 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Immunoglobulins and T cell receptors (TCRs) share common sequences and structures. With the goal of creating novel bispecific antibodies (BsAbs), we generated chimeric molecules, denoted IgG_TCRs, where the Fv regions of several antibodies were fused to the constant domains of the α/β TCR. Replacing CH1 with Cα and CL with Cβ, respectively, was essential for achieving at least partial heavy chain/light chain assembly. Further optimization of the linker regions between the variable and constant domains, as well as replacement of the large FG loop of Cβ with a canonical β-turn, was necessary to consistently obtain full heavy chain/light chain assembly. The optimized IgG_TCR molecules were evaluated biophysically and shown to maintain the binding properties of their parental antibodies. A few BsAbs were generated by co-expressing native Fabs and IgG_TCR Fabs within the same molecular construct. We demonstrate that the IgG_TCR designs steered each of the light chains within the constructs to specifically pair with their cognate heavy chain counterparts. We did find that even with complete constant domain specificity between the CH1/CL and Cα/Cβ domains of the Fabs, strong variable domain interactions can dominate the pairing specificity and induce some mispairing. Overall, the IgG_TCR designs described here are a first step toward the generation of novel BsAbs that may be directed toward the treatment of multi-faceted and complex diseases.
Abbreviations
| DSC | = | differential scanning calorimetry |
| Ha | = | heavy chain containing Ca in place of CH1 |
| Hb | = | heavy chain containing Cb in place of CH1 |
| HC | = | heavy chain |
| La | = | heavy chain containing Ca in place of CL |
| Lb | = | heavy chain containing Cb in place of CL |
| LC | = | light chain |
| RU | = | resonance units |
| SDS-PAGE | = | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SEC | = | size exclusion chromatography |
| SPR | = | surface plasmon resonance |
| TCR | = | T cell receptor |
Introduction
Researchers in both industry and academia utilize a plethora of bispecific antibody (BsAb) platforms to address complex and multi-faceted biological problems.Citation1,2 There are many ways to generate BsAbs based on combining the recognition domains of monoclonal antibodies (mAbs) or mAb-like scaffold proteins.Citation1,2,3,4 Many have drawbacks, which include instability, insolubility, or restrained geometry. One of the most commonly used BsAb platforms is the IgG-scFv. The IgG-scFv platform combines the activities of 2 mAbs while maintaining IgG-like pharmacokinetics and the potential for immune effector functions. First described in 1997,Citation5 the IgG-scFv platform did not meet with rapid success due to manufacturing issues relating to the scFvs.Citation6 In scFvs, the VH and VL domain stabilities and interdomain interaction are attenuated by the loss of the CH1/CL domains, often leading to aggregation or insolubility.Citation6 Multiple reports have shown that stability/solubility engineering of scFvs within IgG-scFvs can reduce these manufacturing issues.Citation7,8,9,10 The engineering requires significant time and resources for every BsAb therapeutic, and can be limiting in terms of the geometry and the choice of mAbs.Citation7,8,9,10
Here, we strive to generate an IgG-Fab BsAb platform that combines 2 mAb specificities within a single therapeutic protein and circumvents the stability/solubility flaws of IgG-scFvs (). Fab regions are fundamentally more stable than scFvs due to the natural scaffolding provided by the CH1/CL domains.Citation6,11 We propose replacing the scFv building block of an IgG-scFv with a more stable Fab-like moiety. The IgG-Fab format is tetravalent and contains 2 putative binding sites for each epitope of the BsAb, which is different than bivalent IgG BsAbs obtained using other protein engineering approaches.Citation12,13,14,15 The challenge with an IgG-Fab platform is that the 2 LCs of an IgG-Fab BsAb can bind heterogeneously to the 2 HC Fd (VH and CH1) regions within the BsAb (). Conceptually, 16 HC/LC pairings are possible, resulting in unacceptable product heterogeneity. There is a need to direct each LC to pair with its cognate HC to produce a homogeneously assembled IgG-Fab BsAb.
Figure 1. Schematic diagram of an IgG-Fab BsAb (A). Diagrams to the right of the correctly assembled IgG-Fab are potential mispairings related to the lack of LC specificity for a particularly HC Fd region. (B) Schematic diagram of the domain architecture of a α/β TCR. The receptor is a heterodimer consisting of 2 chains that each comprise a V-class and a C-class Ig-fold, much like an immunoglobulin Fab. (C) Superposition of the structures of an IgG1 CH1/Cκ heterodimer (pdb id: 3HC0) and the constant domains of an α/β TCR (pdb id: 3ARB). The structural homology between Cβ and Cκ as well as that between Cα and CH1 is apparent. (D) Diagram demonstrating the exchange of the CH1/Cκ domains with the TCR Cα/Cβ domains within an IgG1 antibody (denoted IgG_TCR).
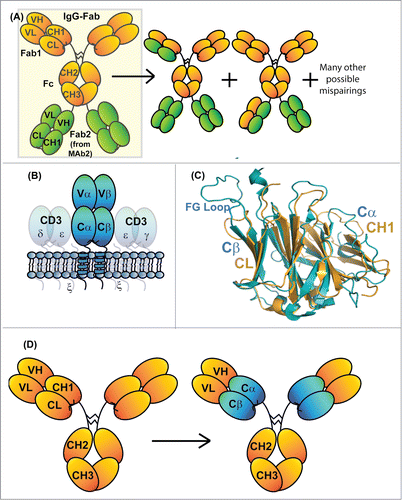
We propose achieving specific HC/LC assembly within IgG-Fab BsAbs by replacing the constant domains (CH1/CL) of one of the Fabs with the constant domains of the α/β T-cell receptor (TCR Cα/Cβ). TCRs maintain a similar heterodimeric quarternary structure as Fabs () and their constant domains are structurally homologous with IgG CH1/CL (). TCR Cα and Cβ have very different primary sequences than Fab constant domains (∼20% identity between the Cα/Cβ and CH1/CL domains) with an orthogonal heterodimeric interface. In both α/β TCRs and IgGs, stable interaction between their 2 chains is based on interactions between variable domains and interactions between constant domains.Citation16 We hypothesized that replacement of the CH1/CL domains of an IgG with those of the Cα/Cβ of a TCR may create a more stable heterodimeric complex than what is observed for VH/VL in isolation or as an scFv (). The goal would be for these synthetic IgG/TCR chimeric Fabs to have immunoglobulin-like stability, solubility, and binding properties. The chimeric HCs and LCs should have unique specificity for one another and not mix with wild-type (WT) antibody HCs and LCs, solving the specificity problem that arises when expressing 2 Fab moieties simultaneously. If specificity were obtained by replacing Fab constant domains with TCR constant domains, a large number of potential BsAb formats would be possible, including IgG-Fab BsAbs and BsAbs with native-like IgG architecture.
Chimeric fusions of antibodies and TCRs have been contemplated previously. Combinations of mAb variable domains with TCR constant domains have been used to generate recombinantly modified T cells directed to the protein antigens recognized by the IgG VH/VL.Citation17,18,19,20 Some of these constructs contained only the TCR β-chain, which recruits CD3 and the rest of the activation machinery. Citation21 Other designs simply fuse mAb Fv regions to the CD3ζ chain to enable TCR-like signaling. Citation22 Decades ago, a report described the individual insertion of the TCR constant domains within antibody kappa LCs (between the VL and CL domains) to serve as soluble immunogens.Citation23 A concept similar to the approach described in this report was also published over 20 y ago. In that report, the TCR constant domains were attached to the Fv region of a mAb (also containing an additional CL domain apparently for solubility); Citation24 however, there was little characterization of the proteins that might suggest whether the concept could be successfully applied.
