ABSTRACT
Binding interactions with the neonatal Fc receptor (FcRn) are one determinant of pharmacokinetic properties of recombinant human monoclonal antibody (rhumAb) therapeutics, and a conserved binding motif in the crystallizable fragment (Fc) region of IgG molecules interacts with FcRn. Surface plasmon resonance (SPR) biosensor assays are often used to characterize interactions between FcRn and rhumAb therapeutics. In such assays, generally either the rhumAb (format 1) or the FcRn protein (format 2) is immobilized on a biosensor chip. However, because evidence suggests that, in some cases, the variable domains of a rhumAb may also affect FcRn binding, we evaluated the effect of SPR assay configuration on binding data. We sought to assess FcRn binding properties of 2 rhumAbs (rhumAb1 and rhumAb2) to FcRn proteins using these 2 biosensor assay formats. The two rhumAbs have greater than 99% sequence identity in the Fc domain but differ in their Fab regions. rhumAb2 contains a positively charged patch in the variable domain that is absent in rhumAb1. Our results showed that binding of rhumAb1 to FcRn was independent of biosensor assay configuration, while binding of rhumAb2 to FcRn was highly SPR assay configuration dependent. Further investigations revealed that the format dependency of rhumAb2-FcRn binding is linked to the basic residues that form a positively charged patch in the variable domain of rhumAb2. Our work highlights the importance of analyzing rhumAb-FcRn binding interactions using 2 alternate SPR biosensor assay configurations. This approach may also provide a simple way to identify the potential for non-Fc-driven FcRn binding interactions in otherwise typical IgGs.
Introduction
The neonatal Fc receptor (FcRn) is a type I membrane glycoprotein consisting of a β2-microglobulin and an α-chain resembling the MHC class I molecules.Citation1,2,3 FcRn is responsible for the transfer of immunoglobulin G (IgG) from the mother to the fetus or newborn,Citation45 and it also plays a critical role in regulating IgG levels in adults.Citation6,7 Studies have shown that FcRn interacts with a binding motif in the crystallizable fragment (Fc) of IgG at the CH2-CH3 domain interface in a pH-dependent manner.Citation6,8 The pH dependency of this interaction is essential for maintaining the long serum half-life of IgG molecules. Specifically, in the endosomes of endothelial cells (∼pH 6.0), IgG internalized through pinocytosis binds to FcRn to form IgG-FcRn complexes; the IgG-FcRn complexes are then trafficked to the cell surface where IgG is released back into the circulation at physiological pH (∼7.4). This prevents lysosomal degradation of the IgG.Citation9,10,11
For recombinant human monoclonal antibody (rhumAb) therapeutics, the FcRn-rhumAb binding interaction is a critical determinant of pharmacokinetic (PK) properties and targeted engineering of the FcRn binding motif may enable less frequent dosing of mAb therapeutics in patients.Citation12 Multiple studies have suggested that there is a correlation between FcRn binding affinity and antibody half-life,Citation13,14,15,16,17 although the absence of such a correlation has also been reported.Citation18,19,20 Technologies used for evaluating FcRn-rhumAb interactions vary extensively. Some of the methods used to assay FcRn-rhumAb interactions include surface plasmon resonance (SPR) (Biacore, ProteOn),Citation13,17,21,22,23,24,25,26 biolayer interferometry (Octet),Citation26 isothermal titration calorimetry,Citation27 enzyme-linked immunosorbent assays (ELISA),Citation28,29 cell-based assays,Citation30,31 AlphaScreen,Citation32 affinity chromatography,Citation33 and asymmetrical flow field flow fractionation.Citation34 Among these technologies, SPR-based biosensor assays are the most commonly used, likely due to the fact that they can provide real-time quality data on binding specificity and kinetics over a wide range of binding affinities.
A wide range of affinity values have been reported from SPR biosensor studies of rhumAb-FcRn binding interactions. For example, the reported equilibrium dissociation constants (KD) for rhumAbs with wild type human IgG1 Fc domains binding to human FcRn vary over 100-fold, from 19 nM to 2.5 µM.Citation13,17,23,24,26,35,36,37,38 This variability in KD values has also been seen in studies of FcRn proteins from other species. At least 15-fold differences in KD values were reported for rhumAbs binding to FcRn proteins derived from cynomolgus monkey and rat (104 nM to 2.4 µM for cynomolgus monkey,Citation17,37,39 and 35 nM to 567 nM for rat,Citation25,26,40). A number of factors may contribute to this broad range of reported binding activities, including the source and quality of the assay reagents (FcRn, rhumAbs), the experimental setup (e.g., assay format, ligand immobilization level, temperature, analyte concentrations), and the data evaluation models used. However, only a few studies have systematically explored the potential cause(s) of these differences.Citation25,41,42
The reported SPR biosensor assays mostly employed only one of the following 2 formats when assessing rhumAb-FcRn interactions: 1) rhumAbs were directly immobilized on the biosensor chip, typically via amine coupling chemistry, and FcRn proteins in solution were injected over the sensor chip; or 2) the orientation was reversed so that the FcRn proteins were directly amine coupled on the biosensor chips, and rhumAbs were injected over the chips.
Amine coupling of ligands onto biosensor chips is widely used, often through lysine residues and the N-terminal amino group. Since the key contact residues on both FcRn and wild type rhumAb Fc do not include lysine,Citation7 it has been assumed that amine coupling of either FcRn or rhumAb to a biosensor chip is unlikely to affect the binding interaction. However, growing evidence suggests that the variable domains of a rhumAb may also affect FcRn binding.Citation24,36,43 Since lysine residues may be present in rhumAb Fab regions, we evaluated the effect of the assay configuration on rhumAb-FcRn binding data from SPR analysis.
We sought to assess the binding interactions between 2 rhumAbs (rhumAb1 and rhumAb2) and FcRn proteins using both of the aforementioned SPR biosensor formats. These two rhumAbs have greater than 99% sequence identity in the Fc domain but differ in the Fab region. rhumAb2 contains a positive charge patch in the variable domainCitation44 that is absent in rhumAb1. The positive charge patch on rhumAb2 consists of 4 basic amino acids, one arginine (R) and one lysine (K) on the heavy chain and 2 lysine residues on the light chain (HC-R57, HC-K65, LC-K31, LC-K67). Our results show that the rhumAb1:FcRn binding data were independent of biosensor assay format, while the rhumAb2:FcRn binding data were highly assay format dependent. Further investigation reveals that the assay format dependency of the rhumAb2 binding data are caused by the basic residues that formed the positive charge patch in the variable domain. Due to the lack of prior knowledge about the presence, and therefore impact, of an IgG charge patch, the use of just one SPR biosensor assay format to assess rhumAb/FcRn binding may have compromised the binding affinity data and the potential to correlate binding affinities with antibody half-lives. This work highlights the importance of using 2 different biosensor assay formats to assess rhumAb-FcRn binding interactions, and provides one possible reason why in vitro FcRn binding data (e.g., affinity, ratio of affinities at different pH conditions, off rate) for antibody therapeutics cannot always be correlated with in vivo terminal half-life data.
Results
Binding of rhumAbs to recombinant rat FcRn
We first analyzed the binding of rhumAb1 and rhumAb2 to recombinant rat FcRn using biosensor assay format 1, where each rhumAb was immobilized on a sensor chip, and FcRn was run in solution. At acidic pH (pH 6.0), rhumAb1 and rhumAb2 had similar FcRn binding profiles (), with average KD values of 2.0 and 1.2 μM, respectively, based on a 1:1 binding model (). As expected, at physiological pH (pH 7.4), rhumAb1 and rhumAb2 did not show significant binding to rat FcRn ().
Figure 1. Representative sensorgrams of rhumAbs binding to recombinant rat FcRn on 2 Biacore assay formats. (A) recombinant rat FcRn bound to immobilized rhumAbs on format 1 at pH = 6.0, and (B) at pH = 7.4; (C) rhumAbs bound to immobilized recombinant rat FcRn on format 2 at pH = 6.0, and (D) at pH = 7.4. The experimental data are represented by colored lines. On format 1, at pH 6.0, concentrations of rat FcRn (from bottom to top) are 0.078, 0.156, 0.313, 0.625, 1.25, 2.5, 5, and 10 µM. The black lines are fitted data using the kinetic monovalent binding model. At pH 7.4 concentrations of tested rat FcRn (from bottom to top) are 2.5, 5, 10, and 20 µM. On format 2, concentrations of rhumAbs (from bottom to top) are 0.039, 0.078, 0.156, 0.313, 0.625, 1.25, 2.5, and 5 µM. The black lines are fitted data using the kinetic bivalent binding model. At pH 7.4 concentrations of rhumAbs (from bottom to top) are 1.25, 2.5, 5, and 10 µM. The sensorgrams were generated after in-line reference cell correction followed by buffer sample subtraction. The experiments were conducted using running buffer containing PBS, 0.05% polysorbate 20, pH 6.0 or pH 7.4.
