 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
A myriad of innovative bispecific antibody (BsAb) platforms have been reported. Most require significant protein engineering to be viable from a development and manufacturing perspective. Single-chain variable fragments (scFvs) and diabodies that consist only of antibody variable domains have been used as building blocks for making BsAbs for decades. The drawback with Fv-only moieties is that they lack the native-like interactions with CH1/CL domains that make antibody Fab regions stable and soluble. Here, we utilize a redesigned Fab interface to explore 2 novel Fab-based BsAbs platforms. The redesigned Fab interface designs limit heavy and light chain mixing when 2 Fabs are co-expressed simultaneously, thus allowing the use of 2 different Fabs within a BsAb construct without the requirement of one or more scFvs. We describe the stability and activity of a HER2×HER2 IgG-Fab BsAb, and compare its biophysical and activity properties with those of an IgG-scFv that utilizes the variable domains of the same parental antibodies. We also generated an EGFR × CD3 tandem Fab protein with a similar format to a tandem scFv (otherwise known as a bispecific T cell engager or BiTE). We show that the Fab-based BsAbs have superior biophysical properties compared to the scFv-based BsAbs. Additionally, the Fab-based BsAbs do not simply recapitulate the activity of their scFv counterparts, but are shown to possess unique biological activity.
Abbreviations
| BiTE | = | bispecific T cell engager |
| BsAb | = | bispecific antibody |
| CD | = | circular dichroism |
| DSC | = | differential scanning calorimetry |
| Fab | = | antigen binding antibody fragment |
| Fv | = | variable domains antibody fragment |
| HC | = | antibody heavy chain |
| LC | = | antibody light chain |
| LCMS | = | liquid chromatography with in-line mass spectrometry |
| mAb | = | monoclonal antibody |
| scFv | = | single chain Fv |
| SEC-LC | = | size exclusion chromatography with in-line static light scattering |
| Tm | = | temperature at the midpoint of thermal unfolding |
Introduction
Genomic and proteomic profiling have provided evidence of the complexity of multifaceted diseases such as cancer and autoimmunity. Targeted therapeutics such as monoclonal antibodies (mAbs) typically intervene in single molecular pathways. Although they have provided substantial benefits in various diseases, many targeted therapeutics have limited efficacy or durability in complex diseases. For example, the multiplicity of genetic alterations and strong mutagenic potential of many cancers allows them to circumvent targeted pathway inhibition.Citation1-3 Therapies that provide more complex mechanisms of disease intervention may enable enhanced efficacy. Bispecific antibodies (BsAbs) are an emerging class of therapeutics that may fulfill this need because they allow the engagement of multiple targets and more complex intervention mechanisms.Citation4,5
Recombinant DNA technologies have enabled the generation of many different types of BsAbs. Some of the common BsAb formats are made possible through the use of single chain Fv (scFv) building blocks Citation6 taken from existing mAb therapeutics or discovered using in vitro screening methodologies. The Fv region contains the antigen recognition domains (VH and VL) of a mAb and tethers them together using a flexible or structured linker. BsAbs formats using scFvs or disulfide bonded Fvs include tandem scFvs (often used as bispecific T cell engagers or ‘BiTEs’), tetravalent IgG-scFvs, diabodies, and many other formats.Citation7-10 An issue with these Fv moieties is that they lack the native Fab (antigen binding fragment) architecture found within the majority of mammalian immunoglobulins. The native architecture provides the stabilizing interactions of heavy and light chain constant domains, CH1 and CL, respectively.Citation11,12 The lack of these stabilizing domains can lead to compromised thermal stability or solubility and an increased potential for aggregation within BsAbs containing isolated Fv fragments.Citation12,13
Replacement of the commonly used scFv moiety with a more stable Fab moiety may make these BsAb formats more stable and well-behaved. Recently, we generated a novel Fab interface that enabled 2 Fab moieties to be expressed simultaneously without heavy chain (HC)/light chain (LC) mixing.Citation14 We demonstrated the use of this methodology for generating BsAbs with native-like IgG architecture.Citation14 Here, we utilize these Fab interface designs to enable the expression and correct assembly of IgG-Fab (instead of IgG-scFv) and tandem Fab (instead of tandem scFv) BsAbs. Such multivalent molecules have been described previously, but only with monospecific constructs.Citation15 The bispecific versions of these formats lead to HC/LC mispairing, but this problem can be mitigated using the Fab interface designs.Citation14 We demonstrate the robust biophysical properties of these BsAbs and their potential utility in various oncology applications.
Results
Construction and assembly of an IgG-Fab BsAb against HER-2
The most common tetravalent bispecific platforms are the IgG-scFv Citation8,16-19 and DVD-Ig.Citation20,21 Both formats contain Fv regions stripped from their natural VH_CH1/VL_CL Fab format. We wished to evaluate an IgG-Fab BsAb similar to the IgG-scFv BsAb (). IgG-Fab BsAbs require a 3 chain transfection (HC, LC1, and LC2) compared to the other tetravalent formats that require an antibody-like 2 chain transfection. For proof-of-concept, we wished to combine the activities of trastuzumab (HerceptinTM) and pertuzumab (PerjetaTM), which were approved for use as combination therapy in 2013 in advanced metastatic HER-2-positive breast cancer.Citation22
Figure 1. (A) Schematic diagrams of pertuzumab and trastuzumab IgG1 and the pertuzumab × trastuzumab IgG-Fab and IgG-scFv BsAbs. Notice the N-terminal Fab of the IgG-Fab contains a fully intact pertuzumab Fab with VH, VL, CH1, and CL, while the N-terminal scFv contains only a VH and VL domain of pertuzumab. (B) Analytical SEC of the IgG-scFv and IgG-Fab BsAbs after expression in 293F cells and a single step protein A purification. (C) DSC results with the HER2 × HER2 IgG-Fab (bottom) and IgG-scFv (top). The unfolding events of the various domains within the 2 molecules are annotated. The Fc (CH2 and CH3) unfolding events for IgG1 are well characterized.Citation47 The trastuzumab and pertuzumab Fab unfolding curves were characterized previously Citation14 and their unfolding events within the IgG-Fab can be deduced based on the data with the monoclonal antibodies. For the IgG-scFv, one of the variable domains of pertuzumab unfolds at a much lower Tm resulting from the lack of the CH1/CL domains, a trait typical of scFvs.Citation12
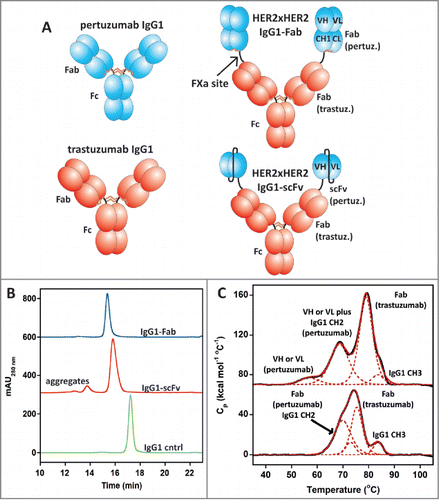
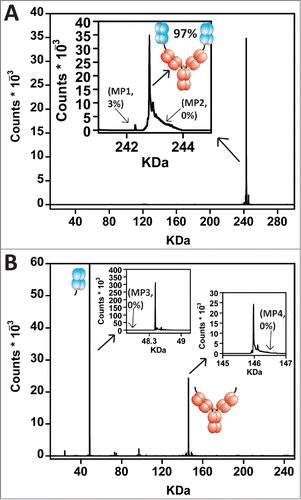
We built an IgG-Fab BsAb containing the LC specificity designs described previously Citation14 and an IgG-scFv BsAb for comparison. We chose to utilize pertuzumab as the N-terminal Fab or scFv and trastuzumab as the IgG based on a previous study that evaluated the orientational effects of N-terminal and C-terminal fusions of pertuzumab and trastuzumab.Citation23 Both molecules expressed at levels comparable to their parental mAbs in the transient HEK293F system (Life Technologies). The BsAbs were purified using a single affinity chromatography step and characterized by analytical size exclusion chromatography (SEC) with in-line static light scattering (SEC-LS). The IgG-scFv BsAb had the expected molecular mass of 200 kDa based on static light scattering measurements and demonstrated a propensity to form aggregates. The IgG-Fab had the expected molecular mass of 250 kDa and was monodisperse with low aggregates similar to IgG control proteins (). Differential scanning calorimetry (DSC) studies of the 2 BsAbs demonstrated that the pertuzumab scFv within the IgG-scFv unfolded at a lower temperature (Tm (midpoint of unfolding) = 57°C) than the pertuzumab Fab within the IgG-Fab (Tm = 70°C, ). To evaluate whether the LCs were assembled properly within the IgG-Fab, we analyzed the protein using intact mass spectrometry (MS) where the molecular mass of the entire oxidized molecule is determined. The protein had the molecular weight expected for 2 HCs, 2 trastuzumab LCs, and 2 pertuzumab LCs (). To specifically demonstrate that the LCs correctly bound their cognate HC Fd regions (i.e., VH_CH1), we engineered a Factor Xa-specific cleavage site between the N-terminal Fab and the IgG of the IgG-Fab BsAb. The IgG-Fab BsAb was cleaved using Factor Xa and analyzed by MS. The Fab and IgG regions of the spectrum were found to contain the correct LC/Fd pairing (), indicating that the LC specificity designs were successful in directing the LCs to the correct HC Fd.
