ABSTRACT
We isolated HIV-1 Envelope (Env)-specific memory B cells from a cow that had developed high titer polyclonal immunoglobulin G (IgG) with broad neutralizing activity after a long duration vaccination with HIV-1AD8 Env gp140 trimers. We cloned the bovine IgG matched heavy (H) and light (L) chain variable (V) genes from these memory B cells and constructed IgG monoclonal antibodies (mAbs) with either a human constant (C)-region/bovine V-region chimeric or fully bovine C and V regions. Among 42 selected Ig+ memory B cells, two mAbs (6A and 8C) showed high affinity binding to gp140 Env. Characterization of both the fully bovine and human chimeric isoforms of these two mAbs revealed them as highly type-specific and capable of binding only to soluble AD8 uncleaved gp140 trimers and covalently stabilized AD8 SOSIP gp140 cleaved trimers, but not monomeric gp120. Genomic sequence analysis of the V genes showed the third heavy complementarity-determining region (CDRH3) of 6A mAb was 21 amino acids in length while 8C CDRH3 was 14 amino acids long. The entire V heavy (VH) region was 27% and 25% diverged for 6A and 8C, respectively, from the best matched germline V genes available, and the CDRH3 regions of 6A and 8C were 47.62% and 78.57% somatically mutated, respectively, suggesting a high level of somatic hypermutation compared with CDRH3 of other species. Alanine mutagenesis of the VH genes of 6A and 8C, showed that CDRH3 cysteine and tryptophan amino acids were crucial for antigen binding. Therefore, these bovine vaccine-induced anti-HIV antibodies shared some of the notable structural features of elite human broadly neutralizing antibodies, such as CDRH3 size and somatic mutation during affinity-maturation. However, while the 6A and 8C mAbs inhibited soluble CD4 binding to gp140 Env, they did not recapitulate the neutralizing activity of the polyclonal antibodies against HIV infection.
Introduction
While there are numerous approved or investigational drugs against HIV infection,Citation1 passive antibody prophylaxis and immunotherapy could hold a valuable place in both the prevention and treatment of human immunodeficiency virus (HIV) infection. Various novel monoclonal antibodies (mAbs) that have been derived from HIV-infected individuals show extremely high potency with neutralization breadth that includes many HIV-1 strains. Furthermore, the efficacy of passive transfer of broadly neutralizing antibodies (BrNAbs) to block HIV-1 transmission in humans was shown in HIV-1 infection studies in humanized mice and simian/HIV infection in macaques.Citation2-5
Eliciting effective antibody responses plays a fundamental role in vaccine development. If a purified therapeutic antibody is protective, then eliciting similar antibodies with a vaccine could protect against the relevant pathogen.Citation6 However, despite three decades of attempts, prophylactic vaccines to prevent HIV transmission have not been successful. The most challenging factor toward achieving an effective vaccine has been eliciting BrNAbs against a wide array of circulating viral strains. An immune response producing BrNAbs is dependent on the suitability of a designed immunogen to present conserved epitopes and the capability of the host immune system to adequately respond to these elusive and highly conformational epitopes.,Citation7,8
Among various configurations of HIV Env, monomeric gp120 is relatively easy to produce, but cannot induce adequately protective antibodiesCitation9 because it dominantly presents non-neutralizing epitopes that are likely to be concealed on native trimeric spikes. These strong non-neutralizing epitopes act as a decoy for the immune system, which produces non-neutralizing antibodies that cannot bind the functional epitopes targeted by genuine neutralizing antibodies.Citation10 In comparison, trimeric soluble Envelope (Env) gp140 has been much better than gp120 monomers in stimulating BrNAbs,Citation11 with powerful responses obtained from vaccination of guinea pigs,Citation8,12 llamaCitation13 and cows.Citation14,15