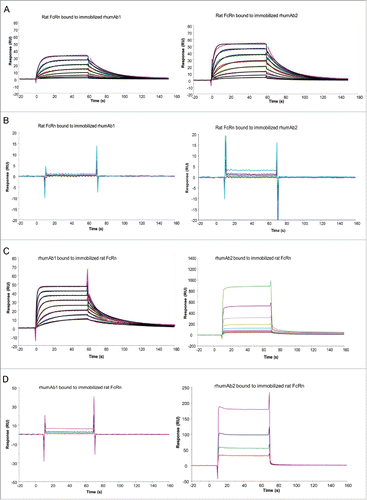
Table 1. Binding affinities of recombinant rat, cynomolgus monkey, and human FcRn proteins to immobilized rhumAbs at pH 6.0.
We then analyzed binding of recombinant rat FcRn to rhumAb1 and rhumAb2 using biosensor assay format 2, where FcRn was immobilized on the sensor chip, and each rhumAb was run in solution. As expected, dose-dependent binding of rhumAb1 to rat FcRn was clearly detected at pH 6.0 (), but not at pH 7.4 (). At pH 6.0, one IgG molecule can bind to one or 2 immobilized FcRn molecules.Citation42,45,46 The interaction could not be fully described by a simple 1:1 binding model (data not shown), despite a very low FcRn immobilization level (∼25 RU). Therefore, we used a bivalent binding model to fit the data. Using this model, rhumAb1 bound to immobilized rat FcRn with an average KD1 of 0.9 µM (), similar to that obtained using format 1 (2.0 µM). However, rhumAb2 gave very different results. At pH 6.0, the binding responses for rhumAb2 were much higher than those observed for rhumAb1 at the same concentrations (). For example, at 5 µM, rhumAb1 bound to rat FcRn with an approximate response of 50 RU, while the same concentration of rhumAb2 had a binding response of 500 RU, even though the 2 antibodies share a greater than 99% amino acid sequence identity in the Fc domain. It is worth noting that the signal obtained from rhumAb2 binding to FcRn far exceeded the theoretical Rmax values of this interaction (45 RU, assuming all IgG binds to FcRn bivalently, or 90 RU, assuming all IgG binds to FcRn monovalently). Unexpectedly, at physiological pH, rhumAb2 also showed some dose-dependent binding to FcRn ().
Table 2. Binding affinities of rhumAbs to immobilized recombinant rat, cynomolgus monkey, and human FcRn proteins at pH 6.0.
The unusually high FcRn binding responses observed with rhumAb2 in format 2 at pH 6.0 () suggested that another type of binding interaction was occurring in addition to the expected specific and saturable FcRn-IgG Fc binding interaction; this prevented accurate calculation of kinetic parameters and affinities. This can be seen in the SPR data prior to reference cell correction: very low background signals were obtained for both antibodies using Format 1. Much higher reference flow cell signals were obtained with Format 2. In addition, the rhumAb2 background signal was much higher than the rhumAb1 background signal. This differential was not observed using Format 1 (). In Format 2 at pH 6.0, the FcRn/rhumAb2 binding profile was not consistent with the stoichiometry commonly seen with FcRn/antibody binding interactions and appeared to be non-saturable. Moreover, in Format 2, rhumAb2 also bound to FcRn at pH 7.4 in a dose-dependent, but non-saturable, manner ().
Table 3. Binding response of recombinant rat FcRn rhumAbs on reference and test flow cells (FCs) in assay formats 1 and 2 at pH 6.0.
In summary, our data showed the expected specific and saturable binding interactions between FcRn and both rhumAbs using assay Format 1, and between FcRn and rhumAb1 using Format 2. However, we observed a non-saturable binding interaction between FcRn and rhumAb2 when Format 2 was used.
Binding of rhumAbs to recombinant cynomolgus monkey FcRn
As binding interactions between IgG and FcRn are species-specific,Citation21,36 we tested binding of the 2 rhumAbs to recombinant cynomolgus monkey FcRn to determine whether we saw a similar assay format dependency as observed for rat FcRn. As shown in , both rhumAb1 and rhumAb2 bound to recombinant cynomolgus monkey FcRn with very fast association and dissociation rates, precluding accurate kinetic analysis. Binding of rhumAb1 and rhumAb2 to recombinant cynomolgus monkey FcRn readily reached steady-states at all tested concentrations during the 1-minute sample injections (). Therefore, the KD values were calculated from the equilibrium analysis using the steady-state affinity binding model provided by the vendor (Biacore T200 Evaluation Software). Using format 1, rhumAb1 and rhumAb2 had very similar FcRn binding profiles at both tested pHs. At pH 6.0, the averaged KD values of recombinant cynomolgus monkey FcRn bound to immobilized rhumAb1 and rhumAb2 were 3.0 and 2.7 μM, respectively ( and ). At pH 7.4, no significant interactions were observed between recombinant cynomolgus monkey FcRn and the tested rhumAbs (), as expected.
Figure 2. Representative sensorgrams of rhumAbs binding to recombinant cynomolgus monkey FcRn on 2 Biacore assay formats. (A) recombinant cynomolgus monkey FcRn bound to immobilized rhumAbs on format 1 at pH = 6.0, and (B) at pH = 7.4; (C) rhumAbs bound to immobilized recombinant cynomolgus monkey FcRn on format 2 at pH = 6.0, and (D) at pH = 7.4. The experimental data are represented by colored lines. On format 1, at pH 6.0, concentrations of cynomolgus monkey FcRn (from bottom to top) are 0.039, 0.078, 0.156, 0.313, 0.625, 1.25, 2.5, 5, 10, and 20 µM. The black lines are fitted data using the steady-state affinity binding model (inserts). At pH 7.4 concentrations of tested recombinant cynomolgus monkey FcRn (from bottom to top) are 2.5, 5, 10, and 20 µM. On format 2, at pH 6.0, concentrations of recombinant cynomolgus monkey FcRn (from bottom to top) are 0.078, 0.156, 0.313, 0.625, 1.25, 2.5, 5, and 10 µM. The black lines are fitted data using the steady-state affinity binding model (inserts). At pH 7.4, concentrations of recombinant cynomolgus monkey FcRn (from bottom to top) are 1.25, 2.5, 5, and 10 µM. The sensorgrams were generated after in-line reference cell correction followed by buffer sample subtraction. The experiments were conducted using running buffer containing PBS, 0.05% polysorbate 20, pH 6.0 or pH 7.4.
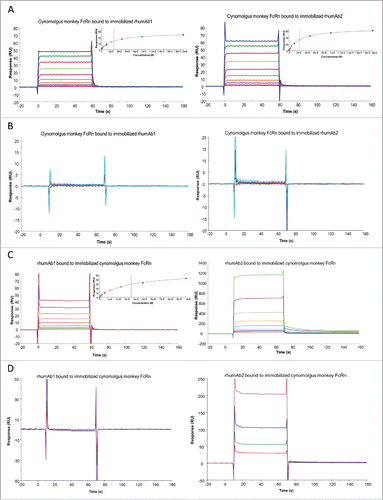
However, when we used format 2 the binding responses of rhumAb1 and rhumAb2 to immobilized recombinant cynomolgus monkey FcRn again showed dramatic differences. While rhumAb1 demonstrated expected binding activity to recombinant cynomolgus monkey at pH 6.0 with an averaged KD of 3.7 µM, similar to what was obtained using format 1 (3.0 µM), rhumAb2 showed an unusually high FcRn binding response ( and ), similar to what was observed under these conditions for rat FcRn. At pH 7.4, rhumAb2 also exhibited significant dose-dependent non-saturable binding to immobilized recombinant cynomolgus monkey FcRn, while rhumAb1 showed no detectable binding ().
