and endometrial microbiome pattern on assisted reproduction outcomes. J Clin Med. 10(18):4063.
- Eggert-Kruse W, Reuland M, Johannsen W, Strowitzki T, Schlehofer JR. 2009. Cytomegalovirus (CMV) infection – related to male and/or female infertility factors? Fertil Steril. 91(1):67–82. https://www.sciencedirect.com/science/article/pii/S0015028207040575.
- Franasiak JM, Werner MD, Juneau CR, Tao X, Landis J, Zhan Y, Treff NR, Scott RT. 2016. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet. 33(1):129–136.
- Gazvani R, Fowler PA, Coyne L, Odds FC, Gow NAR. 2013. Does Candida albicans play a role in the etiology of endometriosis? J Endometr Pelvic Pain Disord. 5(1):2–9.
- Hirata K, Kimura F, Nakamura A, Kitazawa J, Morimune A, Hanada T, Takebayashi A, Takashima A, Amano T, Tsuji S, et al. 2021. Histological diagnostic criterion for chronic endometritis based on the clinical outcome. BMC Womens Health. 21(1):94.
- Hollier LM, Scott LL, Murphree SS, Wendel GDJ. 1997. Postpartum endometritis caused by herpes simplex virus. Obstet Gynecol. 89(5 Pt 2):836–838.
- Ichiyama T, Kuroda K, Nagai Y, Urushiyama D, Ohno M, Yamaguchi T, Nagayoshi M, Sakuraba Y, Yamasaki F, Hata K, et al. 2021. Analysis of vaginal and endometrial microbiota communities in infertile women with a history of repeated implantation failure. Reprod Med Biol. 20(3):334–344.
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. 2015. Role of the normal gut microbiota. World J Gastroenterol. 21(29):8787–8803.
- Jang JE, Hwang DY, Chung H, Kim S-J, Eom J-I, Jeung H-K, Song J, Kim JS, Cheong J-W, Min YH. 2019. Early cytomegalovirus reactivation and expansion of CD56brightCD16dim/−DNAM1+ natural killer cells are associated with antileukemia effect after haploidentical stem cell transplantation in acute leukemia. Biol Blood Marrow Transplant. 25(10):2070–2078.
- Kleinman D, Sarov I, Insler V. 1984. Infection of endometrial cells with human cytomegalovirus. Gynecol Obstet Invest. 17(2):89–95.
- Kyono K, Hashimoto T, Kikuchi S, Nagai Y, Sakuraba Y. 2019. A pilot study and case reports on endometrial microbiota and pregnancy outcome: an analysis using 16S rRNA gene sequencing among IVF patients, and trial therapeutic intervention for dysbiotic endometrium. Reprod Med Biol. 18(1):72–82.
- Li Y, Xu S, Yu S, Huang C, Lin S, Chen W, Mo M, Lian R, Diao L, Ding L, et al. 2021. Diagnosis of chronic endometritis: how many CD138(+) cells/HPF in endometrial stroma affect pregnancy outcome of infertile women? Am J Reprod Immunol. 85(5):e13369.
- Liu N-N, Zhao X, Tan J-C, Liu S, Li B-W, Xu W-X, Peng L, Gu P, Li W, Shapiro R, et al. 2022. Mycobiome dysbiosis in women with intrauterine adhesions. Microbiol Spectr. 10(4):e0132422.
- Liu Y, Ko EY-L, Wong KK-W, Chen X, Cheung W-C, Law TS-M, Chung JP-W, Tsui SK-W, Li T-C, Chim SS-C. 2019. Endometrial microbiota in infertile women with and without chronic endometritis as diagnosed using a quantitative and reference range-based method. Fertil Steril. 112(4):707–717.e1.
- Lozano FM, Bernabeu A, Lledo B, Morales R, Diaz M, Aranda FI, Llacer J, Bernabeu R. 2021. Characterization of the vaginal and endometrial microbiome in patients with chronic endometritis. Eur J Obstet Gynecol Reprod Biol. 263:25–32.
- Miles SM, Hardy BL, Merrell DS. 2017. Investigation of the microbiota of the reproductive tract in women undergoing a total hysterectomy and bilateral salpingo-oopherectomy. Fertil Steril. 107(3):813–820.e1.
- Mitchell CM, Haick A, Nkwopara E, Garcia R, Rendi M, Agnew K, Fredricks DN, Eschenbach D. 2015. Colonization of the upper genital tract by vaginal bacterial species in nonpregnant women. Am J Obstet Gynecol. 212(5):611.e1–9–611.e9.
- Moreno I, Codoñer FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazán J, Alonso R, Alamá P, Remohí J, Pellicer A, et al. 2016. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol. 215(6):684–703.
- Moreno I, Garcia-Grau I, Perez-Villaroya D, Gonzalez-Monfort M, Bahçeci M, Barrionuevo MJ, Taguchi S, Puente E, Dimattina M, Lim MW, et al. 2022. Endometrial microbiota composition is associated with reproductive outcome in infertile patients. Microbiome. 10(1):1.
- Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, Bonazzi V, McEwen JE, Wetterstrand KA, Deal C, et al. 2009. The NIH human microbiome project. Genome Res. 19(12):2317–2323. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2792171/
- Reschini M, Benaglia L, Ceriotti F, Borroni R, Ferrari S, Castiglioni M, Guarneri D, Porcaro L, Vigano’ P, Somigliana E, et al. 2022. Endometrial microbiome: sampling, assessment, and possible impact on embryo implantation. Sci Rep. 12(1):8467.
- Riganelli L, Iebba V, Piccioni M, Illuminati I, Bonfiglio G, Neroni B, Calvo L, Gagliardi A, Levrero M, Merlino L, et al. 2020. Structural variations of vaginal and endometrial microbiota: hints on female infertility. Front Cell Infect Microbiol. 10:350.
- Rodriguez M, Okagaki T, Richart RM. 1972. Mycotic endometritis due to Candida. A case report. Obstet Gynecol. 39(2):292–294.
- Romero R, Espinoza J, Mazor M. 2004. Can endometrial infection/inflammation explain implantation failure, spontaneous abortion, and preterm birth after in vitro fertilization? Fertil Steril. 82(4):799–804.
- Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14(8):e1002533–e1002533.
- Tissier H. 1900. Recherches sur la flore intestinale des nourrissons:(état normal et pathologique). [place unknown].
- Tsypurdeeva ND, Shipitsyna EV, Savicheva AM, Gzgzyan AM, Kogan IY. 2018. Composition of endometrial microbiota and chronic endometritis severity in patients with in vitro fertilization failures. Zakuszenbolezn 67(2):5–15.
- Vitagliano A, Laganà AS, De Ziegler D, Cicinelli R, Santarsiero CM, Buzzaccarini G, Chiantera V, Cicinelli E, Marinaccio M. 2022. Chronic endometritis in infertile women: impact of untreated disease, plasma cell count and antibiotic therapy on IVF Outcome & mdash: a systematic review and meta-analysis. Diagnostics 12(9):2250.
- Walker SP, Palma-Dias R, Wood EM, Shekleton P, Giles ML. 2013. Cytomegalovirus in pregnancy: to screen or not to screen. BMC Pregnancy Childbirth. 13:96.
- Walther-António MRS, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, Keeney GL, Creedon DJ, Nelson H, Mariani A, et al. 2016. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med. 8(1):122.
- Winters AD, Romero R, Gervasi MT, Gomez-Lopez N, Tran MR, Garcia-Flores V, Pacora P, Jung E, Hassan SS, Hsu C-D, et al. 2019. Does the endometrial cavity have a molecular microbial signature? Sci Rep. 9(1):9905.