Potent antibody binding to key pathogen infectivity determinants relies on the evolution of Ig CDRH3 toward high affinity interactions with an antigen.Citation16 The CDRH3 domain has the highest amino acid (aa) variability in IgG and plays the most critical role in the antigen binding interaction. Diversity arises by DNA rearrangement between the variable (IGHV), diversity (IGHD) and joining (IGHJ) genes to create CDRH3 with diverse gene sequences.Citation17,18 In contrast to human and mice, the bovine antibody diversity occurs through Ig gene somatic hypermutation (SHM), which may also happen without antigen contact at the fetal stage.Citation19,20 The ultimate result is an affinity-maturation of the variable (V) region, especially the CDRH3 domain, and this increases the IgG neutralizing activity of HIV Env –targeting antibodies.Citation21
The emergence of HIV-1 BrNAbs exhaustively extends the diversity and maturation mechanisms of human immune system compared with the IgG antibodies effective against other pathogens or the common type-specific neutralizing HIV antibodies.Citation22-24 Extensive maturation by SHM of the VH region is considered crucial for BrNAbs to HIV because un-mutated germline ancestors of BrNAbs have low or absent reactivity, let alone neutralizing potency.Citation25,26 In addition, it appears that B cells producing antibodies with long CDRH3 (20 – 34 residues) are selected in BrNAbs that target the deep epitopes of HIV-1 Env, such as the CD4 binding site (CD4bs),Citation27,28 the glycan-related V1/V2 and V3 epitopes,Citation29 the gp120/gp41 bridging region Citation24,30 and the gp41-membrane-proximal external region ( MPER).Citation31-41 Mice and rabbits have a genetic bias toward shorter CDRH3 antibodies,.Citation42,43 restricting their utility for HIV vaccine trials,Citation44 but cattle have very long CDRH3 domains of up to 67 aa,Citation20,45-48 making them a novel animal model to assess HIV vaccines.Citation14,15
The Bos taurus V gene is almost limited to one family (boVH1).Citation20,46,47 However, the bovine immune system has a novel diversification mechanism that duplicates short segments of DNA coding for aromatic aa in CDRH3, and this helps the IgG V regions to expand for effective targeting of epitopes from diverse complex pathogens. High-level SHM of bovine Ig is the main strategy for antibody evolution and efficient antigen recognition.Citation49 Bovine IgG is exceptional among other species, including human, with the long length of CDRH3 compared with other mammals.Citation20,45-48 In bovine, CDRH3 is elongated to contain frequent cysteine (Cys) residues that become engaged in disulfide bonds at the antigen binding sites.Citation48 The long CDRH3, high level of SHMs and presence of Cys residues that are normal in bovine Ig are features also identified in human HIV-neutralizing IgG. The evolution in humans of antibodies with these characteristics, however, requires a long time and is uncommon.Citation50
Previous studies of human IgG illustrate that, although the presence of Cys and aromatic residues are rare, these residues play a crucial role in epitope binding or neutralization of the virus in HIV human monoclonal BrNAbs. The substitution of aromatic residues, mainly tryptophan (Trp) and tyrosine (Tyr) in MPER-binding mAbs, reduces their neutralization activity.Citation31,51 Furthermore, the importance of Cys and aromatic residues such as Trp is shown in neutralization activity of CD4bs BrNAbs antibodies,Citation52,53 highlighting the critical function of these residues in either a direct antigen-binding interaction, or in steric effects upon the required structure for epitope access and engagement.
Vaccination of cows with uncleaved HIV AD8 strain gp140 Env (HIVAD8 gp140, AD8 clone of ADA) resulted in a high titer of BrNAbs in serum that collected in very large quantities in colostrum samples of the immunized cows.Citation14,15 The colostrum IgG had broad neutralizing activity and was able to inhibit anti-CD4bs mAbs such as b12 and VRC01,Citation14 and had antibody-dependent cell-mediated cytotoxicity activity.Citation54 Here, we describe the isolation of HIV-specific bovine memory B cells from a gp140 vaccinated cow producing BrNAbs and construction of fully bovine and chimeric human/bovine mAb antibodies that exemplify how bovine IgG engages conformation-dependent epitopes on trimeric Env gp120. We assessed the CDRH3 length and hypermutation levels of bovine anti-HIV binding antibodies and the role of Cys and other aromatic aa in HIV antigen binding activity. This study opens a path toward humanized bovine mAbs and examines the potential to harness the bovine immune system against HIV.