Binding of rhumAbs to recombinant human FcRn
We also examined rhumAb1 and rhumAb2 binding to recombinant human FcRn using both biosensor assay formats. As shown in , rhumAb1 and rhumAb2 had very similar binding interactions with recombinant human FcRn when assay format 1 was used. At pH 6.0, the average KD values for recombinant human FcRn binding to immobilized rhumAb1 and rhumAb2 were 3.5 and 3.2 μM, respectively ( and ). At pH 7.4, no significant interactions were observed between recombinant human FcRn and the tested rhumAbs (), as expected.
Figure 3. Representative sensorgrams of rhumAbs binding to recombinant human FcRn on 2 Biacore assay formats. (A) recombinant human FcRn bound to immobilized rhumAbs on format 1 at pH = 6.0, and (B) at pH = 7.4; (C) rhumAbs bound to immobilized recombinant human FcRn on format 2 at pH = 6.0, and (D) at pH = 7.4. The experimental data are represented by colored lines. On format 1, at pH 6.0, concentrations of recombinant human FcRn (from bottom to top) are 0.039, 0.078, 0.156, 0.313, 0.625, 1.25, 2.5, 5, 10, and 20 µM. The black lines are fitted data using the steady-state affinity binding model (inserts). At pH 7.4 concentrations of tested recombinant human FcRn (from bottom to top) are 2.5, 5, 10, and 20 µM. On format 2, at pH 6.0, concentrations of recombinant human FcRn (from bottom to top) are 0.078, 0.156, 0.313, 0.625, 1.25, 2.5, 5, and 10 µM. The black lines are fitted data using the steady-state affinity binding model (inserts). At pH 7.4, concentrations of recombinant human FcRn (from bottom to top) are 1.25, 2.5, 5, and 10 µM. The sensorgrams were generated after in-line reference cell correction followed by buffer sample subtraction. The experiments were conducted using running buffer containing PBS, 0.05% polysorbate 20, pH 6.0 or pH 7.4.
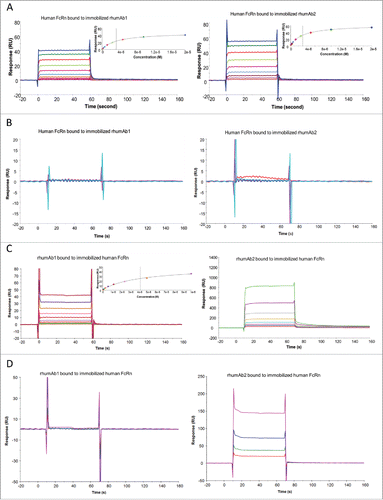
However, as had been seen with recombinant rat and cynomolgus monkey FcRn proteins, the binding activities of rhumAb1 and rhumAb2 to immobilized recombinant human FcRn differed using assay format 2. rhumAb1 demonstrated dose-dependent binding to recombinant human FcRn with an averaged KD of 4.3 µM at pH 6.0, similar to the value obtained from format 1 (3.5 µM), and no detectable binding at pH 7.4; rhumAb2 again showed unusually high FcRn binding responses at both pHs, which are very different from the results obtained from format 1 ().
Binding of F(ab’)2 to FcRn using format 2—FcRn immobilized on chip, F(ab’)2 (rhumAbs) as analytes
To investigate whether the unexpected binding of rhumAb2 to FcRn proteins observed in format 2 was mainly contributed by the Fc of rhumAb2 or not, we generated F(ab′)2 of rhumAb2 and analyzed the binding of the F(ab′)2 and the intact antibody to immobilized FcRn proteins. The F(ab′)2 of rhumAb1 was also generated and served as a control.
Binding responses of full-length rhumAbs and their F(ab′)2 to immobilized FcRn proteins at pH 6.0 and 7.4 are shown in . As expected, at pH 6.0, F(ab’)2 derived from rhumAb1 showed minimal binding to immobilized FcRn proteins (). However, under similar conditions, F(ab’)2 derived from rhumAb2 showed significant binding responses to FcRn proteins from all 3 species (). It is worth pointing out that the binding response seen with the F(ab’)2 of rhumAb2 is greater than 50% of the full-length antibody binding signal under the same conditions. Because the binding response signal is directly proportional to the total mass of bound analyte, this suggests that the Fab region of rhumAb2 is heavily involved in the unexpectedly high levels of FcRn binding that are seen with this antibody.
Figure 4. Binding response levels of full-length rhumAb (in blue) and the F(ab’)2 (in red) of rhumAb binding to immobilized recombinant rat, cynomolgus monkey, and human FcRn. (A) at acidic pH (pH = 6.0); (B) at physiological pH (pH = 7.4). Concentration of tested full-length and F(ab’)2 of rhumAb samples is 5 µM. The in-line reference subtracted response was corrected by buffer sample, and was reported 5 seconds before the end of each injection. The reported data was averaged from 2 individual experimental runs. The experiments were conducted using running buffer containing PBS, 0.05% polysorbate 20, pH 6.0 or pH 7.4.
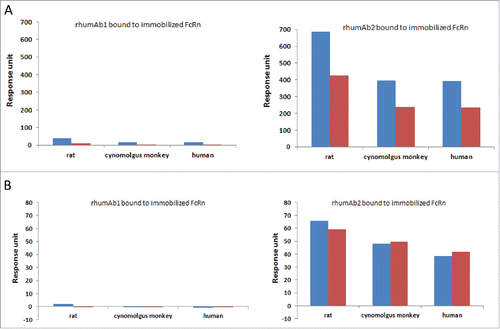
At pH 7.4, both rhumAb1 and its F(ab’)2 showed no or minimal binding to immobilized FcRn molecules as expected (), while rhumAb2 and its F(ab’)2 again exhibited significant binding (). At pH 7.4, the binding response seen with F(ab’)2 derived from rhumAb2 is comparable to that seen with the whole molecule, suggesting that most of the binding observed at neutral pH can be attributed to Fab/FcRn binding interactions.
Binding of F(ab’)2 (rhumAbs) to FcRn on format 2 in the presence of additional salt
To understand the nature of the unexpected binding of rhumAb2 to immobilized FcRn at pH 6.0, increasing concentrations of NaCl were added to the assay running buffer. As shown in , addition of NaCl resulted in a dose-dependent decrease in the binding response for both rhumAbs and the F(ab’)2 of rhumAb2 at pH 6.0; however, the effect of the added salt was much greater for rhumAb2 than for rhumAb1. For example, when the NaCl concentration in the running buffer was increased from 0 to 0.05 M, the binding responses of intact rhumAb1 to immobilized rat, cynomolgus monkey, and human FcRn were reduced by 22%, 40%, and 43%, respectively; while for intact rhumAb2 the binding responses were reduced by 79%, 77%, and 76%, respectively ().
Figure 5. Binding response levels of the full-length molecules and F(ab’)2 of rhumAbs to Immobilized recombinant rat, cynomolgus monkey, and human FcRn in the presence of additional salt (0–0.5 M) in running buffers (PBS, 0.05% polysorbate 20, pH 6.0 or pH 7.4) (A) at acidic pH (pH = 6.0); (B) at physiological pH (pH = 7.4). F(ab’)2 of rhumAbs were only tested only in PBS buffers with and without additional 0.1 M NaCl. Concentration of tested the full-length molecules and F(ab’)2 of rhumAb samples is 5 µM. The in-line reference subtracted response was corrected by buffer sample, and was reported 5 seconds before the end of each injection.
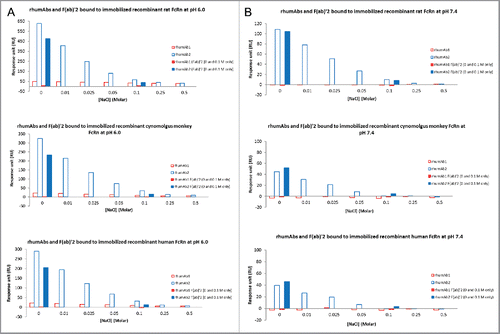
Table 4. Change in binding of rhumAbs to immobilized recombinant rat, cynomolgus monkey, and human FcRn proteins at pH 6.0 in the presence of 0.05 M NaCl.