Here, we describe our protein engineering efforts to stably generate Fab-like moieties with antibody VH/VL domains linked to TCR α/β constant domains. We take advantage of advances in molecular biology and expression technologies to explore this space in a more exhaustive fashion than was previously possible. We explore the domain orientation of the IgG/TCR chimeric proteins, the linkages between the domains, and additional designs to stabilize these Fab-like moieties. We go further to describe specific examples where we utilize the specificity afforded using the IgG_TCR Fabs to generate IgG-Fab BsAbs with novel activities.
Results
Generation of Chimeric IgG_TCR proteins
We first evaluated the ability of Cα/Cβ to replace CH1/CL within an anti-IL-17 IgG4/κ mAb (an in-house, Eli Lilly asset). Based on sequence homology, it was not clear whether CH1 or CL should be replaced with Cα or Cβ. The variable domain of the β-chain maintains V-D-J joining similar to antibody VH; however, its constant domain is structurally more homologous to CL based on published Fab and TCR structures (3HC4 and 3QEU, respectively). Additionally, the linker lengths between the variable and constant domains of IgGs and TCRs are somewhat different. Therefore, we generated 8 initial IgG_TCR constructs to investigate the possibility of replacing the CH1/CL domains of a Fab with the Cα/Cβ from the TCR (see Table S1 for the Cα and Cβ sequences used in each construct). The eight constructs included testing Cα and Cβ in place of CH1 and CL (both orientations), as well as varied linker lengths including either all residues of Cα and Cβ or N-terminal truncated versions to match the IgG linker lengths between variable and constant domains.
The new constructs were expressed at the 2 mL scale in HEK293F cells. The constructs were purified using protein G magnetic beads. All constructs expressed; however, only those containing Cβ in the LC and Cα in the HC demonstrated measurable HC/LC assembly (, ). Reduced and non-reduced SDS-PAGE gels indicated that all purified constructs containing Cα in the LC and Cβ in the HC lacked LCs attached to the purified HC components. The proteins containing Cβ in the LC and Cα in the HC appear to be secreted with at least some HC/LC assembly. IgG HCs have a stringent secretion mechanism involving the LC displacement of BiP chaperone bound to unfolded CH1.Citation25 Replacement of CH1 with Cα or Cβ appeared to release the HC from this quality control mechanism.
Table 1. Biochemical characterization of initial IgG_TCRs
Figure 2. Characterization of initial IgG_TCR constructs. (A, B) SDS-PAGE analysis of IgG_TCR constructs under reducing, 10 mM DTT, (A) and non-reducing conditions (B). 15 μL of protein G magnetic bead purified protein from 2 mL 293F purifications was added to each lane. β1 and β2 differ in the N-terminal residues of the β-domain (β1 starts with E117, while β2 starts with K121). Similarly, α1 and α2 differ in the N-terminal residues of the α-domain (α1 starts with P116, while α2 starts with I118). (C) Non-reduced (left side of gel) and reduced (right side of gel) SDS-PAGE analysis of protein G pull-downs from supernatants expressed using mismatched and matched pairs of IgG and IgG_TCR heavy and light chains. (D) Cation exchange separation of IgG_TCR proteins secreted with 0 (1st peak), 1 (middle peak), or 2 (3rd peak) associated LCs. The inset shows the SDS-PAGE analysis of the 3 cation exchange fractions. (E) Binding activity of the protein fractions separated in (D), demonstrating the importance of LC association for binding to antigen. The association of the IgG_TCR protein can be observed between 300–600 s, while the antigen (IL-17 in this case) association can be observed between 800–1000 s. (F) Improvement in the uniform expression of fully paired (HC2LC2) IgG_TCR proteins after truncating the C-terminal tail of the β-constant domain of the LC. HC2LC2 elution time was at 13.5 minutes based on static light scattering analysis.
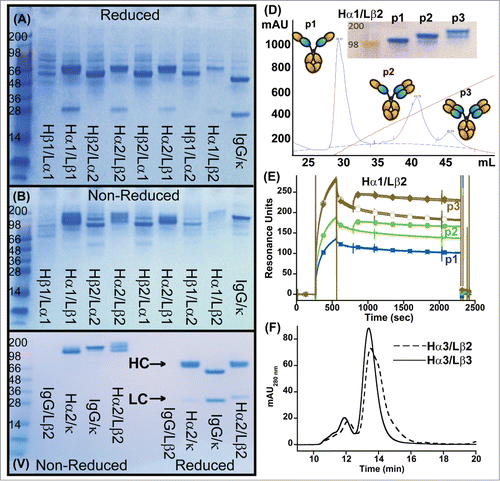
We subsequently narrowed our evaluation of IgG_TCR proteins only to those with Cα in the HC and Cβ in the LC. To confirm the identity of the different protein components of the IgG proteins containing Cα in the HC and Cβ in the LC, we scaled up all 4 of the Hα/Lβ IgG_TCR constructs. At the 1 L scale, all Hα/Lβ constructs expressed at near equivalent levels and were purified by protein A chromatography. Indiscriminately, we chose Hα1/Lβ2 (Table S1) for further separation/analysis by monoS chromatography. The protein separated into 3 distinct peaks on a monoS column (). Using SDS-PAGE analysis, the 3 peaks appeared to be HC2, HC2LC1, and HC2LC2 (). The three peaks were individually evaluated for their ability to bind ligand (IL-17 in this case) using surface plasmon resonance (SPR). The lower molecular weight protein (putatively HC2) bound no ligand; the middle molecular weight protein (putatively HC2LC1) bound ligand with high affinity; and the high molecular weight protein (putatively HC2LC2) bound approximately 2-fold more ligand (based on RUs) with high affinity (). The IL-17 mAb apparently needs intact HC/LC pairing to interact with IL-17. The SPR assay was then used to approximate the level of HC/LC assembly of each of the IgG_TCRs using the following:
where ‘%Activity’ relates to the amount of intact HC/LC in an IgG_TCR. The WT IgG is used as a positive control for 100%. Based on these measurements, we determined that Hα2Lα2 (Table S1) had the highest HC/LC assembly level of all the constructs expressed at the 1 L scale (). This construct had Cα/Cβ in the HC/LC orientation and contained truncated N-termini for both the Cα and Cβ domains. The HC/LC assembly of Hα2Lβ2 was still only 54%, indicating that additional modification will be necessary to insure all HCs consistently have a LC attached to them.
We next sought to determine if Cα/Cβ-containing IgG_TCR HCs and LCs could discriminate from binding to natural IgG HCs and LCs. We expressed a LC containing Cβ with a WT IgG HC and expressed a WT LC with an IgG HC containing Cα. Interestingly, we found that no antibody was expressed in the former case, presumably because the Cβ-containing LC could not associate with a WT HC and release it from BiP for secretion (). In the latter case, the HC-containing Cα could secrete on its own, but only vanishingly small amounts of the WT kappa LC could be found associated with the HC (), demonstrating that Cα and CL do not interact with one another. Assembly could be observed for the fully WT IgG HC/LC pair and the IgG HC/LC containing both Cα and Cβ, although the TCR-containing construct appeared to be a mixture of HC2, HC2LC1, and HC2LC2 (fully assembled).
Optimization of IgG_TCR chimeric proteins
We evaluated the effect of modifying the C-termini of the Cα and Cβ domains within the IgG_TCR context. We used Hα2Lβ2 as a basis for optimization since it was found to provide the highest level of HC/LC assembly of the various IgG_TCR chimeric proteins produced initially. First we replaced the C-terminus of Cα that makes the disulfide with the LC with the upper hinge of IgG1 and converted the remainder of the HC to an IgG1 receptacle antibody (Hα3, Table S1). Additionally, we wished to utilize the activity of the anti-HER-2 mAb trastuzumab (Herceptin®) Citation26,27 to later investigate the activity of the IgG_TCRs on HER-2+ tumor cells and therefore swapped in the trastuzumab variable domains. After expressing and purifying IgG_TCR containing Hα3 at the 2 mL scale, the protein was tested for LC assembly. Modification led to a modest increase in LC assembly (∼75%); however, this increase in HC/LC assembly may be due to stronger VH/VL interactions within trastuzumab compared to the IL-17 mAb. Modification to IgG1 led to a decrease in aggregates based on analytical size exclusion chromatography (SEC), likely because IgG1 is less pH sensitive than IgG4 and suffers less from the acid elution off protein G.Citation28 Next, we trimmed the C-terminal residues of the LC Cβ back to the cysteine that makes the disulfide with the HC (Lβ3, Table S1). Interestingly, the C-terminal residues in this newly truncated Cβ domain strongly resemble those at the C-terminus of human Cκ. The transient HEK293F expression level of Hα3Lβ3 increased (>2-fold) over that of Hα3Lβ2. Additionally, full HC/LC assembly was observed based on the uniform analytical SEC trace () and solution-based SPR measurements (described below).