Figure 2. (A) Intact mass analysis of the IgG-Fab BsAb (aglycosyl). The expected molecular weight was observed for containing 2 pertuzumab LCs, 2 trastuzumab LCs, and 2 HCs. The inset is a zoomed in view of the intact IgG-Fab peak. Visible is a 3% impurity of material containing 1 pertuzumab LC and 3 trastuzumab LCs denoted mispair#1 or ‘MP1.’ Also shown is the expected molecular weight for material containing 3 pertuzumab LCs and 1 trastuzumab LC denoted ‘MP2’with an arrow pointing to where it would be expected on the graph. (B) To show that the LCs bound the proper HC Fd region, we utilized an engineered FXa site between the N-terminal pertuzumab Fab and the trastuzumab IgG to cleave them apart. Intact mass analysis found only properly HC/LC assembled fragments. The insets are zoomed in views of the intact pertuzumab Fab and intact trastuzumab IgG fragments to demonstrate their purity. The arrow attached to ‘MP3’ points to the expected molecular weight of a pertuzumab Fd fragment with a mispaired trastuzumab LC. The arrow attached to ‘MP4’ shows the expected molecular weight of a trastuzumab IgG with a mispaired pertuzumab LC. No evidence of MP3 nor MP4 was observed.
Activity of anti-HER-2 IgG-Fab BsAb
To confirm the tetravalent binding activity of the IgG-Fab and IgG-scFv BsAbs, we performed solution-based competition studies using surface plasmon resonance (SPR). Since the BsAbs both comprise mAbs directed toward HER-2, we separately tested the BsAbs’ ability to inhibit HER-2 binding to sensorchip surfaces labeled with pertuzumab IgG1 or trastuzumab IgG1. The assay format enables measurement of the stoichiometry of inhibition, which is a gauge of the valency of each BsAb toward each epitope. The pertuzumab and trastuzumab IgG1s could only block binding to matched sensorchip surfaces and actually led to increased signal when combined with HER-2 antigen and tested on the orthogonal surface, likely the result of simultaneous binding of soluble IgG and surface-bound IgG to HER-2 antigen (). The pertuzumab IgG appeared to inhibit HER-2 binding to its own surface at a valency slightly above the BsAbs (∼11 nM), but within experimental error. Both the IgG-Fab and IgG-scFv BsAbs demonstrated a slope of inhibition of ∼17 nM in the presence of 40 nM HER-2 on both the pertuzumab () and trastuzumab () sensorchip surfaces indicative of tetravalent activity. The fact that all inhibition curves hit the x-axis at competitor concentrations slightly less than 20 nM is likely due to the HER-2 concentration being slightly off or the HER-2 protein not being fully intact. Both the IgG-Fab and IgG-scFv BsAbs also demonstrated a shallow increase in signal on the sensorchip surfaces with increased concentration. When titrating the IgG-Fab and IgG-scFv BsAbs onto the sensorchip surfaces in the absence of HER-2, we observed a shallow association with the pertuzumab and trastuzumab surfaces (and not to a control IgG1 surface) that increased with BsAb concentration and roughly matched the high concentration signals observed in the presence of HER-2 ().
Figure 3. (A and B) Solution-based binding and stoichiometry studies with the HER2xHER2 BsAbs. (A) Titration of IgGs and BsAbs evaluating their ability to block soluble HER-2 binding to a sensor chip surface labeled with pertuzumab. The initial velocity (Vi, y-axis) is the initial slope of binding (initial 40 seconds) of HER-2 in the presence of mAb or BsAb and is a measure of the free HER-2 concentration. (B) Same experiment except the sensorchip surface was labeled with trastuzumab. (C) Inhibition of N87 gastric cancer cell growth by the parental trastuzumab and pertuzumab IgGs, a combination of the IgGs, the IgG-Fab BsAb and the IgG-scFv BsAb.
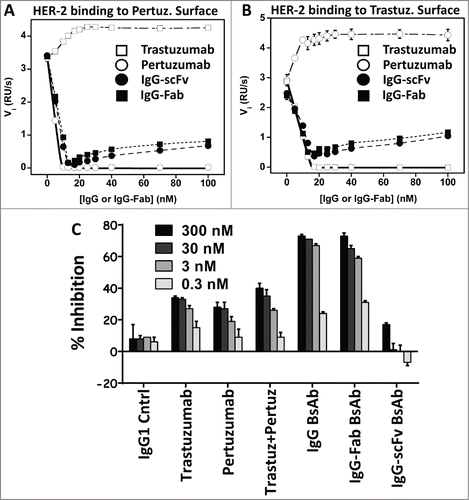
The BsAbs were tested for their ability to inhibit the in vitro growth of the HER-2 positive N87 gastric cancer cell line in the presence of cell culture media and 10% fetal bovine serum (FBS) over a 5 day period. Both pertuzumab and trastuzumab led to modest (30–40%) decreases in N87 cell growth and the mAb combination enhanced the inhibition (50%, ). The IgG-scFv BsAb did not inhibit cell growth, although in repeat experiments we observed signals for the IgG-scFv as high as the parental mAbs. The IgG-Fab dramatically decreased cell growth (∼80%) at the saturated dose as did a described previously IgG BsAb that combined the pertuzumab and trastuzumab parental IgGs ().Citation14
Construction and assembly of a tandem Fab BsAb against epidermal growth factor receptor and CD3
Next, we utilized the Fab redesigns Citation14 to generate a novel tandem Fab molecule. Tandem scFvs have been used for nearly 20 years to redirect T cells to target and kill other cell types using antigens on their surfaces.Citation24,25 These tandem scFvs (called bispecific T cell engagers or BiTEs) have been validated in the clinic with the most advanced being blinatumomab (BLINCYTO), a CD19 × CD3 BiTE approved in the US for the treatment of Philadelphia chromosome-negative relap-sed or refractory B-cell precursor acute lymphoblastic leukemia and in Phase 3 clinical trials for refractory non-Hodgkin lymphoma.Citation26,27 We wished to see if a tandem Fab molecule would: 1) be more stable/manufacturable than a tandem scFv containing 2 separate scFvs each carrying the liabilities of scFv moieties; and 2) maintain the activity of the tandem scFv format that has been proposed to work well by ideally bridging the immune synapse between the T cell and tumor cell through its relatively small molecular size.Citation25 As a proof-of-concept, we utilized the anti-epidermal growth factor receptor (EGFR) mAb matuzumab Citation28 and an in-house anti-human CD3ε mAb to generate both a tandem Fab and a tandem scFv that could be compared and contrasted against one another (). The tandem scFv was built with the anti-EGFR scFv at the N-terminus connected to the anti-CD3 scFv via a 5 amino acid G4S linker designed to disfavor inappropriate VH/VL mispairing. The tandem Fab was built to assemble as a 3 chain construct with 2 LCs expressed separately and the 2 HC Fds (VH and CH1) expressed as a single chain with a (G4S)3 linker between the 2 Fd regions. The EGFR Fd was at the N-terminus. The upper hinge (IgG1) was included in both Fd regions of the tandem Fab to allow covalent linkage with the LCs. Both contained C-terminal 8×histidine tags (on the HC for the tandem Fab).