Results
Frequency of total and HIV-specific memory B cells in non-immune and immune peripheral blood mononuclear cells
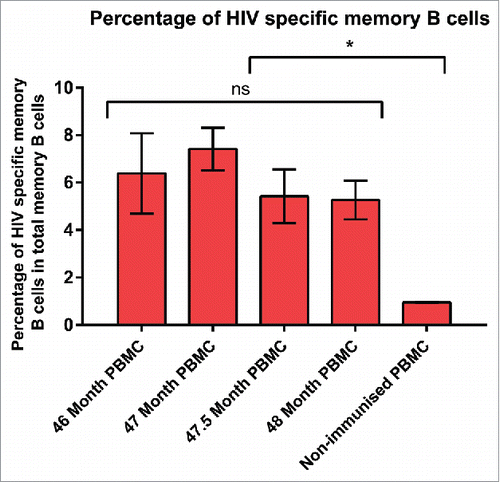
A sustained HIVAD8 Env gp140-specific reciprocal IgG titer was confirmed in serum samples from a cow (7004) that was the subject of an extended vaccination with HIVAD8 Env gp140 oligomers (Fig. S1). To further study the frequency of HIVAD8 Env gp140-specific memory B cells, an Enzyme-Linked ImmunoSpot (ELISPOT) assay was performed. Peripheral blood mononuclear cells (PBMCs) of the immunized cow (collected at 46, 47, 47.5 and 48.5 months) and the non-immunized cow were stimulated for 6 days, then incubated with AD8 gp140 Env. As shown in Fig. S2, the total number of IgG memory B cells (of 106 cultured PBMC) in PBMC collected at 46 months was 5464 ± 2050, 4084 ± 910 at 47 months, 1800 ± 709 at 47.5 months, and 1814 ± 465 at 48.5 months (nonimmune cow). HIVAD8 Env gp140-specific IgG memory B cells (of 106 cultured PBMC) was 348 ± 92, 302 ± 36, 97 ± 20 and 95 ± 14 for PBMCs collected at 46, 47, 47.5 and 48.5 months, respectively (immune cow 7004). The percentage of HIV-specific B cells in total memory B cells was consistent including 6.38 ± 1.69, 7.41 ± 0.9, 5.42 ± 1.13, and 5.26 ± 0.8 in PBMC collected at 46, 47, 47.5 and 48 months, respectively (). The average percentage of assay background in total memory B cells of non-immune PBMC was 0.95 ± 0.01. The presence of anti-AD8 HIV antibodies in serum samples and confirming AD8-specific memory B cells in ELISPOT assay was the essential step to start isolating AD8-specific memory B cells.
Figure 1. Percentage of HIVAD8 Env gp140-specific memory (B) cells in total memory (B) cells. Antibody secreting cell number was calculated according to the number of spots formed on the plates coated with AD8 gp140. Cut-off spot forming unit was five per well. Data represents mean of 2 replicates (intra-variability) and error bars show standard deviation (SD). P values were calculated using one-way ANOVA followed by a Tukey honest significant difference (HSD) test post-test (ns, not significant; *P ≤ 0.05).
Detection of Env-specific memory B cells from vaccinated cow 7004 by flow cytometry
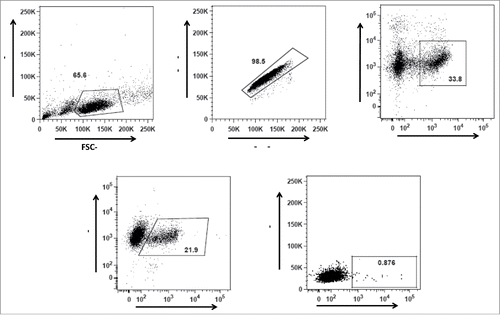
After biotinylation of gp140, the activity of epitope sites was confirmed as mentioned in text S1 and Fig. S3. Then, gp140 reactive memory IgG B cells were identified as viable CD21+ IgG+ gp140+ cells within the lymphocyte gate. The frequency of AD8 gp140-specific memory B cells in the vaccinated PBMC sample of cow 7004 was 0.87% of IgG+ CD21+ cells (). In brief, 30 million PBMC were stained and analyzed using flow cytometry. In total, 95 CD21+ IgG+ gp140+ cells were gp140 AD8+ and therefore sorted into single wells.
Figure 2. Gating strategy for gp140 binding IgG+ (B)cells from the vaccinated and unvaccinated cows. (A) Gating strategy for lymphocytes according to their size and granularity, (B) Gating on singlet cells, (C) Gating for viable CD21+ B cells, and (D) Gating of IgG+ CD21+ cells. Gp140 reactive memory IgG B cells (E) were identified as viable CD21+ IgG+ gp140+ cells within the lymphocyte gate.