At pH 7.4, rhumAb1 showed no detectable binding to immobilized FcRn regardless of the amount of additional NaCl added (). Consistent with , rhumAb2 exhibited significant binding to FcRn at pH 7.4, and the binding decreased with increasing amounts of salt in the assay running buffer. Binding was completely eliminated when the salt concentration was increased to 0.5 M NaCl. Similar results were obtained for each of the FcRn tested, suggesting the phenomenon was not FcRn-species specific.
Binding of rhumAb2 and its mutants to rat FcRn on format 2
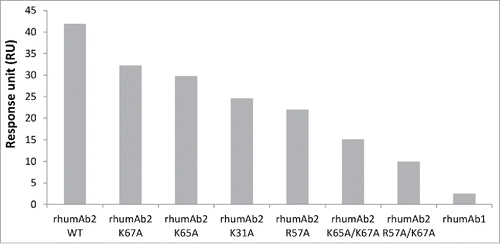
To test whether the unusually high FcRn binding responses observed for rhumAb2 were related to the 4 basic residues that form the positive charge patch on this antibody (HC-R57, HC-K65, LC-K31, LC-K67), six rhumAb2 variants were created where one or two of these residues were substituted with alanine. The binding of these variants to recombinant rat FcRn was evaluated at pH 6.0 using assay format 2. As shown in , all variants showed decreased FcRn binding response levels compare with the wild type rhumAb2. Not surprisingly, the antibodies containing 2 mutations had greater signal reductions than the wild type antibody or the antibodies containing a single mutation. It is also interesting to note that changing arginine 57 to alanine reduced the FcRn binding response the most, either individually (mutant R57A) or simultaneously with lysine 67 (mutant R57A/K67A), as compare with other residues forming the charge patch.
Figure 6. Binding response levels of wild type rhumAb2 and its variants to immobilized recombinant rat FcRn at acidic pH (pH 6.0). Concentration of tested rhumAb is 3 nM. The in-line reference subtracted response was corrected by buffer sample, and was reported 5 seconds before the end of each injection. The reported data was averaged from 2 individual experimental runs. The experiments were conducted using running buffers containing PBS, 0.05% polysorbate 20, pH 6.0.
FcRn Chromatography Analysis on rhumAbs
Interactions between rhumAbs and human FcRn were further evaluated by FcRn chromatography analysis, where the FcRn was immobilized onto solid support of the column.Citation33 A pH gradient from pH 6 to 8.5 was applied for elution during the analysis. As shown in Fig. S1, rhumAb2 had a longer retention time than rhumAb1; rhumAb2 was eluted at pH 8.2 as compare with pH 7.6 for rhumAb1.
Discussion
FcRn-IgG binding interactions play a critical role in regulating circulating IgG levels in vivo,67 and affect the PK properties of therapeutic antibodies.Citation47 In vitro FcRn binding assays, when designed appropriately, are highly valuable in providing insightful information in understanding PK behavior of mAb therapeutics.
SPR-based biosensor assays have been widely used to characterize FcRn-rhumAb binding interactions.Citation13,17,18,21,22,23,24,25,26,35,36,39,40 Like many other methodologies that require direct immobilization of proteins, the SPR biosensor assays involve immobilization of either the rhumAb or the FcRn on sensor chips, typically via an amine coupling reaction. Numerous studies have shown that results derived from SPR biosensors correlate with those obtained in solution.Citation48,49,50 However, not all proteins retain their biological activities after being immobilized on a sensor chip. Therefore, it is critical to use more than one assay configuration to assess binding interactions. When different assay formats yield consistent results, this suggests that the biological activity of the assay reagents has not been compromised by immobilization. Although discrepant results from biosensor assays using different immobilization formats have been reported,Citation51,52,53 in their meta-analysis benchmarking study of optical biosensor data from over 1200 publications, Rich and MyszkaCitation54 state that only a few investigators have directly compared multiple immobilization and capture methods to address the potential caveats associated with protein immobilization methods.
Typically an optical biosensor method that has been developed to assess the binding of one antibody to FcRn is assumed to be applicable to other similar antibodies.Citation13,20,24,36 A 2013 review of SPR-derived binding affinities for interactions between mAb therapeutics and FcRnCitation55 reported that the majority of the researchers applied a single CM5 sensor chip-based assay format to examine FcRn binding properties of antibodies with similar structures, and that assay format 1 was used more frequently than assay format 2. We believe that the use of a single rhumAb-FcRn assay configuration for SPR studies may generate misleading data. Moreover, the use of different SPR biosensor assay formats makes it difficult to directly compare FcRn binding results from different studies. For example, different groups had conducted studies to investigate whether there is a correlation between in vitro FcRn binding affinity and in vivo PK data for rhumAbs with wild type Fc sequences. The results, however, are mixed. Two groups that used assay format 2 observed a relationship between in vitro FcRn binding affinity and in vivo PK data (Suzuki et al,Citation24 Wang et alCitation36), and reported that the Fab domain of a rhumAb can affect binding to FcRn. However, a third group, which used assay format 1, was unable to confirm these observations (Hotzel et alCitation20).
To determine whether the choice of the biosensor assay format can potentially influence the observed FcRn/rhumAb binding interaction, we characterized the binding of 2 rhumAbs to FcRn proteins using 2 alternate assay formats. The rhumAbs used for the current study share greater than 99% sequence identity in the Fc regions, and have very similar overall hydrophobicities and PI values. However, rhumAb2 has an anomalous charge distribution profile that has been mapped to a positive charge patch caused by a cluster of 4 surface-exposed basic amino acid residues. Results from a comprehensive evaluation of rhumAb1 binding to FcRn showed that binding interactions were independent of biosensor assay format: similar affinities and binding characteristics were obtained using assay formats 1 and 2. At acidic pH, the averaged KD (KD1) values of rhumAb1 to recombinant rat, cynomolgus monkey, and human FcRn proteins were 2.0, 3.0, and 3.5 uM, respectively, using format 1; and were 0.9, 3.7, and 4.3 uM, respectively, using format 2, similar to those obtained from format 1. These values are generally in good agreement with those reported for human IgG1 binding to FcRn under similar experimental conditions. Citation13,23,24,37,38,39,40 In contrast to rhumAb1, the binding interactions between rhumAb2 and FcRn were highly dependent on the assay format used. In format 1, the average KD values of rhumAb2 binding to recombinant rat, cynomolgus monkey, and human FcRn were 1.2, 2.7, and 3.2 uM, respectively. However, in format 2 when FcRn was immobilized on sensor chips, rhumAb2 showed unexpectedly high binding to all FcRn proteins under both acidic and physiological pH conditions. Although exact affinities cannot be obtained for format 2, the estimated binding affinities ought to be much higher than those of rhumAb1. This observation is consistent with orthogonal data generated using an FcRn affinity chromatography assay that suggest that rhumAb2 had stronger binding to immobilized human FcRn than rhumAb1. Further investigation indicated that the unusually high binding response of rhumAb2 to immobilized FcRn resulted from both the F(ab)’2 and Fc regions of the molecule, with F(ab)’2 being a greater contributor. Moreover, the high level of binding between the FcRn protein and the rhumAb2 is believed to be mainly driven by electrostatic interactions between F(ab’)2 and FcRn proteins since it can be readily disrupted by the addition of salt, while salt had an impact on the binding of rhumAb1 to FcRn to a less degree. This hypothesis is consistent with the observation that rhumAb2 had a higher binding response to the reference flow cell on format 2, which is expected to result from the interaction between the positively charged residues on rhumAb2 and negatively charged dextran surface on the reference flow cell.
Because there are 4 strongly basic residues in the Fab region of rhumAb2, we hypothesized that this cluster of basic residues may be responsible for the unexpectedly high levels of binding seen between this antibody and FcRn proteins. Under acidic conditions at pH 6.0, arginine (pKa 12.5) and lysine (pKa 10.5) can form stronger salt bridges with acidic residues on FcRn than histidine (pKa 6.0), the key basic residue in the Fc region accountable for a typical FcRn-Fc interaction.Citation56 Moreover, under physiological conditions at pH 7.4, unlike histidine, both arginine and lysine remain positively charged and can still form salt bridges with FcRn, although the resulting interactions between IgG and FcRn are weaker due to loss of protons.