Figure 4 (see previous page). (A) Schematic diagrams of the matuzumab (anti-EGFR) IgG1 mAb, in-house anti-CD3 IgG1agly (N297Q mutation) mAb, and the EGFR × CD3 tandem Fab and tandem scFv BsAbs. (B) Analytical SEC of the purified BsAbs along with an IgG1 control mAb. The tandem Fab was purified using a single step NiCitation2+-NTA affinity chromatography step, while the tandem scFv required the affinity step along with a preparative SEC step to remove aggregates and lower molecular weight cleavage products. (C and D) Intact mass analyses of the tandem Fab and tandem scFv, respectively. The inserts show a zoomed in view of the intact proteins demonstrating the purity. A 6% impurity within the tandem Fab, which contained 2 anti-CD3 LCs. (E) Thermal unfolding profiles of the EGFR×CD3 tandem Fab (•) and tandem scFv (♦) BsAbs monitored by CD. Even though both BsAbs could have multiple unfolding transitions, they both displayed only one that could be fit using a 2-state approximation to obtain Tm. The tandem Fab exhibited a Tm of 73.6°C while the tandem scFv exhibited a Tm of 56.7°C.
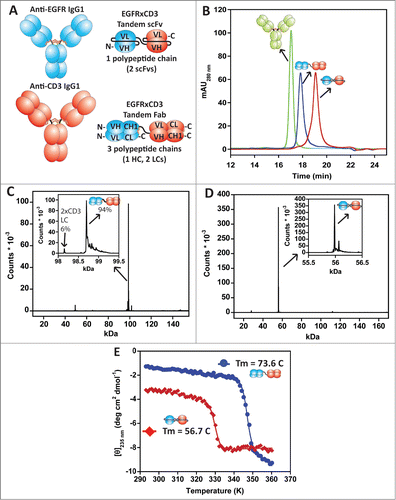
The tandem Fab and tandem scFv production was very different. Both were expressed transiently in HEK293F. The tandem Fab expressed well and was >98% monomer directly off a histag affinity resin with a 35 mg/L purified yield that needed no further purification. The tandem scFv expressed modestly and contained 17% aggregates (mostly dimer/trimer) directly off the histag affinity resin. Lower molecular weight peaks were also apparent – some of them putatively cleavage products of the tandem scFv. The tandem scFv was further purified by preparative SEC with a final purified yield of 4.4 mg/L (8-fold lower than the tandem Fab). Analytical SEC chromatograms of the final purified forms of the tandem Fab and tandem scFv are shown in . MS analyses of the tandem Fab and tandem scFv were performed to confirm that they were both intact. Both were found to have their expected molecular weights (). Roughly 6% of the tandem Fab (based on detector counts) was found to contain 2 anti-CD3 LCs instead of both the anti-EGFR and anti-CD3 LCs. The thermal stability of the tandem scFv was also significantly lower than what was observed for the tandem Fab based on temperature-induced unfolding monitored by circular dichroism (CD) (). Both BsAbs exhibited apparent single unfolding transitions comprising the unfolding of all the protein domains. The Tm of the tandem scFv was 56.7°C, and the Tm of the tandem Fab was 73.6°C if fit to a 2-state transition,Citation29 although the unfolding was almost certainly not reversible or 2-state. Thus, the expression, purity, and thermal stability of the tandem Fab was superior to that of the tandem scFv.
Activity of EGFR×CD3 tandem Fab and tandem scFv BsAbs
Next, we evaluated the binding activity of the EGFR×CD3 tandem Fab and tandem scFv BsAbs. Kinetic SPR was used to evaluate the binding to CD3 and EGFR separately. Each was found to have equivalent binding kinetics to the 2 antigens (, ). Therefore, binding kinetics are unlikely to be a discriminating factor between the biological activity of the 2 formats. A low level of heterogeneous binding was observed for the EGFR kinetics, which led to imperfections in the fits with both BsAbs. Possible explanations could be non-specific binding or differential folding/epitope exposure within the EGFR extracellular domain protein (). Similar, but exaggerated, heterogeneity was observed when using commercial human EGFR-Fc (R&D systems, data not shown).
Figure 5. (A–D) Binding of soluble, monomeric CD3εγ and EGFR extracellular domains (3 to 50 nM) to immobilized tandem Fab (A, B) and tandem scFv (C, D), respectively, using SPR. Each concentration was run in duplicate and the curves were fit to a 1:1 binding model to derive ka, kd, and KD.
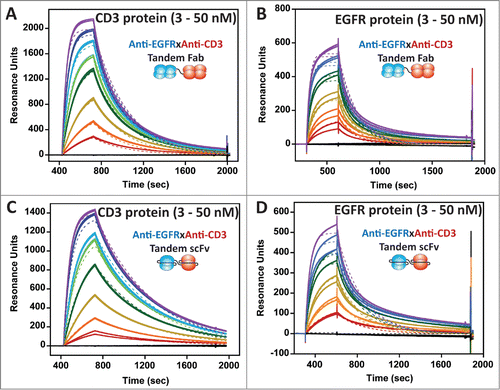
Table 1. Kinetic EGFR and CD3 protein binding data for the tandem Fab and tandem scFv BsAbs
The tandem Fab and tandem scFv BsAbs were tested for their ability to redirect activated T cells to kill tumor cells. T cells from a healthy human donor were purchased and cultured in the presence of anti-CD3, anti-CD28 and IL-2 for at least 5 days for activation. The CD4/CD8 profile of the activated T cells as well as markers of their activation (CD69/CD25) is provided in . The BsAbs, the parental mAbs (anti-CD3 and anti-EGFR), as well as the combination of the parental mAbs were separately mixed with activated T cells and applied to A431 tumor cells, which were measured using a QuantiBRITE assay to have an average of 160,000 cell surface EGF receptors per cell.Citation14 The ability of the tandem Fab BsAb to redirect the activated T cells to kill the tumor cells was assessed using 2 separate methods. The first method used CSFE labeling of the tumor cells followed by incubation with the T cells/tandem Fab BsAb or control mAbs for 12 hours followed by FACS analysis using 7-aminoactinomycin (7-AAD) to visualize dead or dying tumor cells. A small trend toward slightly higher killing was observed with the anti-CD3 mAb (alone or in combination) over the control IgG, perhaps the result of non-BsAb-mediated T cell killing (). The EGFR×CD3 tandem Fab resulted in significantly greater tumor cell killing (∼80%) compared to the controls demonstrating its ability to redirect activated T cells to kill the tumor cells().