Amplification of bovine constant/variable gene
The bovine heavy gamma (1010 bp) and light lambda (330 bp) constant genes were amplified by PCR (). Genes were cloned into pFUSE gamma and lambda expression vectors to construct bovine constant region expression vectors. The accuracy of cloning was confirmed by sequencing.
Figure 3. Bovine genes amplification and expression. (A) Representative of bovine heavy and light constant gene amplification. (B) Representative of amplification of Igγ and Igλ genes in single cell RT-PCR. The fragments amplified in the second round PCR were heavy Igγ (350–500bp) and light Igλ (320–340bp). (C) Representative cell culture supernatants of chimeric mAbs on a 12% reducing gel. Negative control includes the supernatant of cells mock transfected. The supernatant of VRC01 transfection was used as positive control for assessment of transfection and western blotting.
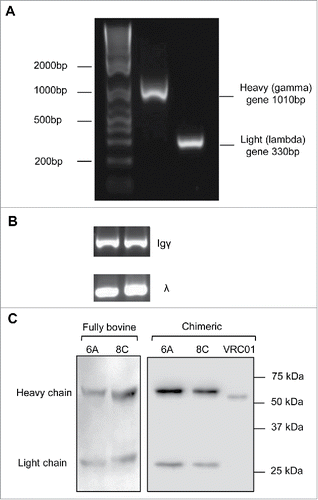
The heavy and light V genes were amplified with nested PCR. The fragments amplified in the first round PCR were 450–700bp, including signal peptide, V gene and some nucleotides from the constant gene. The second round PCR resulted in shorter fragments, in the range of 300–500bp (). The Ig heavy reverse external primer was designed to amplify only Igγ1 and Igγ2 genes since Igγ3 is not expressed at the protein level.Citation55 In addition, because the bovine immune system expresses Ig lambda chain preferentially,Citation56 in this study, the Ig light reverse external primer targets all lambda genes (Ig λ 1–4) but not kappa chain genes. Out of 95 cells, 42 cells showed an Ig heavy V gene band, but the Ig lambda V gene for only 39 cells could be amplified (positive for both heavy and light V gene) and selected for sub-cloning and expression cloning.
Expression of Chimeric/full bovine antibodies
The bovine V genes were digested and cloned into expression vectors with human/bovine light/heavy constant genes. 293T cells were co-transfected with the recombinant expression vectors and the cell culture supernatants were harvested for further analysis by western blotting () and enzyme-linked immunosorbent assay (ELISA). Out of 39 co-transfections of paired heavy/light expression vectors, 32 mAbs were expressed efficiently. Two mAbs (6A and 8C) showed anti-AD8 gp140-specific binding activity in ELISA assay. Chimeric and bovine versions of 6A and 8C were then expressed in large scale and purified using Protein G (prt G) agarose.
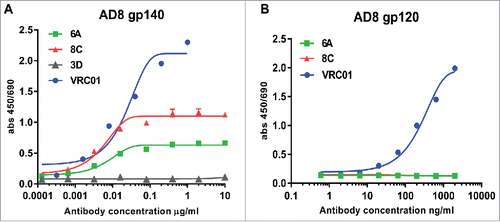
6A and 8C chimeric mAbs bind to conformation-dependent epitopes on trimeric AD8 gp120 Env but not AD8 gp120 or gp41 monomers: The harvested supernatants were assessed in a direct ELISA to determine HIV-1 AD8-specific reactivity. Only two mAbs (6A and 8C) showed anti-AD8 gp140-specific binding activity assessed by ELISA (). The end-point concentration of 6A and 8C mAb was 3.2 ng/ml and 0.64 ng/ml, respectively. Though 6A and 8C chimeric mAbs bind to AD8 gp140 Env, they do not show any binding activity against YU-2, MW and PSC 89 gp140 Env, which indicates that 6A and 8C mAbs are strain-specific antibodies (data not shown). In contrast to VRC01 mAb, 6A and 8C chimeric mAbs exhibited no binding activity to monomeric gp120 in ELISA () and immunoprecipitation (IP) (Fig. S4) protein, suggesting that the epitopes are not present on monomeric gp120. In addition, our result showed that 6A and 8C mAbs do not bind to linear epitopes on AD8 gp140 Env (Fig. S5) showing that 6A and 8C target conformation-dependent epitopes.