Because the positive charge patch is in the Fab region of rhumAb2, F(ab)’2 derived from this antibody should also bind to FcRn and may also bind to the negatively charged surface of a CM5 chip. This was verified experimentally. When we used rhumAb2 variants where the 4 positively charged residues were replaced with alanine either individually or simultaneously, this significantly reduced the binding of rhumAb2 to immobilized recombinant rat FcRn. In addition, the higher pKa value of arginine than lysine explains why mutating arginine 57 to alanine had the greatest effect.
The unusual interaction between rhumAb2 and FcRn is observed only when using format 2, but not format 1, of the biosensor assay. This result can be explained again by the residues in the positive charge patch. Since the amine coupling immobilization method used in many biosensor-based assays often modifies the lysine residues of the immobilized protein, and the positive charge patch in rhumAb2 contains 3 lysine residues, binding between the Fab charge patch and FcRn will likely be disrupted by the antibody's immobilization in assay format 1. Therefore, the detected binding using format 1 is driven by the expected specific Fc-FcRn interaction. The only non-lysine residue in the positive charge patch on the Fab region of rhumAb2, HC-R57, might account for the slightly higher FcRn binding response at both pH 6.0 and pH 7.4 using format 1 as compare with rhumAb1.
Recently, Abdiche et al observed that amine coupling an IgG antibody on a CM5 biosensor chip resulted in a 2- to 4-fold weaker affinity (mainly driven by a slower association rate constant) than those obtained using 2 other alternate biosensor assay formats,Citation25 although the exact mechanism for the difference was not given. In our study, the extent of the format-dependent difference for rhumAb2-FcRn far exceeded any SPR-based data reported so far (), suggesting that a fundamentally different mechanism that accounts for the atypical behavior of rhumAb2.
It is worth mentioning that Bjorkman and coworkers previously reported that the binding of IgG to FcRn was influenced by SPR biosensor assay format.Citation41,42 They observed that binding affinities of IgG to FcRn were over 100-fold higher when FcRn, instead of IgG, was immobilized on biosensor chips. The observed assay format dependence in those studies was attributed to FcRn dimerization on the biosensors;Citation41 there was no indication of additional contributing factors (e.g., positive charge patch in the Fab region) discussed in their studies. In their studies, FcRn was immobilized at very high densities (several thousands of RU) that likely promoted FcRn dimerization. In our study, a much lower density (around 25 RU) of FcRn was used so dimerization was not expected to contribute to the observed affinity difference for rhumAb2 using different assay formats. This assumption is further supported by the rhumAb1 binding data: similar binding affinities were observed for rhumAb1 to FcRn proteins regardless of the assay format.
It is well recognized that every biosensor assay format has pros and cons in studying FcRn-rhumAb binding, and so far no caveat has been reported that precludes use of any format. Previously, concerns about avidity effects associated with the bivalent nature of antibody binding to FcRn on format 2 have been discussed.Citation13 The avidity effects can result in enhanced affinity values compare with those measured from a 1:1 interaction,Citation57 which offers an explanation for generally higher rhumAb-FcRn binding affinities measured on format 2 than those from format 1 in the literature.Citation55 This avidity concern, however, can be addressed through careful experimental design such as lowering the immobilization level. Therefore, the choice of assay formats seems to be mainly driven by a combination of practical considerations (e.g., reagent availability, ease of molecule comparison) and the preference of each individual laboratory. Some may choose format 1Citation13,20,23,39 because it most likely offers monovalent binding affinity, and some may prefer format 2Citation17,18,24,36 because it better mimics the rhumAb-FcRn interaction orientation in endosomes.Citation13,55 Others may opt for an indirect formatCitation25,43 or measuring solution affinity based on FcRn active concentration determination using an antibody-immobilized assay format.Citation25 The latter methods bypass the need of direct modification of lysine residues during the amine coupling immobilization; however, there are other limitations associated with them. Depending on the nature of the interaction between a rhumAb and the capture molecule (usually antigen or anti-Fab antibodies), it is possible that a positive charge patch in the paratope of a rhumAb can be occluded. In addition, rhumAbs, when complexed with capturing molecules, may exhibit different FcRn binding due to allosteric effect. Our study provided, for the first time, clear evidence of a fundamental caveat associated with format 1 due to the potential disruption of a positive charge patch in the Fab region. It is reasonable to further speculate that the solution affinity determination using the antibody-immobilized format may suffer a similar limitation.
As rhumAb-FcRn interactions are mainly governed by charge-charge interactions, it is crucial to assess physicochemical attributes of therapeutic mAb candidates early in drug development. Our study will have a general impact on decisions regarding how to best evaluate FcRn-rhumAb interactions using SPR biosensor assays because the positive charge patch observed with rhumAb2 is not an isolated case. Schoch et alCitation58 recently reported that a rhumAb with a short half-life in human, briakinumab, also possesses a large positive charge patch in the Fab region, which contributed to the antibody's binding to FcRn. Interestingly, we found that rhumAb2 had faster clearance than rhumAb1 in the cynomolgus monkey (14.2 ± 3.4 vs. 5.0 ± 0.6 mL/day/kg) (4 animals were used in each PK study; details not shown). This faster clearance could potentially be attributed to multiple causes, such as off-target binding due to the positive charge patchCitation59 or FcRn interactions. The unexpected binding characteristic of rhumAb2 to FcRns observed using Biacore assay format 2, which would be missed if only assay format 1 was used, might potentially offer insightful information in better understanding the PK behavior of this antibody. Despite the fact that many methods have been developed to evaluate FcRn-rhumAb interactions, there remains a need for a robust and reliable methodology that can provide biologically meaningful FcRn binding information. As reported here and elsewhere, multiple mechanisms can be involved in rhumAb-FcRn binding.Citation24,36,58 Therefore, a sensitive technology that provides relevant information and the ability to delineate different contributions from various interactions (e.g., FcRn binding that involves Fc vs. non-Fc regions) can be of great value. SPR biosensor assays offer a unique feature in that different formats can be readily constructed and rich information such as binding affinities and kinetics can be obtained, but data can be confusing because the commonly used immobilization method may alter actual binding events. Therefore, careful experimental design is needed in studying bimolecular interactions by biosensor assays.
Here, we described the binding of a closely related pair of monoclonal antibodies to the FcRn. One of this pair shows the expected canonical binding profile while the second, which shares 99% sequence identity in the Fc region with the first, shows an additional, non-saturable binding component when FcRn is coupled to the biosensor chip. This additional binding can be linked to a patch of positively charged residues in the Fab region of rhumAb2; SPR analysis of variants created by site directed mutagenesis showed that removal of this charge patch eliminated non-saturable binding. The difference in binding profiles for the 2 antibodies can clearly be seen using one SPR biosensor assay format, but not using a more commonly used alternate format. For therapeutic monoclonal antibodies in early development, surface charge distribution is often unknown. The use of 2 independent SPR biosensor assay formats is highly desirable and should be applied whenever possible. Subsequent studies are ongoing to further understand the effect of charge distribution on rhumAbs binding to FcRn.
Materials and methods
FcRn expression and purification
Recombinant human, cynomolgus monkey, and rat FcRn proteins were expressed in Chinese hamster ovary (CHO) cells. Plasmids were transiently transfected into CHO cells using PEI (Polyplus) as previously described.Citation60 In all cases, harvested cell culture fluid (HCCF) was ultrafiltered (UF) 10-fold using a tangential flow filtration (TFF) system equipped with 10 kDa molecular weight cutoff (MWCO) membranes. The resulting UF HCCFs were all purified by affinity chromatography.
UF HCCF was pH adjusted to 6.5 with 1 M phosphoric acid. The pH adjusted UF HCCF was purified as a series of cycles on an IgG Sepharose 6 Fast Flow affinity chromatography column (GE Healthcare). The column was equilibrated with 50 mM Tris, 150 mM NaCl, 0.02% sodium azide, pH 6.5. Following loading of the UF HCCF, the column was washed with the same equilibration buffer, and then eluted with 50 mM Tris, 150 mM NaCl, 0.02% sodium azide, pH 8.0. Fractions containing the FcRn reagents were pooled for formulation.
The IgG Sepharose pools were concentrated using Amicon Ultra 10 kDa MWCO units (Millipore Corporation), and then formulated using Sephadex G-25 size-exclusion chromatography (SEC) (GE Healthcare). The SEC column was cleaned with one column volume (CV) of 0.5 M NaOH, and then equilibrated with 3 CVs of phosphate-buffered saline (PBS), pH 7.2. Following loading, the SEC column was developed with PBS, pH 7.2. Fractions containing the FcRn reagents were pooled for final concentration.