Figure 6. (A) Flow cytometry histograms of the population of dead EGFR + A431 tumor cells (quadrant 2, Q2) after a 12 hour incubation in the presence of 100 nM of each article, mAb or BsAb, using a 15:1 E:T cell ratio. (B) Redirected tumor cell lysis activity after a 5 hour incubation of A431 tumor cells with pre-activated T cells (1:10 ratio) monitored with a cell viability assay. The BsAb curves were fit to a standard 4 parameter sigmoidal equation described in the methods.
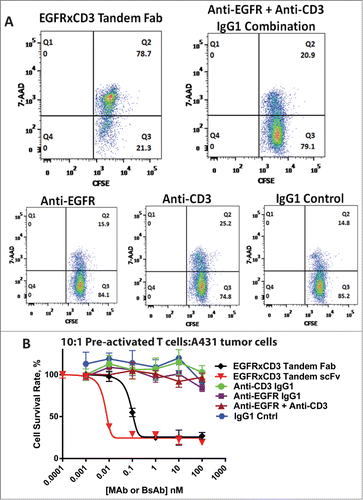
Table 2. T cell activation by the BsAbs in the presence of tumor cells
Next, the tandem Fab and tandem scFv BsAbs were titrated in the presence of the activated T cells and their redirected lysis activity was assessed using a tumor cell viability assay. Both BsAbs redirected T cells to kill the tumor cells with high potency; however, the apparent potency was roughly 10-fold better for the tandem scFv (EC50 < 10 pM for the tandem scFv vs. <100 pM for the tandem Fab, ). One small difference between the tandem scFv and tandem Fab was the length of the linker sequence between the scFvs (G4S)1 and the Fabs (G4S)3. We shortened the tandem Fab linker to (G4S)1 and expressed, purified and tested its redirected lysis activity. Modification of the linker length within the tandem Fab had no effect on the potency of the tandem Fab redirected lysis activity (data not shown).
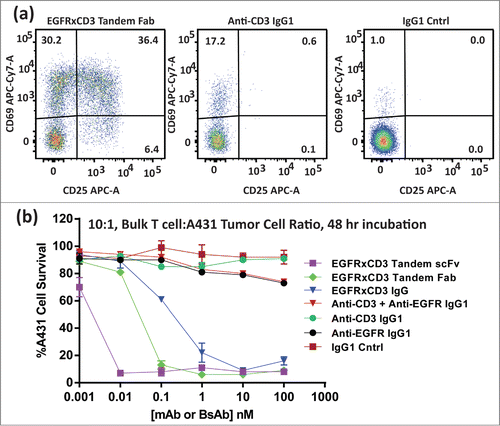
The tandem Fab and tandem scFv BsAbs were further tested for their ability to recruit and activate bulk non-stimulated T cells in the presence of tumor cells. In this study, untreated T cells from a healthy human donor were thawed and immediately mixed with either the BsAbs or control mAbs just prior to their addition to A431 cells. After 48 hours of incubation, the viability of the tumor cells and the activated status of the T cells were evaluated. Both BsAbs led to strong activation in the majority of T cells based on increased CD69/CD25 expression (, ). A minority of the T cells demonstrated CD69 upregulation (early activation) within both the anti-CD3 and anti-CD3/anti-HER-2 combination arms, likely due to the bivalent nature of the aglycosyl anti-CD3 mAb (, ). Only the BsAbs led to significant tumor cell lysis via T cell recruitment and activation (). Low level T cell activation by anti-CD3 alone did not lead the T cells to kill A431 tumor cells. We included an EGFR×CD3 IgG BsAb described previously Citation14 into the assay. While each of the BsAbs demonstrated equivalent binding kinetics to both EGFR and CD3, the BsAb format appeared to significantly effect BsAb potency.
Figure 7. (A) Flow cytometry histograms of CD69 and CD25 activation marker levels on CD8 positive T cells after a 48 hour incubation in the presence of EGFR + A431 cells and 1 nM EGFR × CD3 tandem Fab (left), Anti-CD3 IgG1 mAb (center), or an IgG1 control mAb (right). Numbers in each quadrant of the histrograms indicate the relative population of cells displaying each marker or both markers (upper right). (B) The percent cell survival of A431 cells incubated for 48 hours in the presence of various test articles and bulk (non-pretreated) T cells. Included in the assay was an IgG BsAb described previously.Citation14 The lines between points were simple interpolations (no fit).
Discussion
One of the goals of the current study was to compare the biophysical properties of BsAbs using Fab moieties versus those using scFvs to gain the multiple specificities. The ability to specifically direct HC/LC assembly when co-expressing 2 Fab moieties Citation14 enables the use of Fabs as the building block of BsAbs compared to the commonly used scFv. The stability and solubility benefit provided to the VH/VL domains by the CH1/CL domains has been described in many reports Citation12 and a dissection of the stabilizing effects was performed by Rothlisberger and coworkers.Citation11 We show that even with substantial changes to the interface within the Fab moiety based on the Fab designs,Citation14 the Fab moieities within both the IgG-Fab and tandem Fab BsAbs are substantially more thermally stable than their scFv counterparts within the IgG-scFv and tandem scFv BsAbs. Like common IgGs, the Fab-based BsAbs showed little to no propensity to form aggregates, while the scFv-based BsAbs both demonstrated significant tendencies toward aggregation. Attenuated stability and a propensity to aggregate is likely linked to the tandem scFv's poor expression. Based on their biophysical superiority over scFv-based BsAbs, we believe Fab-based BsAbs will be an attractive avenue as therapeutic agents.
Many new anti-CD3-based BsAbs with varied formats are demonstrating the ability to redirect and activate T cells to target tumors expressing the tumor antigen of the BsAb. The most common BsAb format for redirecting T cells to lyse target cells is the tandem scFv because it is thought to recapitulate the immune synapse endogenously formed between T cells and target cell MHC proteins.Citation30,31 Other dual Fv-based moieties like Dual-Affinity Re-Targeting (DART) molecules are thought to derive their activity and potency through a similar mechanism.Citation32 New BsAb formats with very different geometries have recently shown their utility for T cell activation/redirected lysis such as bivalent, bispecific IgGs and others.Citation33,34 Here, we show that tandem Fab anti-CD3 BsAbs have the capacity to redirect and activate T cells to kill antigen presenting cells. T cell redirection and tumor cell lysis are not necessarily linked as shown by anti-CD8 or anti-CD4-based BsAbs described recently that can redirect activated T cells to kill tumor cells, but cannot activate them on their own.Citation35 The ability to activate the T cells along with redirecting them appears to be a property of targeting CD3ε, which is an invariant subunit of the T cell receptor complex.