Figure 4. Env-binding of 6A and 8C chimeric mAbs against HIV Env. (A) HIV AD8 Env gp140 and (B) HIV AD8 Env. gp120 was immobilized on 96-well plates and serial dilutions of chimeric 6A and 8C mAbs were incubated with the coated wells. The binding of antibodies was detected with anti-human IgG-HRP conjugated.
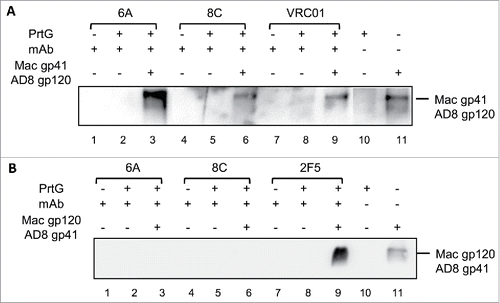
6A and 8C mAbs were incubated with the supernatant of chimeric soluble gp140, which was constructed from oligomers of simian immunodeficiency virus (SIV) and AD8 Env (SIV gp41-AD8 gp120 and SIV gp120-AD8 gp41) in an IP assay. This experiment was performed to investigate if the tested chimeric mAbs bind to HIV AD8 gp41 or HIV AD8 gp120 in an oligomeric context with SIV oligomeric gp120 or gp41, respectively. The results show that 6A and 8C mAbs bind to trimeric gp120 in SIV gp41-AD8 gp120, whereas no binding to trimeric gp41 was detected in SIV gp120-AD8 gp41 IP assay ().
Figure 5. Binding of mAbs to the chimeric (HIV/SIV) AD8 gp140. (A) SIVgp41-AD8 gp120 IP assay: 6A/8C/VRC01-prt G controls (lanes: 2, 5, 8) were used to confirm the specific Env protein capturing in 6A/8C/VRC01-gp140-prt G samples (lanes: 3, 6, 9). gp41-AD8 gp120-prt G (lane: 10 show the non-specific background of chimeric gp140 binding to prt G in IP assay. 6A/8C/VRC01 controls (lanes: 1, 4, 7) included purified mAb in western blot assay to discriminate any possible western blot related background from real captured chimeric gp140 bands. Gp140 control (lane: 11) was used in western blotting assay to confirm the correct size of captured Env proteins in IP assay. (B) SIVgp120-AD8 gp41 IP assay; the control included: western blot control of 2F5, 6A and 8C (lanes 12, 15, 18), IP assay controls (13, 16, 19). The captured chimeric gp140 Env from 2F5, 6A and 8C IP assay could be detected in lanes 14, 17 and 20. Lane 21 represents the background on chimeric SIV gp120-AD8 gp41 non-specific binding to prt G. Lane 22 is the control samples which shows the position of SIV gp120-AD8 gp41.
6A and 8C chimeric mAbs bind to soluble non-immobilized AD8 gp140 and covalently stabilized, soluble cleaved trimeric gp140 (SOSIP gp140)
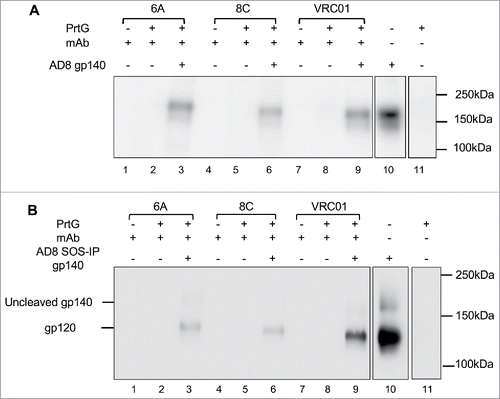
As shown in , mAbs 6A and 8C not only bind to immobilized AD8 gp140 Env (ELISA assay), but are also able to target their epitopes on a soluble non-immobilized form of Env () in IP assay. The binding of 6A and 8C chimeric mAbs to SOSIP AD8 gp140 Env () confirms that the bovine antibodies detect an HIV Env epitope that is present on SOSIP AD8 gp140 trimers that closely mimic the structure on the virion-associated surface Env.