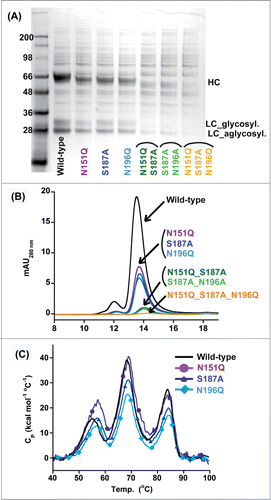
Cα and Cβ harbor 3 and one N-linked glycosylation motifs, respectively. In-house peptide mapping experiments indicate that all 3 Cα sites were fully occupied, while the Cβ site was only partially occupied (). Unlike the glycosylation within the IgG-Fc region (N297), the Cα sites are highly derivatized with terminal galactose and neuraminic acid. Others have shown that certain mouse soluble TCRs can be deglycosylated by mutation and maintain expression and activity.Citation29 We evaluated how mutating each of the Cα glycosylation motifs individually or in tandem would affect protein expression and stability. Single mutation of any one of the 3 Cα motifs reduced transient expression by roughly 2 thirds; double mutation of 2 sites decreased expression by roughly 2 thirds more; and triple mutation of all the sites nearly eliminated all expression (). The singly mutated proteins were monodisperse and as thermally stable as the fully glycosylated protein based on analytical SEC and differential scanning calorimetry (DSC), respectively (). Drastic decreases in expression of the double and triple mutants precluded detailed analyses on these proteins. Due to the overall loss of protein expression observed when removing the glycosylation within the Ca domain, the N-linked glycosylation motifs were preserved in all remaining construct within this report.
Table 2. Occupancy of predicted N-linked glycosylation sites
Figure 3. Effect of deleting N-linked glycosylation on HEK293 expression of IgG_TCR proteins. Reduced SDS-PAGE analysis (A) and analytical SEC (B) of fully glycosylated (WT) IgG_TCR, single N-linked glycosylation deletion mutants, double mutants, and a triple mutant after protein G pull-down from 2 mL HEK293 expression supernatants. (C) DSC analysis of fully glycosylated (WT) and single N-linked glycosylation deletion mutants of IgG_TCR proteins after 100 mL HEK293 scale-up and protein A purification.
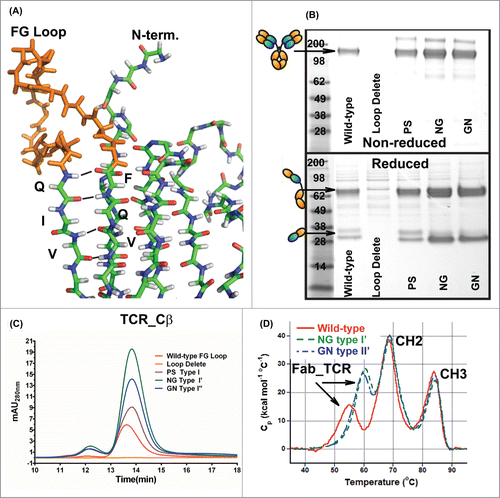
Lastly, we evaluated the effect of replacing the large FG loop of Cβ with canonical β-turn amino acid motifs. In a comparison of Fab and TCR structures (3HC4 and 3QEU, respectively), the large FG loop within Cβ () sterically overlaps with the linkage between Vκ and Cκ. The FG loop is thought to be important for interactions with the TCR-associated protein CD3, but it is inconsequential for our purposes. We therefore replaced the FG loop with canonical Type I (amino acids ‘PS’), I’ (amino acids ‘NG’), and II’ β-turns (amino acids ‘GN’).Citation30 All β-turn replacements improved expression of the IgG_TCR. Replacement with the most common turns, Type I’ or II’, resulted in greatly improved expression and increased thermal stability by DSC ().
Figure 4. Replacement of the FG loop from the β-constant domain with common β-turn motifs. (A) Stick diagram of the structure of the β-constant domain (from pdb 3QEU) where the FG loop is colored orange. Non-reduced (top) and reduced (bottom) SDS-PAGE analyses (B) and analytical SEC (C) of WT IgG_TCR, FG loop-deleted IgG_TCR, and IgG_TCRs with the FG loop replaced with a PS (proline_serine, Type I), NG (Type I’), and GN (Type II’) β-turn. The analyses in (B) and (C) were performed on IgG_TCR proteins expressed at the 2 mL scale in HEK293 and pulled down using protein G magnetic beads. (D) DSC analyses of WT IgG_TCR and FG loop replaced IgG_TCR after scale-up and protein A purification.
Characterization of the IgG_TCR proteins
We generated 2 fully optimized (i.e., truncated N- and C-terminal linkages and FG-loop replaced) IgG_TCRs using both the pertuzumabCitation26,31 and trastuzumab variable domains. The molecules expressed well and were primarily monodisperse by SEC/LS (data not shown). We compared the soluble HER-2 binding kinetics of the IgG_TCRs to the parental IgGs using SPR. The pertuzumab IgG and IgG_TCR proteins were measured to have equilibrium KDs of 3.3 and 2.2 nM, respectively, with similar binding kinetics (Fig. S1A, B). The trastuzumab IgG and IgG_TCR proteins were both measured to have equilibrium KDs of 1.1 nM, again with similar binding kinetics (Fig S1C, D). Importantly, the IgG_TCRs were found to have near identical Rmax values as the IgGs (within 6%), suggesting that both binding arms of the IgG_TCRs had fully assembled HC/LC pairs.
Using the pertuzumab IgG_TCR, we compared the thermal stability of the optimized IgG_TCR Fab with that of the pertuzumab scFv and the traditional Fab from an IgG. Based on DSC measurements, the lowest thermal transition of the IgG_TCR had a midpoint of thermal unfolding (Tm) of 60°C, while that of the scFv was 55°C (Fig. S2). Addition of Cα/Cβ appeared to stabilize the pertuzumab variable domains over what was observed within the scFv without constant domains. However, the stability of the IgG_TCR was still much less than that of the WT Fab, whose Tm was 77°C (Fig. S2). Thus, the Cα/Cβ domains appear to marginally stabilize antibody Fv; however, not to the same extent as the WT CH1/Cκ domains.
We evaluated the pharmacokinetic (PK) profiles of trastuzumab, pertuzumab, and matuzumab Citation32 human IgG1s and compared them to IgG1_TCR versions of the immunoglobulins in Balb/c mice using intravenous injections. While all 3 human IgG1s had long half-lives in Balb/c mice, the IgG_TCRs uniformly demonstrated a rapid initial (α-phase) clearance (Fig. S3). Interestingly, the IgG_TCR remaining in circulation after the initial clearance event appeared to have a long-lived, IgG-like β-phase (Fig. S3). Many hypotheses could explain these observations. One likely hypothesis is that the IgG_TCR bound glycoprotein receptors, potentially in the liver considering the high level of terminal galactose we found to exist within the carbohydrate structures.Citation33,34 The material observed during the long-lived β-phase may represent the amount of IgG_TCR remaining after saturation of the glycoprotein receptors, a slow release from the glycoprotein receptors, or a small population of the IgG_TCR proteins with glycoforms that do not bind strongly to the glycoprotein receptors. The data indicate that IgG_TCR proteins can be long lived in serum, although with decreased overall serum concentrations compared to normal IgG. An interesting future study will be to evaluate the PK properties of Chinese hamster ovary-derived IgG_TCR where there are known differences in N-linked glycan modifications. It is possible there would be a reduction in galactose and neuraminic acid and a change in the initial clearance.