Lastly, the Fab-based BsAbs did not simply recapitulate the activity of scFv-based BsAbs. In both approaches investigated here, the Fab-based BsAbs had different biological activity than their scFv-based counterparts. For the HER-2×HER-2 combination, the IgG-Fab inhibited N87 tumor cell growth to a significantly greater extent than the IgG-scFv. The ability to inhibit HER-2-driven cell growth has been shown to have a strong dependence on the geometry and valence of engagement.Citation36 The HER-2×HER-2 IgG-Fab was not the first BsAb to show enhanced efficacy over the mAb combination; Citation14,37,38 however, it does highlight the value of having access to multiple BsAb formats such as IgG BsAbs or IgG-Fab for empirical testing of the optimal geometry, valency, and mAb combinations that provide the desired activity within a therapeutic. The EGFR×CD3 BsAbs demonstrated significantly different potencies for redirecting T cells to lyse EGFR-expressing tumor cells with the tandem scFv BsAb (50 kDa) being almost an order of magnitude more potent than the tandem Fab BsAb (100 kDa) and almost 2 orders of magnitude more potent than the IgG BsAb (150 kDa) even though the BsAb binding kinetics to EGFR and CD3 proteins were equivalent. This does lead one to speculate whether the BsAb molecular weight affects the intercellular distance and immune synapse formation between T cells and target cells.Citation39 More data with varied epitopes and perhaps different antigens would help test this hypothesis. Others have recently recognized the value of Fab-based BsAbs as well. A recent study highlights the ability of the CrossMab approach to generate IgG-Fabs targeting TNF and IL-17.Citation40,41 New methods of attaining specificity of heavy chain/light chain pairing have recently been reported with the potential to create IgG-Fab or tandem Fab BsAbs.Citation42,43 Overall, both the IgG-Fab and tandem Fab BsAb formats have biophysical advantages over their scFv-based counterparts and can be tested in parallel with these BsAbs or other formats such as IgG BsAbs to more thoroughly and empirically test for the most desired therapeutic outcomes.
Methods
Construction of test articles
The pertuzumab (anti-HER-2), trastuzumab (anti-HER-2) and matuzumab (anti-EGFR) sequences were obtained from the National Center for Biotechnology information (NCBI) and the Rutgers Center for Structural Biology Protein Data Bank (RCSB-PDB). The anti-human CD3 sequence was obtained from an in-house clone. The IgG-Fab heavy chain coding DNA sequence was generated in 2 pieces (Fab and IgG) using overlapping PCR with pertuzumab and trastuzumab IgG constructs already published.Citation14,44 The 2 pieces were cloned into a mammalian expression vector (Lonza) using a 3-way ligation using the ClonablesTM Kit (Novagen) and existing HindIII (5’) and EcoRI (3’) expression cassette restriction sites and an internal BamHI site designed within the (G4S)5 linker region. The HER-2×HER-2 IgG-scFv heavy chain was cloned the same way into the same mammalian expression vector. For construction of the EGFR×CD3 tandem Fab heavy chain and the entire EGFR×CD3 tandem scFv, the entire coding DNA sequences were generated using overlapping PCR and cloned directly into the same mammalian expression construct using the HindIII and EcoRI restriction sites. The pertuzumab, trastuzumab, matuzumab, and anti-CD3 light chains containing the Fab specificity mutations were also synthesized via overlapping PCR and cloned into the same mammalian expression plasmid using the same restriction sites. All constructs utilized a murine kappa leader signal sequence that is cleaved upon secretion. All ligated constructs were transformed into chemically competent TOP 10 E. coli cells (Life Technologies). Colonies were picked, cultured, and the plasmids were prepped (Qiagen MiniPrep kit). Correct sequences were confirmed by in-house DNA sequencing.
Expression, purification, and biophysical characterization
The IgG-Fab and IgG-scFv plasmids were scaled up by transformation in TOP10 E. coli, mixed with 100 mL Luria broth in a 250 mL baffled flask and shaken overnight at 220 r.p.m. Large-scale plasmid purifications were done using the BenchPro 2100 (Life Technologies) according to the manufacturer's protocols. Proteins were expressed in HEK293F cells using Freestyle transfection reagents and protocols provided by the manufacturer (Life Technologies). For the HER-2×HER-2 IgG-Fab, one HC plasmid and 2 LC plasmids were co-transfected at 3 μg DNA per 1 mL culture using a 1:1:1 ratio. For the HER-2×HER-2 IgG-scFv, one HC plasmid and one LC plasmid were co-transfected at 2 μg DNA per 1 mL culture using a 1:2 ratio. For the EGFR×CD3 tandem Fab, one HC plasmid and 2 LC plasmids were co-transfected at 3 μg DNA per 1 mL culture using a 1:1:1 ratio. For the EGFR×CD3 tandem scFv, one plasmid containing both scFvs was transfected using 2 μg DNA. Transfected cells were grown at 37°C in a 5% CO2 incubator while shaking at 125 r.p.m. for 5 days. Supernatants were harvested by centrifugation at 10 K r.p.m. for 5 minutes followed by passage through 2 μm filters.
Purification of the IgG-Fab and IgG-scFv was performed using standard protein A affinity chromatography methods as described previously.Citation14 No second step of purification was performed. Purification of the tandem Fab and tandem scFv BsAbs was performed using standard Ni2+-nitrilo acidic acid-agarose affinity chromatography on an AKTA Explorer (GE Healthcare). The resin was equilibrated in PBS. Supernatants were directly loaded onto the affinity column, washed with PBS, then eluted with 10 column volumes of PBS + 500 mM imidazole, pH 7.4. The tandem Fab was highly monodisperse after affinity chromatography and required no further purification steps. For the tandem scFv, the eluants were concentrated using a 10 kDa centrifugal concentrator (Millipore) and loaded onto a PBS-equilibrated 120 mL Superdex 75 column for aggregate and low molecular weight cleavage product removal.
Analytical size exclusion chromatography with in-line light scattering (SEC-LS), differential scanning calorimetry (DSC), and mass spectrometry was performed as described previously.Citation14
Factor Xa cleavage of the HER-2×Her-2 IgG-Fab
The HER2×HER-2 IgG-Fab was dialyzed in dialyzing buffer containing 20 mM Tris-HCl, 100 mM NaCl, 2 mM CaCl2 at pH 8.0. Factor Xa (1 μL per 50 μg of IgG-Fab protein – New England BioLabs) was added and incubated at 4°C overnight to cleave the Fab from the IgG. Factor Xa was removed by adding 100 μl of Xarrest agarose (EMD Millipore) per 8 μL Factor Xa and incubated at room temperature for 1 hr. The resin was spun down and the supernatant was collected. The resulting passed through a 0.2 mm filter and analyzed directly by mass spectrometry as described.Citation44 The 2 proteins were further separated using an IgSelect column (GE Healthcare) that specifically binds IgG-Fc. Fab eluted in the flow-through. The IgG fraction was eluted from the column with 100 mM glycine buffer at pH 3.2 and neutralized using 1 M Tris buffer. The solution was dialyzed against PBS prior to mass spectrometry analysis.
Stability measurements using automated circular dichroism
All CD experiments were performed on a Chirascan Plus (Applied Photophysics) with a peltier sample heater, external water bath heat sink, and an autosampling unit. The stability of the EGFR×CD3 tandem Fab and tandem scFv were assessed by thermally unfolding the proteins over a temperature range of 20–100°C. Wavelength scans from 250 to 198 nm were performed every 1°C of the temperature gradient. Scans used a 0.5 mm cuvette, with a 1 nm step size and bandwidth and an averaging time of 0.7 seconds. Both proteins were at 5 μM in PBS. Temperature-dependent protein unfolding parameters were either analyzed using the Global3 module of the dynamic multimode spectroscopy software of the manufacturer or fit at a single wavelength (235 nm) using a 2-state approximation.