The Sephadex G-25 pools were concentrated to a protein concentration of 10–15 mg/mL using Amicon Ultra 10 kDa MWCO units. Final concentrated, recombinant FcRn reagents in the formulation buffer were characterized for freeze-thaw stability by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and high performance liquid chromatography size-exclusion chromatography (HPLC-SEC). The reagents were further characterized by N-terminal sequence analysis and peptide mass fingerprinting (PMF) to confirm identify and purity.
Generation of rhumAb 1 and rhumAb 2 and their F(ab)’2
rhumAb1 and rhumAb2, which are both IgG1 antibodies, were manufactured in-house at Genentech (South San Francisco, CA). To generate the F(ab)′2, 30 mg of rhumAbs at 10 mg/ml in PBS pH 7.4 were digested with 1000U of Fabricator (Genovis) for 2 hours at 37°C. The F(ab)’2 digest was then separated on an S200 (GE Healthcare) size exclusion column in 200 mM argi9, 137 mM succinic acid, 1 mM sodium azide, pH 5.0; fractions corresponding to the F(ab)’2 peak were pooled and buffer exchanged into PBS. Mass spectrometry (Agilent 6210 Time-of-Flight LC/MS) confirmed the predicted mass of the F(ab)’2. The formulated proteins were greater than 95% pure and contained no high molecular weight species as determined by SDS-PAGE gel and SEC analysis using OD280 and multi-angle static light scattering (Wyatt Optilab rex, Dawn Helios-II, Agilent HPLC).
Kinetics affinity measurement of rhumAbs binding to FcRn
Binding kinetics and affinities between FcRn and rhumAbs were evaluated on 2 different assay formats by SPR technology on a Biacore T200 instrument (GE Healthcare; Piscataway, NJ).
The first assay format involved immobilizing rhumAbs directly onto the sensor chip. Briefly, rhumAbs manufactured in-house were coupled onto 2 of the 4 different flow cells, designated here as FC1–4, of a Series S CM5 sensor chip (GE Healthcare), FC2 and FC4, using a standard amine coupling and blocking procedure recommended by the manufacturer. The immobilization levels were ∼200 response units (RU). The remaining 2 flow cells, FC1 and FC3, were treated in a similar manner except that no proteins were immobilized before blocking. FC1 and FC3 were used as in-line reference cells for FC2 and FC4, respectively. FcRn at various concentrations diluted into running buffer (PBS, 0.05% polysorbate 20, pH 6.0 or pH 7.4) were injected over all 4 flow cells at a flow rate of 50 μL/min for 1 minute, and the dissociation of FcRn was allowed to proceed for 2 minutes. Surfaces were regenerated using PBS (pH 7.4) between cycles at a flow rate of 50 μL/minute twice for 35 seconds each. This protocol takes advantage of the unique pH dependency of FcRn: they typically bind to IgGs at acidic pH (< 6.5) and release IgG at physiological pH (∼7.4). The experiments were performed at 37°C.
Experiments measuring binding interactions between FcRn and rhumAbs on the second assay format were performed as described above, except for the changes described below. FcRn were immobilized onto FC2 and FC4 of the sensor chip at densities of ∼25 RU. rhumAbs diluted in running buffer were injected over all 4 flow cells at a flow rate of 50 μL/minute for 1 minute and the dissociation of rhumAbs from immobilized FcRn on the CM5 chip was allowed to proceed 2 minutes.
Binding affinity and kinetic parameters (association and dissociation rate constants, and dissociation equilibrium constant) were calculated with the Biacore T200 Evaluation Software (version 1.0; GE Healthcare) using either a 1:1 binding model or a steady-state equilibrium model. All sensorgrams were generated after in-line reference cell correction followed by buffer sample subtraction.
rhumAb and F(ab)’2 binding to FcRn in the presence of salt
Experiments measuring the binding interactions between FcRn and rhumAb F(ab)’2 were performed essentially as described above, except that each injection consisted of a 2-minutes association phase at a flow rate of 5 µL/minute. rhumAb and F(ab)’2 samples were diluted to 5 µM in running buffer with different amount of NaCl added (0–0.5 M). The reference-corrected binding responses were reported 5 seconds before the end of each injection.
Binding of rhumAb2 and its mutants to FcRn
rhumAb2 mutants with residues replaced with alanine at the following 4 positions in rhumAb2 (HC-R57, HC-K65, LC-K31, LC-K67) were created in-house at Genentech: R57A, K65A, K31A, K67A, K65A/K67A, and R57A/K67A. Binding interactions between rhumAb2 mutants and FcRn were evaluated with rat FcRn being immobilized on biosensor chip. Wild type rhumAb2 and its mutants diluted in running buffer were injected over all 4 flow cells at a flow rate of 50 μL/minute for 90 seconds. The binding activity of mutants to immobilized rat FcRn was reference flow cell and buffer subtracted, and was reported as the response 5 seconds before the end of each injection. The experiments were performed at 37°C.
Disclosure of potential conflicts of interest
All authors are employees of Genentech, a member of the Roche group.
Supplemental_Data.docx
Download MS Word (386.2 KB)Acknowledgements
The authors would like to thank Patricia Siguenza for her support, Liangyi Zhang for generating rhumAb2 mutants, Meina Tang, Amrita Kamath for helpful discussions, and several Genentech colleagues for providing their helpful reviews of this manuscript.
References
- Simister NE, Rees AR. Isolation and characterization of an Fc receptor from neonatal rat small intestine. Eur J Immun 1985; 15:733-38; PMID:2988974; http://dx.doi.org/10.1002/eji.1830150718
- Simister NE, Mostov KE. Cloning and expression of the neonatal rat intestinal Fc receptor, a major histocompatibility complex class I antigen homolog. Cold Spring Harb Symp Quant Biol 1989; 54 Pt 1:571-80; PMID:2534798; http://dx.doi.org/10.1101/SQB.1989.054.01.068
- Bjorkman PJ, Parham P. Structure, function, and diversity of class I major histocompatibility complex molecules. Annu Rev Biochem 1990; 59:253-88; PMID:2115762; http://dx.doi.org/10.1146/annurev.bi.59.070190.001345
- Rodewald, R. Intestinal transport of antibodies in the newborn rat. J Cell Biol 1973; 58:198-211; PMID:4726306; http://dx.doi.org/10.1083/jcb.58.1.189
- Story CM, Mikulska JE, Simister NE. A major histocompatibility complex class I-like Fc receptor cloned from human placenta: possible role in transfer of immunoglobulin G from mother to fetus. J Exp Med 1994; 180:2377-81; PMID:7964511; http://dx.doi.org/10.1084/jem.180.6.2377
- Ghetie V, Ward ES. Multiple roles for the major histocompatibility complex class I– related receptor FcRn. Annu Rev Immunol 2000; 18:739-66; PMID:10837074; http://dx.doi.org/10.1146/annurev.immunol.18.1.739
- Roopenian DC, Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immunol 2007; 7:715-25; PMID:17703228; http://dx.doi.org/10.1038/nri2155
- Raghavan M, Bonagura VR, Morrison SL, Bjorkman PJ. Analysis of the pH dependence of the neonatal Fc receptor/immunoglobulin G interaction using antibody and receptor variants. Biochemistry 1995; 34:14649-57; PMID:7578107; http://dx.doi.org/10.1021/bi00045a005