Assembly and activity of IgG-Fab bispecific antibodies containing Cα/Cβ domains
We next evaluated the ability to express 2 different Fabs simultaneously within IgG-Fab BsAbs. The anti-HER-2 mAbs pertuzumab and trastuzumab represent an ideal pair to evaluate in the context of a BsAb. The two mAbs were approved by the Food and Drug Administration in 2013 for co-administration in HER-2 positive metastatic breast cancer, and we sought to evaluate the effect of bonding the 2 antibodies into BsAb molecules with varied antigen-binding geometries. We generated 2 tetravalent IgG-Fab BsAbs for extensive characterization where the additional Fab was placed either N-terminal (N-BsAb) or C-terminal (C-BsAb) to the HC position (Fig. S4). The resulting BsAbs require the co-expression of an extended HC and 2 LCs. The TCR Cα/Cβ domains in one of the Fabs should enable each LC to bind to its correct HC partner. The additional Fab arms were connected via a (G4S)5 or a (G4S)4 linker to the IgG portions of the N-BsAb and C-BsAb, respectively. All the BsAbs contained the optimized V/C linker sequences, the IgG1 upper hinge in place of the HC Cα C-terminus, the trimmed LC Cβ C-terminus, and the FG loop replaced with the Type I’ β-turn.
After expression and purification, initial characterization of the BsAbs included SDS-PAGE, SEC/LS, and binding studies. Both BsAbs had the expected molecular weights of ∼250 kDa based on SDS-PAGE and static light scattering (). The BsAbs were tested for their ability to bind both the pertuzumab and trastuzumab epitopes on HER-2 using a solution-based SPR experiment. Citation35 In the experiment, varying concentrations of the BsAbs were individually pre-mixed with 40 nM HER-2. These solutions were injected over a surface labeled with pertuzumab or trastuzumab IgG. Both BsAbs could inhibit binding of HER-2 to both surfaces, while the monospecific pertuzumab and trastuzumab IgG_TCRs could only block HER-2 from binding to matched surfaces (). The stoichiometry of binding to HER-2 (2:1) for the BsAbs was nearly identical to the individual stoichiometries of the IgG_TCRs.
Figure 5. Biophysical characterization of HER2×HER2 IgG-Fab BsAbs produced using IgG_TCR modalities to direct LC assembly. Panels A and B are non-reduced (left) and reduced (right) SDS-PAGE analysis and analytical SEC, respectively, of C-BsAb and N-BsAbs. Panels E and F are an evaluation of the HER-2 binding properties of trastuzumab IgG_TCR, pertuzumab IgG_TCR, C-BsAb, and N-BsAb by analyzing their ability to block 40 nM HER-2 from binding surfaces labeled with IgG1 trastuzumab (C) or pertuzumab (D). Panels E and F are intact mass spectrometry analyses of C-BsAb and N-BsAb, respectively under reducing conditions. The N-BsAb contained the VL_Y36F mutation and VL_Q38D/VH_Q39K to reduce the affinity of the trastuzumab LC for the pertuzumab Fd containing Cα/Cβ. The spectra show the levels of LC within the IgG-Fab BsAbs. The HC was heavily N- and O-glycosylated; therefore, non-reduced spectra were complex.
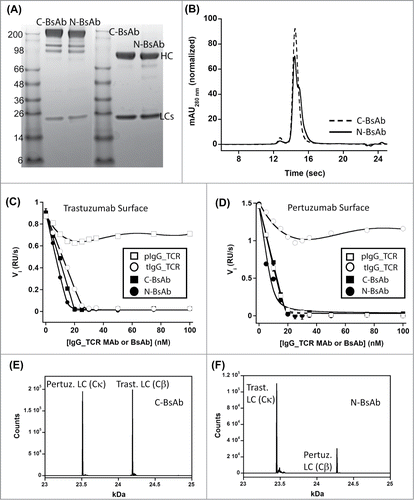
The BsAbs were characterized for their assembly using intact and denatured liquid chromatography/mass spectrometry (LCMS). The molecules were treated with both N- and O-linked glycanases prior to LCMS analyses under reducing conditions. The C-BsAb demonstrated an equal level of each LC liberated from the HC suggesting that the 2 Fd arms of the IgG-Fab HC had their appropriate LCs bound to the HC (). The N-BsAb demonstrated issues with correct assembly. The WT trastuzumab LC dominated binding to all Fd sites on the IgG-Fab HC, including the pertuzumab Fd harboring Cα instead of CH1 (Fig. S5). The C-BsAb and N-BsAb differ in the antibody Fd region that harbors the Cα/Cβ domains. In the C-BsAb, the pertuzumab Fab had the natural CH1/CL domains and the trastuzumab domain harbored Cα/Cβ. We believe the natural pertuzumab LC did not displace the Cα/Cβ-containing trastuzumab LC in the C-BsAb because the pertuzumab VL interaction with VH is weak.Citation12 In the N-BsAb, the trastuzumab LC harbored CH1/CL while the pertuzumab LC harbored Cα/Cβ. The trastuzumab Fab region has a Tm of 85°C measured by DSC (Demarest et al., unpublished data), which is extremely high compared to most human/humanized MAbs Citation36 and suggests a strong VH/VL interaction. We believe the strong VH/VL interaction and high expression level of the natural trastuzumab LC enables it to compete for binding to the pertuzumab Fd containing Cα even with a mismatched constant domain interaction.
To investigate this phenomenon further, we generated a host of additional IgG-Fab BsAbs that use pertuzumab and trastuzumab as the parental mAbs. There are a total of 8 possible N- and C-terminal IgG-Fab BsAb configurations. These configurations depend on (i) which mAb is used as the N- or C-terminal Fab domain and (ii) which antibody Fd region harbors the Cα/Cβ domains (Fig. S4). We constructed the remaining 6 BsAbs, expressed them using transient transfection in HEK293F cells, and purified them by protein A chromatography. The BsAbs containing Cα/Cβ in the trastuzumab Fab region were shown to contain roughly equal levels of trastuzumab and pertuzumab LC (regardless of the N-terminal or C-terminal orientation) suggestive of proper assembly. All BsAbs containing Cα/Cβ in the pertuzumab Fab region were shown to contain a vast majority of trastuzumab LC, much like the N-BsAb that was originally characterized (data not shown).
We hypothesized that weakening trastuzumab VL's ability to bind VH moieties might improve our correct IgG-Fab BsAb assembly. To weaken the trastuzumab VL/pertuzumab VH interaction, trastuzumab's VL was mutated at an interface position (Y36F). To provide additional HC/LC specificity, a charge-charge interaction was added to the trastuzumab Fv (VL_Q38D/VH_Q39K).Citation12 With these modifications, newly generated material had improved pertuzumab LC assembly within the N-BsAb (). The correct assembly, however, did not appear to be complete. The N-BsAb still likely contains a mixture of (i) 2 trastuzumab LCs and 2 pertuzumab LCs (correctly assembled), 3 trastuzumab LCs and 1 pertuzumab LC (incorrectly assembled), and 4 trastuzumab LCs (incorrectly assembled). The N-BsAb preparation was challenging to fully deglycosylate and non-reduced spectra were complex and poorly resolved.