Binding and stoichiometry measurements for HER2 (pertuzumab)×HER2(trastuzumab) IgG-Fab and IgG-scFv BsAbs
Solution Biacore experiments were designed as described previously.Citation18,45 All experiments were performed on a Biacore3000 (GE Healthcare). Pertuzumab and trastuzumab (generated in-house) Citation14 were diluted to 20 μg/mL in 10 mM Acetate, pH 5 and immobilized to separate CM5 sensorchip surfaces at a high level (∼12000 resonance units, RUs) using standard amine coupling protocols provided by the manufacturer (GE Healthcare). Binding of 25 nM soluble HER-2 (speed BioSystems) was performed at 2 μL/min to fall within the mass transfer arena and a strong signal (∼500 RU) was achieved after 3 minutes of injection. The concentration of unbound HER-2 in the presence of varying concentrations of soluble pertuzumab, trastuzumab, IgG-Fab BsAb or IgG-scFv BsAb was measured by comparing the linear slope (RUs/second) of first 40 seconds of the injection. The stoichiometry of binding to each HER-2 epitope was determined based on the amount of mAb or BsAb necessary to fully block HER-2 binding to either the pertuzumab or trastuzumab surface. The pertuzumab and trastuzumab surfaces were both regenerated by a double injection (5 μL) of 0.1 M glycine, pH 2 at a flow rate of 50 μL/min.
Binding kinetics for EGFR×CD3 BsAbs
Human CD3 and EGFR extracellular domain proteins were prepared in-house. The CD3 construct was made as a single chain ε/γ heterodimer as described,Citation46 except it was expressed in HEK293F, not bacteria, and contained a 25 amino acid (G4S)5 linker between the ε and γ domains. The scCD3εγ and sEGFR proteins were diluted to 20 mg/mL in 10 mM acetate, pH 5.0 and immobilized to a CM5 chip surface (∼2000 and ∼6000 RU, respectively) using the standard amine coupling protocols. The EGFR×CD3 tandem scFv and tandem Fab molecules were diluted into HBS-EP buffer (100, 75, 50, 37.5, 25, 12.5, 6.25 and 3.12 nM) and passed over the CD3εγ or EGFR surfaces at 15 μL/min for 5 minutes. Dissociation was allowed to occur for another 20 minutes. Surfaces were regenerated by raising the flow rate to 50 μL/min and injecting 5 μL 0.1 M Glycine pH 2.0. All the curves were fit to a 1:1 Langmuir binding model to obtain binding kinetics and equilibrium KDs. All experiments were performed at 25°C using HBS-EP as the running buffer.
Cell culture
N87 and A431 cells were obtained from the ATCC, tested negative for mycoplasma, and cultured as described previously.Citation14 Untreated, purified T cells from a human donor were purchased from ALLCELLS (Cat#PB009–1F). The cells were thawed and washed in 10% FBS (heat inactivated, Corning) plus 50 μg/ml gentamicin (Gibco/Life Technologies) in RPMI 1640 media (Corning). For pre-activated T cell assays, the T cells were added to a 6 well plate or cell culture flask pre-coated with anti-CD3 at 5 μg/mL (BD Biosciences, clone UCHT1). After overnight incubation at 37°C in a 5% CO2 incubator, the T cells were treated with 2.5 μg/mL anti-CD28 (BD Biosciences) and 5 ng/mL IL-2 (R&D Systems). Every week the anti-CD3/anti-CD28/IL-2 treatment was repeated to insure T cell activation, while IL-2 added to all media during the expansion phase.
Flow cytometry
T cells were counted using a Countess II (Life Technologies). They were spun down at by 1200 rpm for 8 minutes. The supernatant was removed and the cells were resuspended in PBS and adjusted to 1 × 107 cells/mL. The cells were spun down again and resuspended in flow cytometry blocking buffer (PBS w/2% FBS, 0.05% NaN3, 10% normal goat serum or NGS, Life Technologies, and Human Fc block, BD Biosciences) for at least 15 minutes. Next, 50 μL cells are added to a 96-well plate (Corning, Cat#3799). Fluorescently labeled mAbs are added directly to the cells at 50 μL/well and incubated for 45 minutes on ice. The detection mAbs included a PE-mouse anti-human CD3 (BD, Cat#555333), an APC-Cy7 mouse anti-human CD69 (BD, Cat#557756), an APC mouse anti-human CD25 (BD, Cat#555434), Pacific Blue mouse anti-human CD8 (BD, Cat#558207), and an Alexa-488 mouse anti-human CD4 (BD, Cat#557695). The plates were spun for 4 minutes at 1500 rpm; supernatants were aspirated; and the cells were resuspended in 150 μL flow cytometry wash buffer (FCWB, PBS w/2% FBS, 0.05% NaN3, and 10% NGS) at least 3 times. After the final supernatant removal, cells were resuspended in 200 μL 1:1000 PI in FCWB, covered with foil and sorted using an LSRII flow cytometer acquiring with Diva version software (Beckton Dickonson). Analysis was performed using FlowJo software. PI was from Molecular Probes. Compensation was performed using OneComp eBeads from eBioscience using the manufacturer's protocol.
N87 gastric tumor cell proliferation assay
N87 cell proliferation studies were performed as described previously with the modification that the IgG-Fab and IgG-scFv were utilized.Citation14
Pre-activated T cell redirected lysis of A431 carcinoma cells
A431 tumor cells were lifted using Accutase® (Innovative Cell Technologies) and pre-stained with CSFE according to the CSFE/7-AAD kit manufacturer's protocols (abcam, Cat#ab133073). After culturing between 4–6 hrs with pre-activated T cells (10:1, Effector:Tumor cells) in the absence or presence of mAb or BsAb test articles, the A431 cells were lifted again and stained with the DNA-intercalating dye 7-aminoactinomycin (7-AAD) according to the kit protocols. Flow cytometry was performed on a LSRII instrument (BD Biosciences) using an FL1 channel for CSFE and FL3 channel for 7-AAD.
Alternately, pre-activated T cell killing was assessed using the Cell Titer Glo® (Promega) cell viability assay. Briefly, A431 tumor cells cultured in RPMI, 10% FBS were lifted using 0.25% trypsin-EDTA (Sigma) and seeded at a density of 5000 cells per well overnight in 96 well tissue culture black isoplates with clear bottoms (Perkin Elmer, cat#6005050) at 37°C, 5% CO2. Pre-activated T cells (described above) in RPMI, 10% FBS, 50 μg/mL gentamicin were mixed with the test articles (mAbs and BsAbs) and added to each well at a 10:1 T cell:tumor cell ratio and incubated for 5 hours at 37°C, 5% CO2. After incubation, the plate is washed 3 times with serum-free RPMI to remove the T cells. Next, CellTiter-Glo reagent is added and luciferase luminescent signal is measured to quantify the number of viable cells remaining based on the amount of ATP present. Luminescence was measured using an Envision instrument (Perkin Elmer). Test articles demonstrating redirected lysis activity were fit to a standard 4 parameter sigmoidal equation to calculate the midpoint concentration (EC50) leading to 50% specific lysis:
where A is the bottom of the curve, B is the top of the curve, and n is the steepness of the sigmoidal slope of the transition also known as the Hill Slope (GraphPad Prism Software).