- Goebl NA, Babbey CM, Datta-Mannan A,Witcher DR, Wroblewski VJ, Dunn KW. Neonatal Fc receptor mediates internalization of Fc in transfected human endothelial cells. Mol Biol Cell 2008; 19:5490-505; PMID:18843053; http://dx.doi.org/10.1091/mbc.E07-02-0101
- Ober RJ, Martinez C, Lai X, Zhou J, Ward ES. Exocytosis of IgG as mediated by the receptor, FcRn: an analysis at the single-molecule level. Proc Natl Acad Sci USA 2004; 101:11076-81; PMID: 15258288; http://dx.doi.org/10.1073/pnas.0402970101
- Ober RJ, Martinez C, Vaccaro C, Zhou J, Ward ES. Visualizing the site and dynamics of IgG salvage by the MHC class I-related receptor, FcRn. J Immunol 2004; 172:2021-9; PMID:14764666; http://dx.doi.org/10.4049/jimmunol.172.4.2021
- Robbie GJ, Criste R, Dall'Acqua WF, Jensen K, Patel NK, Losonsky GA, Griffin MP. A Novel Investigational Fc-Modified Humanized Monoclonal Antibody, Motavizumab-YTE, Has an Extended Half-Life in Healthy Adults. Antimicrob Agents Chemother 2013; 57:6147-53; PMID:24080653; http://dx.doi.org/10.1128/AAC.01285-13
- Yeung YA, Leabman MK, Marvin JS, Qiu J, Adams CW, Lien S, Starovasnik MA, Lowman HB. Engineering human IgG1 affinity to human neonatal Fc receptor: impact of affinity improvement on pharmacokinetics in primates. J Immunol 2009; 182:7663-71; PMID:19494290; http://dx.doi.org/10.4049/jimmunol.0804182
- Dall'Acqua WF, Kiener PA, Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn). The J Biol Chem 2006; 281:23514-24; PMID:16793771; http://dx.doi.org/10.1074/jbc.M604292200
- Hinton PR, Xiong JM, Johlfs MG, Tang MT, Keller S, Tsurushita N. An engineered human IgG1 antibody with longer serum half-life. J Immunol 2006; 176:346-56; PMID:16365427; http://dx.doi.org/10.4049/jimmunol.176.1.346
- Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IW, Sproule TJ, Lazar GA, Roopenian DC, Desjarlais JR. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol 2010; 28:157-9; PMID:20081867; http://dx.doi.org/10.1038/nbt.1601
- Deng R, Loyet KM, Lien S, Iyer S, DeForge LE, Theil FP, Lowman HB, Fielder PJ, Prabhu S. Pharmacokinetics of humanized monoclonal anti-tumor necrosis factor-{alpha} antibody and its neonatal Fc receptor variants in mice and cynomolgus monkeys. Drug Metab Dispos 2010; 38:600-5; PMID:20071453; http://dx.doi.org/10.1124/dmd.109.031310
- Datta-Mannan A, Witcher DR, Tang Y, Watkins J, Wroblewski VJ. Monoclonal antibody clearance. Impact of modulating the interaction of IgG with the neonatal Fc receptor. J Biol Chem 2007; 19;282:1709-17; PMID:17135257; http://dx.doi.org/10.1074/jbc.M607161200
- Gurbaxani B, Dela Cruz LL, Chintalacharuvu K, Morrison SL. Analysis of a family of antibodies with different half-lives in mice fails to find a correlation between affinity for FcRn and serum half-life. Mol Immunol 2006; 43:1462-73; PMID:16139891; http://dx.doi.org/10.1016/j.molimm.2005.07.032
- Hötzel I, Theil F-P, Bernstein LJ, Prabhu S, Deng R, Quintana L, Lutman J, Sibia R, Chan P, Bumbaca D, et al. A strategy for risk mitigation of antibodies with fast clearance. mAbs. 2012; 4(6):753-60; PMID:23778268; http://dx.doi.org/10.4161/mabs.22189
- Ober RJ, Radu CG, Ghetie V, Ward ES. Differences in promiscuity for antibody-FcRn interactions across species: implications for therapeutic antibodies. Int Immunol 2001; 13:1551-9; PMID:11717196; http://dx.doi.org/10.1093/intimm/13.12.1551
- Kelley RF, Meng YG. Methods to engineer and identify IgG1 variants with improved FcRn binding or effector function. Methods Mol Biol 2012; 901:277-93; PMID:22723108; http://dx.doi.org/10.1007/978-1-61779-931-0_18
- Dall'Acqua WF, Woods RM, Ward ES, Palaszynski SR, Patel NK, Brewah YA, Wu H, Kiener PA, Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J Immunol 2002; 169:5171-80; PMID:12391234; http://dx.doi.org/10.4049/jimmunol.169.9.5171
- Suzuki T, Ishii-Watabe A, Tada M, Kobayashi T, Kanayasu-Toyoda T, Kawanishi T, Yamaguchi T. Importance of neonatal FcR in regulating the serum half-life of therapeutic proteins containing the Fc domain of human IgG1: a comparative study of the affinity of monoclonal antibodies and Fc-fusion proteins to human neonatal FcR. J Immunol 2010; 184:1968-76; PMID:20083659; http://dx.doi.org/10.4049/jimmunol.0903296
- Abdiche YN, Yeung YA, Chaparro-Riggers J, Barman I, Strop P, Chin SM, Pham A, Bolton G, McDonough D, Lindquist K, Pons J, Rajpal A. The neonatal Fc receptor (FcRn) binds independently to both sites of the IgG homodimer with identical affinity. mAbs 2015; 7:331-43; PMID: 25658443; http://dx.doi.org/10.1080/19420862.2015.1008353
- Neuber T, Frese K, Jaehrling J, Jäger S, Daubert D, Felderer K, Linnemann M, Höhne A, Kaden S, Kölln J, et al. Characterization and screening of IgG binding to the neonatal Fc receptor. mAbs 2014; 6:928-42; PMID:24802048; http://dx.doi.org/10.4161/mabs.28744
- Huber AH, Kelley RF, Gastinel LN, Bjorkman PJ. Crystallization and stoichiometry of binding of a complex between a rat intestinal Fc receptor and Fc. J Mol Biol 1993; 230:1077-83; PMID:8478919; http://dx.doi.org/10.1006/jmbi.1993.1220
- Lu Y, Vernes J, Chiang N, Ou Q, Ding J, Adams C, Hong K, Truong B, Ng D, Shen A, et al. Identification of IgG(1) variants with increased affinity to Fcγ RIIIa and unaltered affinity to FcγRI and FcRn: Comparison of soluble receptor-based and cell-based binding assays. J Immunol Methods 2011; 365:132-41; PMID:21185301; http://dx.doi.org/10.1016/j.jim.2010.12.014
- Shields RL, Namenuk AK, Hong K, Meng YG, Rae J, Briggs J, Xie D, Lai J, Stadlen A, Li B, et al. High resolution mapping of the binding site on human IgG1 for Fc gamma RI, Fc gamma RII, Fc gamma RIII, and FcRn and design of IgG1 variants with improved binding to the Fc gamma R. J Biol Chem 2001; 276:6591-604; PMID:11096108; http://dx.doi.org/10.1074/jbc.M009483200
- Hinton PR, Johlfs MG, Xiong JM, Hanestad K, Ong KC, Bullock C, Keller S, Tang MT, Tso JY, Vásquez M, et al. Engineered human IgG antibodies with longer serum half-lives in primates. J Biol Chem. 2004; 279:6213-6; PMID:14699147; http://dx.doi.org/10.1074/jbc.C300470200
- Mathur A, Arora T, Liu L, Crouse-Zeineddini J, Mukku V. Qualification of a homogeneous cell-based neonatal Fc receptor (FcRn) binding assay and its application to studies on Fc functionality of IgG-based therapeutics. J Immunol Methods 2013; 390:81-91; PMID:23384837; http://dx.doi.org/10.1016/j.jim.2013.01.011
- Wu Q, Lee HY, Wong PY, Jiang G, Gazzano-Santoro H. Development and applications of AlphaScreen-based FcRn binding assay to characterize monoclonal antibodies. J Immunol Methods 2015; 420:31-7; PMID:25837414; http://dx.doi.org/10.1016/j.jim.2015.03.012
- Schlothauer T, Rueger P, Stracke JO, Hertenberger H, Fingas F, Kling L, Emrich T, Drabner G, Seeber S, Auer J, et al. Analytical FcRn affinity chromatography for functional characterization of monoclonal antibodies. mAbs 2013; 5:576-86; PMID:23765230; http://dx.doi.org/10.4161/mabs.24981