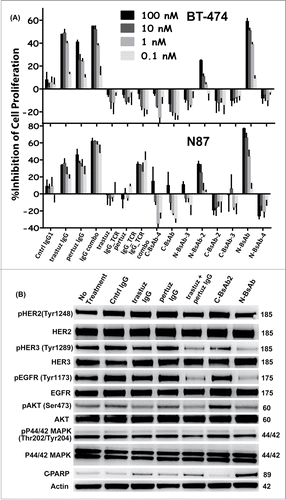
The BsAbs were tested for their ability to inhibit the proliferation of HER2+ BT-474 breast cancer cells and N87 gastric cancer cells. The parental IgGs, monospecific IgG_TCRs, and combinations of the IgGs or IgG_TCRs were used as controls in the cell proliferation assays. In both BT-474 and N87 cells, the majority of BsAbs had little activity or even agonistic activity. Only the BsAb configurations with pertuzumab as an N-terminal Fab moiety demonstrated tumor cell inhibition (). Interestingly, the presence of Cα/Cβ in place of CH1/Cκ in the IgG_TCR trastuzumab or pertuzumab controls led to a change in the behavior, turning them into agonists (). Adding Cα/Cβ must modify the hinge geometry, which modifies trastuzumab's and pertuzumab's activity on HER-2. Similar differences were apparent between BsAbs of similar orientation, but with Cα/Cβ in the other Fab. The best example of this within the BsAbs was between N-BsAb and N-BsAb-2. Both were antagonistic, but N-BsAb led to significantly greater tumor cell inhibition over N-BsAb-2 particularly at high protein concentration () We looked for differences in the signaling behavior of the N-BsAb compared to the other test articles that might help elucidate its superior anti-tumor activity in N87 cells. The N-BsAb mirrored the trastuzumab/pertuzumab combination's ability to decrease EGFR, HER3, Erk and Akt phosphorylation, and to induce some HER-2 downregulation (). The N-BsAb was also found to induce apoptosis as judged by PARP cleavage ().
Figure 6. Biological activity of IgG_TCR BsAbs. The effect of IgG_TCR BsAbs on the FBS-driven proliferation of (A) BT-474 breast cancer cells (top) and N87 gastric cancer cells (bottom). (B) Western blot analyses of the phosphorylated state of EGFR, HER-2, HER-3, Akt, and Erk from N87 tumor cells grown for 48 hours in FBS in the presence of various anti-HER-2 monoclonal and bispecific antibodies. Additionally, the presence of cleaved PARP was evaluated on the blot. Actin was probed to demonstrate the normalized amount of protein loaded into each well.
Discussion
More than 2 decades ago, chimerization of antibody Fabs and TCRs based on their predicted structural similarities was originally described.Citation23,24 These nascent attempts were not pursued further, possibly because the antibody and TCR domains were not fully compatible when making fusions and the task of remolding the IgG and TCR domains to ‘fit’ with one another was a monumental effort given the molecular biology tools at the time. Advances in chemical and PCR-based gene syntheses, as well as optimized expression and purification methodologies, allowed us to perform a ‘next generation’ evaluation of the IgG/TCR chimeras. Clearly, Cα/Cβ orientation, optimization of linker lengths (both N- and C-terminal), and replacement of the FG loop was important to achieve complete HC/LC association in multiple IgG_TCR proteins (i.e., using trastuzumab, pertuzumab, or matuzumab Fv regions). The IgG_TCR format provides a HC/LC interface orthogonal to native HC/LC interfaces and enables the co-expression of 2 Fabs within a single molecule. Other approaches have been described recently for achieving a similar goal, including swapping the orientation of CH1/CL within an IgG (CrossMab approach) Citation15 or the redesign of the HC/LC interface.Citation12 The data presented here demonstrates the first description of IgG_TCR chimeras to achieve a similar goal.
It was our goal to design the IgG_TCR Fabs to have biophysical properties superior to smaller antibody fragments like scFvs that are stripped from their native Fab context. After optimization, the pertuzumab Fv/TCR constant domain fusion did have improved thermal stability over what we observed with the pertuzumab scFv (Fig. S2). The thermal stability improvement was modest and did not approach the optimal stability observed for the native Fab. Additional modifications, either to optimize the Fv/TCR constant domain interface or to stabilize the Cα/Cβ domains themselves would be necessary to obtain a substantial benefit over scFvs. Interestingly, the CH1/CL heterodimer (with Cκ or Cλ) is known to be highly stable,Citation11,12 while very little is known regarding the intrinsic stability of Cα/Cβ. We generated a Cα/Cβ-Fc construct lacking any variable domains and evaluated its thermal stability by DSC (data not shown). Strangely, no unfolding transition was observed for Cα/Cβ even though the construct expressed with Cα/Cβ correctly disulfide-bonded as judged by SDS-PAGE analyses and with an expected molecular weight of 100 kDa based on static light scattering experiments (data not shown). Further experiments evaluating the Cα/Cβ heterodimer expressed in isolation may help shed light on its stability properties and provide a path forward for stabilizing it through protein design.
The utility of IgG_TCR BsAbs as a platform is represented by the novel activity of the HER-2 × HER-2 BsAbs. The concept allows for the conversion of validated antibody therapeutics into BsAbs with varied geometry. These varied geometries can be crucial to achieving the desired function as evidenced by the HER-2×HER-2 BsAbs described here. Engagement of HER-2 using different geometries is known to lead to a spectrum of agonistic and antagonistic activities.Citation37 Only one IgG-Fab BsAb (N-BsAb) described here had equivalent (or even a little better) activity compared to the pertuzumab/trastuzumab combination. Interestingly, the N-BsAb had a much steeper dose-response curve than the mAb combination. We hypothesize that the steep dose-response curve results from the complex nature of tetravalent binding that changes with the BsAb concentration. As the concentration of BsAb increases, it likely competes with itself for binding HER-2 and leads to weaker crosslinking on the surface of the cell, which may in turn result in improved inhibition of HER-2 signaling. However, the results of the experiments with the BsAbs were empirical and highlight how access to multiple BsAb formats can be critical for achieving the desired activity.
The IgG_TCR approach for making BsAbs ideally allows for multiple BsAb formats, such as the IgG-Fabs described here or even IgG BsAbs if combined with a CH3 heterodimer.Citation38 Other approaches for making BsAbs, such as IgG-scFvs require engineering the scFvs for stability and solubility. The use of novel scaffolds that can be appended to IgG or HSA to generate BsAbs requires the discovery and often challenging optimization of these building blocks toward their targets before the BsAbs can be produced. Still, the IgG_TCR BsAbs are not without their flaws. As described above, the stability of IgG_TCR Fabs at present is only marginally improved over IgG-scFvs. Future modifications that improve Cα/Cβ stability would be universal for the platform as opposed to scFv engineering, which is typically performed on a case-by-case basis. The HER2 × HER2 BsAbs described here also revealed the strong influence the variable domains can have on correct HC/LC assembly. Based on sequence and structure, Cα/Cβ and CH1/CL should have no affinity for one another; however, the affinity of trastuzumab's VL for VH domains drove strong HC/LC mispairing that was only partially alleviated through VH/VL specificity designs and VH/VL destabilization. In a recent study,Citation12 we found a similar influence of VH/VL interaction on HC/LC specificity. Variable domain designs such as those described in that studyCitation12 could be applied to reduce HC/LC mispairing in IgG_TCR BsAbs. Like the Fab redesigns described in that study,Citation12 the IgG_TCR chimerics could theoretically be used to generate IgG BsAbs instead of IgG-Fabs. Overall, the work described here provides a first step toward the use of IgG_TCR chimerics to co-express 2 Fab moieties simultaneously for the development of BsAbs for research and therapeutic purposes.