Bulk T cell activation and redirected lysis of A431 cells
Non-pre-treated T cell redirected lysis experiments were performed in a very similar fashion as the pre-activated T cells redirected lysis experiments with the following exceptions. T cells were thawed in RPMI/10% FBS/50 μg/mL gentamicin, quantified, and directly mixed with the test articles (mAbs and BsAbs) before adding them to A431 tumor cells (previously seeded overnight) as described above at a 10:1 T cell:A431 cell ratio. The cells were incubated for 48 hours. After incubation, soluble T cells were collected for flow cytometric analysis of CD4, CD8, CD69, and CD25 by washing with serum-free RPMI (at least 3 times). A431 lysis was measured by adding CellTiter-Glo (Promega) and measuring the luciferase activity as described above for the pre-activated T cell redirected lysis experiments.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.docx
Download MS Word (15.5 KB)Acknowledgments
We thank B. Gutierrez for his help with transient transfections and S. Jeffries for his advice with assays.
Funding
This work was supported by the Lilly Research Laboratories and the Lilly Research Award Program (LRAP).
Supplementary Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Ellis LM, Hicklin DJ. Resistance to Targeted Therapies: Refining Anticancer Therapy in the Era of Molecular Oncology. Clin Cancer Res 2009; 15: 7471-8; PMID: 20008847; http://dx.doi.org/10.1158/1078–0432.CCR-09-1070
- Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res 2008; 14: 2895-9; PMID: 18483355; http://dx.doi.org/10.1158/1078-0432.CCR-07-2248
- Shannon KM. Resistance in the land of molecular cancer therapeutics. Cancer Cell 2002; 2: 99-102; PMID: 12204529
- Byrne H, Conroy PJ, Whisstock JC, O'Kennedy RJ. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends Biotechnol 2013; 31: 621-32; PMID: 24094861; http://dx.doi.org/10.1016/j.tibtech.2013.08.007
- Demarest SJ, Hariharan K, Dong J. Emerging antibody combinations in oncology. MAbs 2011; 3: 338-51; PMID: 21697653; http://dx.doi.org/10.4161/mabs.3.4.16615
- Bird RE, Walker BW. Single chain antibody variable regions. Trends Biotechnol 1991; 9: 132-7; PMID: 1367550; http://dx.doi.org/10.1016/0167-7799(91)90044-I
- Brinkmann U, Reiter Y, Jung SH, Lee B, Pastan I. A recombinant immunotoxin containing a disulfide-stabilized Fv fragment. Proc Natl Acad Sci U S A 1993; 90: 7538-42; PMID: 8356052; http://dx.doi.org/10.1073/pnas.90.16.7538
- Coloma MJ, Morrison SL. Design and production of novel tetravalent bispecific antibodies. Nat Biotechnol 1997; 15: 159-63; PMID: 9035142; http://dx.doi.org/10.1038/nbt0297-159
- Perisic O, Webb PA, Holliger P, Winter G, Williams RL. Crystal structure of a diabody, a bivalent antibody fragment. Structure 1994; 2: 1217-26; PMID: 7704531; http://dx.doi.org/10.1016/S0969-2126(94)00123-5
- Chames P, Baty D. Bispecific antibodies for cancer therapy: the light at the end of the tunnel? MAbs 2009; 1: 539-47; PMID: 20073127; http://dx.doi.org/10.4161/mabs.1.6.10015
- Rothlisberger D, Honegger A, Pluckthun A. Domain interactions in the Fab fragment: a comparative evaluation of the single-chain Fv and Fab format engineered with variable domains of different stability. J Mol Biol 2005; 347: 773-89; PMID: 15769469; http://dx.doi.org/10.1016/j.jmb.2005.01.053
- Demarest SJ, Glaser SM. Antibody therapeutics, antibody engineering, and the merits of protein stability. Curr Opin Drug Discov Devel 2008; 11: 675-87; PMID: 18729019
- Mabry R, Snavely M. Therapeutic bispecific antibodies: The selection of stable single-chain fragments to overcome engineering obstacles. IDrugs 2010; 13: 543-9; PMID: 20721825
- Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nat Biotechnol 2014; 32: 191-8; PMID: 24463572; http://dx.doi.org/10.1038/nbt.2797
- Miller K, Meng G, Liu J, Hurst A, Hsei V, Wong WL, Ekert, R, Lawrence, D, Sherwood, S, DeForge, L, et al. Design, construction, and in vitro analyses of multivalent antibodies. J Immunol 2003; 170: 4854-61; PMID: 12728922; http://dx.doi.org/10.4049/jimmunol.170.9.4854
- Dimasi N, Gao C, Fleming R, Woods RM, Yao XT, Shirinian L, Kiener PA, Wu H. The design and characterization of oligospecific antibodies for simultaneous targeting of multiple disease mediators. J Mol Biol 2009; 393: 672-92; PMID: 19699208; http://dx.doi.org/10.1016/j.jmb.2009.08.032
- Michaelson JS, Demarest SJ, Miller B, Amatucci A, Snyder WB, Wu X, Huang F, Phan S, Gao S, Doern A, et al. Anti-tumor activity of stability-engineered IgG-like bispecific antibodies targeting TRAIL-R2 and LTbetaR. MAbs 2009; 1: 128-41; PMID: 20061822; http://dx.doi.org/10.4161/mabs.1.2.7631
- Dong J, Sereno A, Snyder WB, Miller BR, Tamraz S, Doern A, Favis M, Wu X, Tran H, Langley, E, et al. Stable IgG-like bispecific antibodies directed toward the type I insulin-like growth factor receptor demonstrate enhanced ligand blockade and anti-tumor activity. J Biol Chem 2011; 286: 4703-17; PMID: 21123183; http://dx.doi.org/10.1074/jbc.M110.184317
- Dong J, Sereno A, Aivazian D, Langley E, Miller BR, Snyder WB, Chan E, Cantele M, Morena R, Joseph IB, et al. A stable IgG-like bispecific antibody targeting the epidermal growth factor receptor and the type I insulin-like growth factor receptor demonstrates superior anti-tumor activity. MAbs 2011; 3: 273-88; PMID: 21393993; http://dx.doi.org/10.4161/mabs.3.3.15188
- Wu C, Ying H, Grinnell C, Bryant S, Miller R, Clabbers A, Bose S, McCarthy D, Zhu RR, Santora L, et al. Simultaneous targeting of multiple disease mediators by a dual-variable-domain immunoglobulin. Nat Biotechnol 2007; 25: 1290-7; PMID: 17934452; http://dx.doi.org/10.1038/nbt1345
- Jakob CG, Edalji R, Judge RA, DiGiammarino E, Li Y, Gu J, Ghayur T. Structure reveals function of the dual variable domain immunoglobulin (DVD-Ig) molecule. MAbs 2013; 5: 358-63; PMID: 23549062; http://dx.doi.org/10.4161/mabs.23977
- Nahta R, Hung MC, Esteva FJ. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res 2004; 64: 2343-6; PMID: 15059883; http://dx.doi.org/10.1158/0008-5472.CAN-03-3856
- Wu X, Sereno AJ, Huang F, Zhang K, Batt M, Fitchett JR, He D, Rick HL, Conner EM, Demarest SJ. Protein design of IgG/TCR chimeras for the co-expression of Fab-like moieties within bispecific antibodies. mAbs 2015; 7(2):364–376; PMID: 25611120