- Pollastrini J, Dillon TM, Bondarenko P, Chou RY. Field flow fractionation for assessing neonatal Fc receptor and Fcγ receptor binding to monoclonal antibodies in solution. Anal Biochem 2011; 414:88-98; PMID:21385563; http://dx.doi.org/10.1016/j.ab.2011.03.001
- Magistrelli G, Malinge P, Anceriz N, Desmurs M, Venet S, Calloud S, Daubeuf B, Kosco-Vilbois M, Fischer N. Robust recombinant FcRn production in mammalian cells enabling oriented immobilization for IgG binding studies. J Immunol Methods 2012; 375:20-9; PMID:21939661; http://dx.doi.org/10.1016/j.jim.2011.09.002
- Wang W, Lu P, Fang Y, Hamuro L, Pittman T, Carr B, Hochman J, Prueksaritanont T. Monoclonal antibodies with identical Fc Sequences Can Bind to FcRn differentially with pharmacokinetic consequences. Drug Metab Dispos 2011; 39:1469-77; PMID:21610128; http://dx.doi.org/10.1124/dmd.111.039453
- Datta-mannan A, Witcher DR, Tang WY, Watkins JD, Jiang W, Wroblewski VJ. Humanized IgG1 Variants With Differential Binding Properties To The Neonatal Fc Receptor: relationship to pharmacokinetics in mice and primates. Drug Metab Dispos 2007; 35:85-94; PMID: 17050651; http://dx.doi.org/10.1124/dmd.106.011734
- Firan M, Bawdon R, Radu C, Ober RJ, Eaken D, Antohe F, Ghetie V, Ward ES. The MHC class I-related receptor, FcRn, plays an essential role in the maternofetal transfer of gamma-globulin in humans. Int Immunol 2001; 13:993-1002; PMID:11470769; http://dx.doi.org/10.1093/intimm/13.8.993
- Yeung YA, Wu X, Reyes AE, Vernes J, Lien S, Lowe J, Maia M, Forrest WF, Meng YG, Damico LA, et al. A Therapeutic Anti–VEGF Antibody with Increased Potency Independent of Pharmacokinetic Half-life. Cancer Res 2010; 70:3269-77; PMID:20354184; http://dx.doi.org/10.1158/0008-5472.CAN-09-4580
- Khawli LA, Goswami S, Hutchinson R, Kwong ZW, Yang J, Wang X, Yao Z, Sreedhara A, Cano T, Tesar DB, et al. Charge variants in IgG1 Isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs 2010; 6:613-24; PMID:20818176; http://dx.doi.org/10.4161/mabs.2.6.13333
- Raghavan M, Wang Y; Bjorkman PJ. Effects of receptor dimerization on the interaction between the class I major histocompatibility complex-related Fc receptor and IgG. Proc Natl Acad Sci USA 1995; 92:11200-04; PMID:7479965; http://dx.doi.org/10.1073/pnas.92.24.11200
- Vaughn DE, Bjorkman PJ. High-affinity binding of the neonatal Fc receptor to its IgG ligand requires receptor immobilization. Biochemistry 1997; 36:9374-80; PMID:9235980; http://dx.doi.org/10.1021/bi970841r
- Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, Nanami M, Sekimori Y, Nabuchi Y, Aso Y, et al. Reduced elimination of IgG antibodies by engineering the variable region. Protein Engineering, Design & Selection 2010; 23:385-92; PMID:20159773; http://dx.doi.org/10.1093/protein/gzq009
- Zhang L, Lilyestrom W, Charlene Li, Thomas Scherer, Robert van Reis, and Boyan Zhang. Revealing a Positive Charge Patch on a Recombinant Monoclonal Antibody by Chemical Labeling and Mass Spectrometry. Anal Chem 2011; 83:8501-08; PMID:22004540; http://dx.doi.org/10.1021/ac2016129
- Sánchez LM, Penny DM, Bjorkman PJ. Stoichiometry of the interaction between the major histocompatibility complex-related Fc receptor and its Fc ligand. Biochemistry 1999; 38:9471-6; PMID:10413524; http://dx.doi.org/10.1021/bi9907330
- Huber AH, Kelley RF, Gastinel LN, Bjorkman PJ. Crystallization and stoichiometry of binding of a complex between a rat intestinal Fc receptor and Fc. J Mol Biol 1993; 230:1077-83; PMID:8478919; http://dx.doi.org/10.1006/jmbi.1993.1220
- Vugmeyster Y, Xu X, Theil F, Khawli LA, Leach MW. Pharmacokinetics and toxicology of therapeutic proteins: Advances and challenges. World J Biol Chem 2012; 3:73-92; PMID:22558487; http://dx.doi.org/10.4331/wjbc.v3.i4.73
- Day YS, Baird CL, Rich RL, Myszka DG. Direct comparison of binding equilibrium, thermodynamic, and rate constants determined by surface- and solution-based biophysical methods. Protein Sci 2002; 11:1017-25; PMID:11967359; http://dx.doi.org/10.1110/ps.4330102
- Matsumoto-Takasaki A, Hanashima S, Aoki A, Yuasa N, Ogawa H, Sato R, Kawakami H, Mizuno M, H. Nakada, Yamaguchi Y, et al. Surface plasmon resonance and NMR analyses of anti Tn-antigen MLS128 monoclonal antibody binding to two or three consecutive Tn-antigen clusters. J Biochem 2011; 151:273-82; PMID:22161472; http://dx.doi.org/10.1093/jb/mvr138
- Drake A, Myszka DG, Klakamp SL. Characterizing high-affinity antigen/antibody complexes by kinetic- and equilibrium-based methods. Anal Biochem 2004; 328:35-43; PMID:15081905; http://dx.doi.org/10.1016/j.ab.2003.12.025
- HoareHL, SullivanLC, Pietra G, Clements CS, Lee EJ, Ely LK, Beddoe T, Falco M, Kjer-Nielsen L, Reid LH, et al. Structural basis for a major histocompatibility complex class Ib–restricted T cell response. Nat Immunol 2006; 7:256-64; PMID:16474394; http://dx.doi.org/10.1038/ni1312
- Trilling AK, Harmsen MM, Ruigrok VJ, Zuilhof H, Beekwilder J. The effect of uniform capture molecule orientation on biosensor sensitivity: dependence on analyte properties. Biosens Bioelectron 2013; 40:219-26; PMID:22878083; http://dx.doi.org/10.1016/j.bios.2012.07.027
- Yang J, Wang X, Fuh G, Yu L, Wakshull E, Khosraviani M, Day ES, Demeule B, Liu J, Shire SJ, et al. Comparison of binding characteristics and in vitro activities of three inhibitors of vascular endothelial growth factor A. Molecular Pharmaceutics 2014; 11:3421-30; PMID:25162961; http://dx.doi.org/10.1021/mp500160v
- Rich RL, Myszka DG. Survey of the year 2006 commercial optical biosensor literature. J Mol Recognit 2007; 20:300-66; PMID:18074396; http://dx.doi.org/10.1002/jmr.862
- Gurbaxani B, Dostalek M, Gardner I. Are endosomal trafficking parameters better targets for improving mAb pharmacokinetics than FcRn binding affinity? Mol Immunol. 2013 Dec; 56:660-74; PMID:23917469; http://dx.doi.org/10.1016/j.molimm.2013.05.008
- Vaughn DE. Identification of critical IgG binding epitopes on the neonatal Fc receptor. J Mol Biol 1997; 274:597-607; PMID:9417938; http://dx.doi.org/10.1006/jmbi.1997.1388
- Kaufman EN1, Jain RK. Effect of bivalent interaction upon apparent antibody affinity: experimental confirmation of theory using fluorescence photobleaching and implications for antibody binding assays. Cancer Res. 1992;52:4157-67.
- Schoch A, Kettenberger H, Mundigl O, Winter G, Engert J, Heinrich J, Emrich T. Charge-mediated influence of the antibody variable domain on FcRn-dependent pharmacokinetics. Proc Natl Acad Sci U S A 2015; 112:5997-6002; PMID:25918417; http://dx.doi.org/10.1073/pnas.1408766112
- Bumbaca Yadav D, Sharma VK, Boswell CA, Hotzel I, Tesar D, Shang Y, Ying Y, Fischer SK, Grogan JL, Chiang EY, et al. Evaluating the Use of Antibody Variable Region (Fv) Charge as a Risk Assessment Tool for Predicting Typical Cynomolgus Monkey Pharmacokinetics. J Biol Chem 2015; 290:29732-41; PMID:26491012; http://dx.doi.org/10.1074/jbc.M115.692434
- Wong AW, Baginski TK, Reilly DE. Enhancement of DNA uptake in FUT8-deleted CHO cells for transient production of afucosylated antibodies. Biotechnology and Bioengineering 2010; 106:751-63; PMID:20069558; http://dx.doi.org/10.1002/bit.22749