Methods
Molecular biology
Oligonucleotides containing the IgG4, IgG1, Cκ, and some VH and VL genes were amplified by PCR from existing in-house DNA templates. Oligonucleotides encoding Cα, Cβ, and some VH and VL genes were synthesized by IDT. All full gene constructs for subcloning were synthesized using PCR-based overlapping oligonucleotide synthesis.Citation39 The inserts were subcloned into a CMV promotor-driven mammalian expression vector (Lonza). Secretion was driven using a common mouse antibody LC signal sequence that is translated in-frame as part of the expressed protein and cleaved prior to secretion. Plasmid ligations, transformations, DNA preparations were performed using standard molecular biology protocols. The anti-IL-17 mAb was from an in-house, Eli Lilly asset.
For expression, LC and HC plasmids were transfected into HEK293F cells (2:1 ratio for the HC and LC with 1 μg DNA/uL cell culture) using Freestyle transfection reagents (Life Technologies. Transfected cells were grown at 37°C in a 5% CO2 incubator while shaking at 125 rpm. Secreted protein was harvested on day 5 by centrifugation for 5 min. Supernatants were passed through 2 μm filters (both large scale and small scale) for purification.
Protein purification
Small scale (1 mL purifications) were performed by directly incubating 1 mL transfected supernatant with 100 μL resuspended, PBS-washed Protein G magnetic beads (Millipore). Beads were washed 2-times with PBST according to the manufacturer's protocols. Protein was eluted from the beads by adding 130 μL 0.01 M Acetate, pH 3.0. After harvesting, the eluants were immediately neutralized by adding 20 μL 0.1 M Tris, pH 9.0.
Large scale purifications were performed by passing the supernatants over a Protein A affinity-chromatography column using an AKTA Explorer. Protein was eluted using 0.01 M acetate, pH 3.0 and immediately neutralized with 0.1 M Tris, pH 8.0. In some cases a cation exchange step was performed by dilution into 20 mM sodium citrate pH 5.0, capture onto a Mono S column (GE Healthcare), and eluted using a 30 column volume salt gradient up to 1 M NaCl. All proteins were dialyzed against PBS, aliquoted, and stored at 4°C until characterized.
Protein characterization
SDS-PAGE analyses were performed using Novex® 4–20% Tris glycine gels or 3–8% Tris-acetate gels according to manufacturer protocols (Life Technologies). Approximately 5 μg protein was loaded in each well and 10% 0.5 M DTT in H2O was added for reduced samples.
Analytical SEC with in-line light scattering (SEC/LS) was performed for each sample. 30–80 μL of each sample purified directly from 1 mL supernatants or after large scale-purification (concentrations ∼0.2–0.8 mg/mL) were injected onto a Sepax Zenix SEC 200 analytical HPLC (7.8 × 300 mm) column or a Phenomenex Yarra G3000 SEC analytical HPLC (7.8 × 300 mm) column equilibrated in 10 mM phosphate, 150 mM NaCl, 0.02% NaN3, pH 6.8, using an Agilent 1100 HPLC system. Static light scattering data for material eluted from the SEC column were collected using a miniDAWN TREOS static light scattering detector coupled to an Optilab T-rEX in-line refractive index meter (Wyatt Technologies). UV data were analyzed using HPCHEM (Agilent). Molecular weights of the complexes were determined by their static light scattering profiles using ASTRA V (Wyatt Technologies).
Kinetic surface Plasmon resonance (SPR) experiments were performed using a Biacore3000. Anti-IL-17 IgG_TCR proteins (constructs described above within Protein Expression) were captured onto CM5 sensorchip surfaces with an immobilized goat anti-human IgG-Fc polyclonal antibody (Jackson Labs). The goat polyclonal antibody immobilization was achieved by injecting the protein (50 μg/mL, pH 5) over an NHS/EDC activated sensorchip surface followed by blocking with ethanolamine (as described by manufacturer). The anti-IL-17 IgG or IgG_TCRs were captured onto the anti-human Fc sensorchip surface by injecting 10 μL of protein at 0.1 and 0.5 mg/mL using a 2 μL/mL flow rate. Flow rates were increased to 10 μL/min followed by secondary 30 μL injections of IL-17 at 10 and 50 nM. Following a 15 minute dissociation period, the flow rate was increased to 60 μL/min and sensorchip surfaces were regenerated using 2 consecutive 10 μL injections of 0.1 M glycine, pH 2.0.
Solution-based SPR experimentsCitation35 were performed to evaluate the activity of the IgG_TCRs and IgG-Fab BsAbs with pertuzumab and trastuzumab Fvs. Both trastuzumab and pertuzumab IgG were directly immobilized to 3500 RUs onto an NHS/EDC activated CM5 sensor chip surface by injection of a 50 μg/mL solution of each IgG in 10 mM Acetate, pH 5.0. To evaluate the activity/assembly of the pertuzumab and trastuzumab IgG_TCR proteins, and the IgG-Fab BsAbs, 40 nM human HER-2 protein (Speed BioSystems) was mixed with varying concentrations (ranging from 0 nM to 100 nM) of each test article. The trastuzumab and pertuzumab sensorchip surfaces measure the concentration of unbound or free HER-2 ([R]F) in solutions containing 40 nM soluble HER-2 and IgG_TCRs or BsAbs. Unbound HER-2 is equal to the total amount of receptor in solution ([R]T) minus the bound concentration ([R]B). The equilibrium dissociation constant, KD, and binding stoichiometry, n, of the IgGs, IgG_TCRs and IgG-Fab BsAbs for HER-2 were determined using the linear relationship between [R]F and linear slope of RU/time (known as the velocity or Vi) within the first 60 seconds of the HER-2 injection experiment:
where m = slope of the hHER-2-Fc concentration-dependent standard curve and [IgG_TCR]T = total IgG_TCR concentration.
Additionally, thermal stability analyses were performed using differential scanning calorimetry (DSC). DSC scans were performed using an automated capillary DSC (capDSC, MicroCal, LLC). Protein solutions were sampled automatically from 96-well plates using the robotic attachment. Prior to each protein scan, 2 buffer scans were performed to define the baseline for subtraction. All 96-well plates containing protein were stored within the instrument at 2–8ºC. Each sample was diluted to 1.0 mg/mL in PBS. Scans were performed from 10–95ºC at 1.5ºC/min using the low feedback mode. Scans were analyzed using the Origin software supplied by the manufacturer. Subsequent to the subtraction of reference baseline scans, nonzero protein scan baselines were corrected using a third-order polynomial. The unfolding parameters for each protein were deconvoluted using the multi-peak fitting routine within the software assuming non-2-state unfolding behavior.
Glycosylation profiling
Protein samples were concentrated by speed-vac followed by the addition of a denaturing buffer. Denaturing buffer was prepared by mixing Trizma hydrochloride buffer pH 7.4 with 8.0 M guanidine HCl at a 1:9 ratio (v:v). Samples were reduced by the addition of 0.1 M DTT to a final concentration of 10 mM DTT followed by incubation at 37ºC for 1 hour. Samples were then alkylated by the addition of 0.2 M iodoacetamide to a final concentration of 20 mM iodoacetamide and incubated at 25°C for 45 minutes in the dark. The samples were desalted and buffer exchanged with ZebaTM spin desalting columns (7K MWCO) after the columns were equilibrated with digestion buffer (25 mM Trizma, 20 mM CaCl2, pH 7.5). Samples were digested with a 1:20 (w:w) ratio of trypsin at 37ºC overnight. Sample digests were split into 2 equal aliquots. To one aliquot, N-Glycanase (PNGase F) was added and the solution was incubated at 37ºC for 1 hour. Both aliquots were quenched with formic acid and transferred to HPLC vials for LC/MS analysis. Samples were analyzed on Waters Synapt G2 coupled with Acquity UPLC, using UPLC HSS T3 column, 2.1 × 100 mm, 1.8 μm. Mobile phases were 0.2% formic acid in water and 0.2% formic acid in acetonitrile. The Synapt G2 was calibrated with Glu-fibrinopeptide prior to use. Mse and survey scan data were acquired with the lock mass m/z of 556.2771 from Leucine Enkephalin. All data were processed using MassLynx and BiopharmaLynx software (Waters). Glycoform structures were estimated using GlycoMod.Citation40
Intact LC/MS
Intact LC/MS of protein samples was performed essentially as described previously.Citation12 Thirty μL aliquots of the proteins were enzymatically deglycosylated after purification and neutralization by the addition of 1 μL N-Glycanase (Prozyme) for 3–14 hours at 37°C prior to being submitted for LCMS.