- Choi BD, Cai M, Bigner DD, Mehta AI, Kuan CT, Sampson JH. Bispecific antibodies engage T cells for antitumor immunotherapy. Expert Opin Biol Ther 2011; 11: 843-53; PMID: 21449821; http://dx.doi.org/10.1517/14712598.2011.572874
- Baeuerle PA, Reinhardt C. Bispecific T-cell engaging antibodies for cancer therapy. Cancer Res 2009; 69: 4941-4; PMID: 19509221; http://dx.doi.org/10.1158/0008-5472.CAN-09-0547
- Topp MS, Kufer P, Gokbuget N, Goebeler M, Klinger M, Neumann S, Horst HA, Raff T, Viardot A, Schmid M, et al. Targeted therapy with the T-cell-engaging antibody blinatumomab of chemotherapy-refractory minimal residual disease in B-lineage acute lymphoblastic leukemia patients results in high response rate and prolonged leukemia-free survival. J Clin Oncol 2011; 29: 2493-8; PMID: 21576633; http://dx.doi.org/10.1200/JCO.2010.32.7270
- Nagorsen D, Kufer P, Baeuerle PA, Bargou R. Blinatumomab: a historical perspective. Pharmacol Ther 2012; 136: 334-42; PMID: 22940266; http://dx.doi.org/10.1016/j.pharmthera.2012.07.013
- Heymach JV, Nilsson M, Blumenschein G, Papadimitrakopoulou V, Herbst R. Epidermal growth factor receptor inhibitors in development for the treatment of non-small cell lung cancer. Clin Cancer Res 2006; 12: 4441s-4445s; PMID: 16857825; http://dx.doi.org/10.1158/1078-0432.CCR-06-0286
- Kuhlman B, Boice JA, Fairman R, Raleigh DP. Structure and stability of the N-terminal domain of the ribosomal protein L9: evidence for rapid two-state folding. Biochemistry 1998; 37: 1025-32; PMID: 9454593; http://dx.doi.org/10.1021/bi972352x
- Mallender WD, Voss EW Jr. Construction, expression, and activity of a bivalent bispecific single-chain antibody. J Biol Chem 1994; 269: 199-206; PMID: 8276795
- Gruber M, Schodin BA, Wilson ER, Kranz DM. Efficient tumor cell lysis mediated by a bispecific single chain antibody expressed in Escherichia coli. J Immunol 1994; 152: 5368-74; PMID: 8189055
- Johnson S, Burke S, Huang L, Gorlatov S, Li H, Wang W, Zhang W, Tuaillon N, Rainey J, Barat B, et al. Effector cell recruitment with novel Fv-based dual-affinity re-targeting protein leads to potent tumor cytolysis and in vivo B-cell depletion. J Mol Biol 2010; 399: 436-49; PMID: 20382161; http://dx.doi.org/10.1016/j.jmb.2010.04.001
- Wang L, He Y, Zhang G, Ma J, Liu C, He W, Wang W, Han H, Boruah BM, Gao B. Retargeting T cells for HER2-positive tumor killing by a bispecific Fv-Fc antibody. PLoS One 2013; 8: e75589; PMID: 24086580; http://dx.doi.org/10.1371/journal.pone.0075589
- Junttila TT, Li J, Johnston J, Hristopoulos M, Clark R, Ellerman D, Wang BE, Li Y, Mathieu M, Li G, et al. Antitumor efficacy of a bispecific antibody that targets HER2 and activates T cells. Cancer Res 2014; 74: 5561-71; PMID: 25228655; http://dx.doi.org/10.1158/0008-5472.CAN-13-3622-T
- Michalk I, Feldmann A, Koristka S, Arndt C, Cartellieri M, Ehninger A, Ehninger G, Bachmann MP. Characterization of a novel single-chain bispecific antibody for retargeting of T cells to tumor cells via the TCR co-receptor CD8. PLoS One 2014; 9: e95517; PMID: 24751697; http://dx.doi.org/10.1371/journal.pone.0095517
- Scheer JM, Sandoval W, Elliott JM, Shao L, Luis E, Lewin-Koh SC, Schaefer G, Vandlen R. Reorienting the Fab domains of trastuzumab results in potent HER2 activators. PLoS One 2012; 7: e51817; PMID: 23284778; http://dx.doi.org/10.1371/journal.pone.0051817
- Jost C, Schilling J, Tamaskovic R, Schwill M, Honegger A, Pluckthun A. Structural basis for eliciting a cytotoxic effect in HER2-overexpressing cancer cells via binding to the extracellular domain of HER2. Structure 2013; 21: 1979-91; PMID: 24095059; http://dx.doi.org/10.1016/j.str.2013.08.020
- Li B, Meng Y, Zheng L, Zhang X, Tong Q, Tan W, Hu S, Li H, Chen Y, Song J, et al. Bispecific antibody to ErbB2 overcomes trastuzumab resistance through comprehensive blockade of ErbB2 heterodimerization. Cancer Res 2013; 73: 6471-83; PMID: 24046294; http://dx.doi.org/10.1158/0008-5472.CAN-13-0657
- Wang JH, Reinherz EL. The structural basis of alphabeta T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev 2012; 250: 102-19; PMID: 23046125; http://dx.doi.org/10.1111/j.1600-065X.2012.01161.x
- Fischer JA, Hueber AJ, Wilson S, Galm M, Baum W, Kitson C, Auer J, Lorenz SH, Moelleken J, Bader M, et al. Combined inhibition of tumor necrosis factor alpha and onterleukin-17 As a yherapeutic opportunity in rheumatoid arthritis: development and characterization of a novel bispecific antibody. Arthritis Rheumatol 2015; 67: 51-62; PMID: 25303306; http://dx.doi.org/10.1002/art.38896
- Schaefer W, Regula JT, Bahner M, Schanzer J, Croasdale R, Durr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A 2011; 108: 11187-92; PMID: 21690412; http://dx.doi.org/10.1073/pnas.1019002108
- Liu Z, Leng EC, Gunasekaran K, Pentony M, Shen M, Howard M, Stoops J, Manchulenko K, Razinkov V, Liu H, et al. A novel antibody engineering strategy for making monovalent bispecific heterodimeric IgG antibodies by electrostatic steering mechanism. J Biol Chem 2015; 290(12):7535–7562
- Mazor Y, Oganesyan V, Yang C, Hansen A, Wang J, Liu H, Sachsenmeier K, Carlson M, Gadre DV, Borrok MJ, et al. Improving target cell specificity using a novel monovalent bispecific IgG design. mAbs 2015; 7(2):377–389; PMID: 25621507
- Casimiro DR, Wright PE, Dyson HJ. PCR-based gene synthesis and protein NMR spectroscopy. Structure 1997; 5: 1407-12; PMID: 9384559; http://dx.doi.org/10.1016/S0969-2126(97)00291-8
- Day ES, Capili AD, Borysenko CW, Zafari M, Whitty A. Determining the affinity and stoichiometry of interactions between unmodified proteins in solution using Biacore. Anal Biochem 2013; 440: 96-107; PMID: 23711722; http://dx.doi.org/10.1016/j.ab.2013.05.012
- Kim KS, Sun ZY, Wagner G, Reinherz EL. Heterodimeric CD3epsilongamma extracellular domain fragments: production, purification and structural analysis. J Mol Biol 2000; 302: 899-916; PMID: 10993731; http://dx.doi.org/10.1006/jmbi.2000.4098
- Garber E, Demarest SJ. A broad range of Fab stabilities within a host of therapeutic IgGs. Biochem Biophys Res Commun 2007; 355: 751-7; PMID: 17321501; http://dx.doi.org/10.1016/j.bbrc.2007.02.042