Tumor cell lines
The HER-2-positive NCI-N87 (N87) gastric cancer, SK-BR-3 (breast cancer), BT474 (breast cancer), and Calu-3 (lung cancer) tumor cell lines were purchased from the ATCC and cultured according to the guidelines provided by the ATCC.
Tumor cell proliferation
N87 cells were seeded on 96-well plates at 1 × 103 cells per well and precultured in RPMI-1640 medium (growth medium) containing 10% FBS overnight. To evaluate the effect of various test articles on cell proliferation, 100, 10, 1, and 0.1 nM solutions of each test article (or 100 nM+100 nM, 10 nM+10 nM, 1 nM+1 nM, and 0.1 nM+0.1 nM combinations of 2 test articles) in RPMI-1640 medium containing 10% FBS were added to the cells. After 5 d treatment, cell viability was determined with a Cell Titer Glo reagent (Promega). The percentage of growth inhibition was calculated according to the formula [1-(signal with mAb (or BsAb))/(signal with FBS only)]*100.
Signaling studies and receptor downregulation (Western Blots)
N87 cells were plated in 2 mL RPMI-1640 medium at 2.5 × 105 cells per well in sterile 12 well (flat bottom) plates and placed at 37°C, 5% CO2 for 24 hours. The media was removed and replaced with fresh media containing 100 nM of each mAb or BsAb and the cells were allowed to grow for another 48 hours. The media was then aspirated off and the cells were washed with refrigerated PBS. Next, 200 μL refrigerated lysis buffer (MesoScale Discovery) was added and allowed to incubate while mixing gently for 30 minutes at 4°C to loosen the cells from the plate surface. The lysis buffer contained phosphatase inhibitor cocktails as well as protease inhibitor (MesoScale Discovery). The lysed cells were gently homogenized by mixing up and down with a pipettor. The lysate was collected by centrifugation at 10,000 g for 8 minutes at 4°C. The lysates were either used directly for western blot analyses or stored at −70°C for future western blot analyses.
Lysates were analyzed by western blot. Lysates were analyzed for their protein concentration using a BCA assay (Pierce). Lysates were applied to 4–12% Bis-Tris protein mini-gels (NuPAGE) according to the protocols provided by the manufacturer (Life Technologies) under reducing conditions. A SeeBlue Plus2 protein ladder (Life Technologies) and a biotinylated protein ladder (Cell Signaling) were added to the blots to help identify protein molecular weights. The proteins were transferred to nitrocellulose membranes using an iBlot (Life Technologies) using the protocols provided by the manufacturer. The blots were blocked for 1 hour at 25°C. The blots were washed with Tris-buffered saline with 0.1% Tween 20 (TBST). Primary antibodies (anti-blotted proteins) in 5% BSA/TBST were incubated overnight at 4°C gently mixing. The plates were washed again with TBST and secondary antibodies along with anti-biotin HRP for detection of the protein ladder were added in 5% BSA/TBST for 1.5 hours at 25°C. If necessary, blots were washed again and incubated for 1 hour with a secondary detection reagent. The blots were washed again and developed using 2 mL of SuperSignal West Femto Maximum Sensitivity Substrate (Pierce). The proteins were imaged using a UVP imager. For subsequent detection of multiple proteins on the blots, the blots were stripped using ThermoPLUS Restore (ThermoScientific) for 30 minutes at 25°C (if necessary). Primary antibodies included Neu (C-18, HER-2, Santa Cruz Biotechnology [SCB] cat#sc-284), pHER-2 (Y1248, Cell Signaling Technologies [CST] cat#2247), ErbB3 (C-17, HER-3, SBC cat#sc-285), pHER-3 (Y1289, CST cat#4791), EGFR (CST cat#2232), pEGFR (Y1173, CST cat#4407), Erk (MAPK, CST cat#9102), pErk (T202,Y204, CST cat#9101), Akt (CST cat#9272), pAkt (473, CST cat#4058), cleaved PARP (D214, CST cat#9541), anti-biotin HRP (CST cat#7075), β-Actin (Sigma cat#A2228), anti-mouse IgG (CST cat#7076), goat anti-rabbit IgG-HRP (Jackson ImmunoResearch cat#111-035-003).
Pharmacokinetic studies
Phamacokinetics of the IgG_TCR proteins were carried out essentially as described.Citation12
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
supplemental_KMAB_A_1007826.zip
Download Zip (2.8 MB)Acknowledgments
The authors would like to thank Ms. Jayd Hanna and Mr. Benjamin Gutierrez for assistance with transient transfection and 293F cell culture.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol 2010; 10:301-16; PMID:20414204; http://dx.doi.org/10.1038/nri2761
- Fischer N, Leger O. Bispecific antibodies: molecules that enable novel therapeutic strategies. Pathobiology 2007; 74:3-14; PMID:17496428; http://dx.doi.org/10.1159/000101046
- Boersma YL, Chao G, Steiner D, Wittrup KD, Pluckthun A. Bispecific designed ankyrin repeat proteins (DARPins) targeting epidermal growth factor receptor inhibit A431 cell proliferation and receptor recycling. J Biol Chem 2011; 286:41273-85; PMID:21979953; http://dx.doi.org/10.1074/jbc.M111.293266
- Emanuel SL, Engle LJ, Chao G, Zhu RR, Cao C, Lin Z, Yamniuk AP, Hosbach J, Brown J, Fitzpatrick E, et al. A fibronectin scaffold approach to bispecific inhibitors of epidermal growth factor receptor and insulin-like growth factor-I receptor. MAbs 2011; 3:38-48; PMID:21099371; http://dx.doi.org/10.4161/mabs.3.1.14168
- Coloma MJ, Morrison SL. Design and production of novel tetravalent bispecific antibodies. Nat Biotechnol 1997; 15:159-63; PMID:9035142; http://dx.doi.org/10.1038/nbt0297-159
- Demarest SJ, Glaser S. M. Antibody therapeutics, antibody engineering, and the merits of protein stability. Curr Opin Drug Discov Devel 2008; 11:675-87; PMID:18729019
- Dimasi N, Gao C, Fleming R, Woods RM, Yao XT, Shirinian L, Kiener PA, Wu H. The design and characterization of oligospecific antibodies for simultaneous targeting of multiple disease mediators. J Mol Biol 2009; 393:672-792; PMID:19699208; http://dx.doi.org/10.1016/j.jmb.2009.08.032
- Dong J, Sereno A, Snyder WB, Miller BR, Tamraz S, Doern A, Favis M, Wu X, Tran H, Langley E, et al. Stable IgG-like bispecific antibodies directed toward the type I insulin-like growth factor receptor demonstrate enhanced ligand blockade and anti-tumor activity. J Biol Chem 2011; 286:4703-17; PMID:21123183; http://dx.doi.org/10.1074/jbc.M110.184317
- Michaelson JS, Demarest SJ, Miller B, Amatucci A, Snyder WB, Wu X, Huang F, Phan S, Gao S, Doern A, et al. Anti-tumor activity of stability-engineered IgG-like bispecific antibodies targeting TRAIL-R2 and LTbetaR. MAbs 2009; 1:128-41; PMID:20061822; http://dx.doi.org/10.4161/mabs.1.2.7631
- Dong J, Sereno A, Aivazian D, Langley E, Miller BR, Snyder WB, Chan E, Cantele M, Morena R, Joseph IB, et al. A stable IgG-like bispecific antibody targeting the epidermal growth factor receptor and the type I insulin-like growth factor receptor demonstrates superior anti-tumor activity. MAbs 2011; 3:273-88; PMID:21393993; http://dx.doi.org/10.4161/mabs.3.3.15188
- Rothlisberger D, Honegger A, Pluckthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol 2005; 347:773-89; PMID:15769469; http://dx.doi.org/10.1016/j.jmb.2005.01.053
- Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat Biotechnol 2014; 32:191-8; PMID:24463572; http://dx.doi.org/10.1038/nbt.2797
- Spiess C, Merchant M, Huang A, Zheng Z, Yang NY, Peng J, Ellerman D, Shatz W, Reilly D, Yansura DG, et al. Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat Biotechnol 2013; 31:753-8; PMID:23831709; http://dx.doi.org/10.1038/nbt.2621
- Labrijn AF, Meesters JI, de Goeij BE, van den Bremer ET, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci U S A 2013; 110:5145-50; PMID:23479652; http://dx.doi.org/10.1073/pnas.1220145110
- Schaefer W, Regula JT, Bahner M, Schanzer J, Croasdale R, Durr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A 2011; 108:11187-92; PMID:21690412; http://dx.doi.org/10.1073/pnas.1019002108
- Richman SA, Aggen DH, Dossett ML, Donermeyer DL, Allen PM, Greenberg PD, Kranz DM. Structural features of T cell receptor variable regions that enhance domain stability and enable expression as single-chain ValphaVbeta fragments. Mol Immunol 2009; 46:902-16; PMID:18962897; http://dx.doi.org/10.1016/j.molimm.2008.09.021
- Brocker T, Karjalainen K. Adoptive tumor immunity mediated by lymphocytes bearing modified antigen-specific receptors. Adv Immunol 1998; 68:257-69; PMID:9505091; http://dx.doi.org/10.1016/S0065-2776(08)60561-1
- Porter DL, Levine BL, Kalos M, Bagg A, June CH. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N Engl J Med 2011; 365:725-33; PMID:21830940; http://dx.doi.org/10.1056/NEJMoa1103849
- Kuwana Y, Asakura Y, Utsunomiya N, Nakanishi M, Arata Y, Itoh S, Nagase F, Kurosawa Y. Expression of chimeric receptor composed of immunoglobulin-derived V regions and T-cell receptor-derived C regions. Biochem Biophys Res Commun 1987; 149:960-8; PMID:3122749; http://dx.doi.org/10.1016/0006-291X(87)90502-X
- Gross G, Waks T, Eshhar Z. Expression of immunoglobulin-T-cell receptor chimeric molecules as functional receptors with antibody-type specificity. Proc Natl Acad Sci U S A 1989; 86:10024-8; PMID:2513569; http://dx.doi.org/10.1073/pnas.86.24.10024
- Brocker T, Riedinger M, Karjalainen K. Redirecting the complete T cell receptor/CD3 signaling machinery towards native antigen via modified T cell receptor. Eur J Immunol 1996; 26:1770-4; PMID:8765019; http://dx.doi.org/10.1002/eji.1830260816
- Brocker T, Peter A, Traunecker A, Karjalainen K. New simplified molecular design for functional T cell receptor. Eur J Immunol 1993; 23:1435-9; PMID:8325320; http://dx.doi.org/10.1002/eji.1830230705
- Traunecker A, Dolder B, Karjalainen K. A novel approach for preparing anti-T cell receptor constant region antibodies. Eur J Immunol 1986; 16:851-4; PMID:3087759; http://dx.doi.org/10.1002/eji.1830160722
- Seimiya H, Naito M, Mashima T, Yasui H, Kuwana Y, Kurosawa Y, Tsuruo T. T cell receptor-extracellular constant regions as hetero-cross-linkers for immunoglobulin variable regions. J Biochem 1993; 113:687-91; PMID:8370665
- Feige MJ, Groscurth S, Marcinowski M, Shimizu Y, Kessler H, Hendershot LM, Buchner J. An unfolded CH1 domain controls the assembly and secretion of IgG antibodies. Mol Cell 2009; 34:569-79; PMID:19524537; http://dx.doi.org/10.1016/j.molcel.2009.04.028
- Nahta R, Hung MC, Esteva FJ. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res 2004; 64:2343-6; PMID:15059883; http://dx.doi.org/10.1158/0008-5472.CAN-03-3856
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature 2003; 421:756-60; PMID:12610629; http://dx.doi.org/10.1038/nature01392
- Demarest SJ, Hopp J, Chung J, Hathaway K, Mertsching E, Cao X, George J, Miatkowski K, LaBarre MJ, Shields M, et al. An intermediate pH unfolding transition abrogates the ability of IgE to interact with its high affinity receptor FcepsilonRIalpha. J Biol Chem 2006; 281:30755-67; PMID:16905745; http://dx.doi.org/10.1074/jbc.M605190200
- Kuball J, Hauptrock B, Malina V, Antunes E, Voss RH, Wolfl M, Strong R, Theobald M, Greenberg PD. Increasing functional avidity of TCR-redirected T cells by removing defined N-glycosylation sites in the TCR constant domain. J Exp Med 2009; 206:463-75; PMID:19171765; http://dx.doi.org/10.1084/jem.20082487
- Gunasekaran K, Ramakrishnan C, Balaram P. Beta-hairpins in proteins revisited: lessons for de novo design. Protein Eng 1997; 10:1131-41; PMID:9488138; http://dx.doi.org/10.1093/protein/10.10.1131
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2004; 5:317-28; PMID:15093539; http://dx.doi.org/10.1016/S1535-6108(04)00083-2
- Schmiedel J, Blaukat A, Li S, Knochel T, Ferguson KM. Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization. Cancer Cell 2008; 13:365-73; PMID:18394559; http://dx.doi.org/10.1016/j.ccr.2008.02.019
- Sethuraman N, Stadheim TA. Challenges in therapeutic glycoprotein production. Curr Opin Biotechnol 2006; 17:341-6; PMID:16828275; http://dx.doi.org/10.1016/j.copbio.2006.06.010
- Lepenies B, Lee J, Sonkaria S. Targeting C-type lectin receptors with multivalent carbohydrate ligands. Adv Drug Deliv Rev 2013; 65:1271-81; PMID:23727341; http://dx.doi.org/10.1016/j.addr.2013.05.007
- Day ES, Cachero TG, Qian F, Sun Y, Wen D, Pelletier M, Hsu YM, Whitty A. Selectivity of BAFF/BLyS and APRIL for binding to the TNF family receptors BAFFR/BR3 and BCMA. Biochemistry 2005; 44:1919-31; PMID:15697217; http://dx.doi.org/10.1021/bi048227k
- Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun 2007; 355:751-7; PMID:17321501; http://dx.doi.org/10.1016/j.bbrc.2007.02.042
- Scheer JM, Sandoval W, Elliott JM, Shao L, Luis E, Lewin-Koh SC, Schaefer G, Vandlen R. Reorienting the Fab domains of trastuzumab results in potent HER2 activators. PLoS One 2012; 7:e51817; PMID:23284778; http://dx.doi.org/10.1371/journal.pone.0051817
- Jost C, Pluckthun A. Engineered proteins with desired specificity: DARPins, other alternative scaffolds and bispecific IgGs. Curr Opin Struct Biol 2014; 27:102-12; PMID:25033247; http://dx.doi.org/10.1016/j.sbi.2014.05.011
- Casimiro DR, Wright PE, Dyson HJ. PCR-based gene synthesis and protein NMR spectroscopy. Structure 1997; 5:1407-12; PMID:9384559; http://dx.doi.org/10.1016/S0969-2126(97)00291-8
- Cooper CA, Gasteiger E, Packer NH. GlycoMod–a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 2001; 1:340-9; PMID:11680880; http://dx.doi.org/10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B