Figure 6. Binding of 6A and 8C mAbs to HIV AD8 Env. Binding to soluble trimeric (A) AD8 gp140 and (B) SOSIP gp140. 6A/8C/VRC01-prt G controls (lanes: 2, 5, 8) were used to confirm the specific Env protein capturing in 6A/8C/VRC01-soluble gp140/SOSIP gp140-prt G samples (lanes: 3, 6, 9). 6A/8C/VRC01 controls (lanes: 1, 4, 7) included 2µg of mAb (100ng for VRC01) in western blot assay. Soluble gp140/ SOSIP gp140 control (lane: 10) was used in western blot assay to confirm the correct size of captured Env proteins in IP assay. Lane 11 includes the sample with prt G only in IP assay.
Soluble CD4 inhibit binding of mAbs 6A and 8C to AD8 gp140: To investigate whether 6A and 8C mAbs share any epitope with known BrNAbs, a competition ELISA was performed between bovine-derived 6A and 8C mAbs and BrNAbs b12 and VRC01 (CD4bs), 2G12 (Mannose dependent epitope on gp120), PG9 and PG16 (N glycan linked V1-V2 region), PGT121, PGT126 and PGT135 (N glycan linked V3 region) and 2F5 and 4E10 (MPER region). The results showed no inhibition of binding for BrNAbs after addition of 6A and 8C mAbs. This means that the mentioned bovine-derived mAbs do not have any overlap epitopes with the tested BrNAbs or they bind with low affinity to the target epitopes and their binding is outcompeted (data not shown).
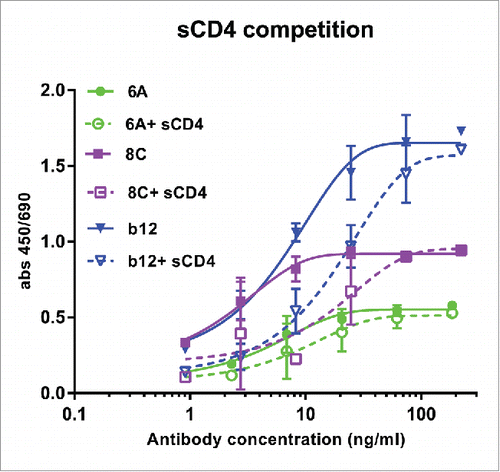
Soluble CD4 (sCD4) competition ELISA showed that the binding of chimeric mAbs (particularly 8C mAb) can be effected negatively by sCD4 (). This result indicates that mAbs 6A and 8C may target an epitope on CD4bs that does not ovelap with those of VRC01 and b12. The other possibillity is that structural alteration of Env gp140 following sCD4 binding may cause the epitopes to be lost or less accessible for 6A and 8C mAbs. However, a competition ELISA between 6A and 8C mAbs showed that these two antibodies do not compete with each other in AD8 gp140 binding (data not shown).
Figure 7. Competition of sCD4 with purified mAbs. Env AD8 gp140 was immobilized, then serial dilution of mAbs 6A, 8C and b12 with constant amount of 20 ug/ml sCD4 was added to each well. Antibody binding was detected as described in Material and Methods. Values represent the mean of two independent replicates.
Despite the strong binding of 6A and 8C to trimeric gp140 AD8, both mAbs did not neutralize AD8, MN and SF162 pseudoviruses using the TZM-bl or CF2th/CD4/CCR5/CXCR4 neutralization assay. This result indicates that both mAbs bind to non-neutralizing epitopes on trimeric gp140 (data not shown).
6A and 8C have average-sized VH and CDRH3
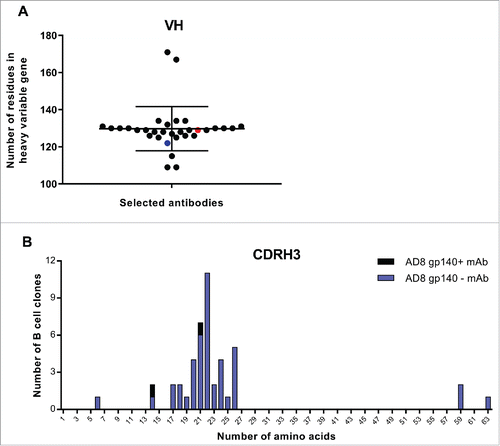
The sequences of VH genes were determined with the conserved residues at the start of framework 1 (FR1) and the end of FR4 of the VH region (Text S2). The size of bovine VH region was 109–171 residues, with the average 129.78 ± 11.91 (). The size of VH region of 6A and 8C mAbs was close to the average of 129 and 122 residues, respectively. The CDRH1 region was flanked by conserved residues: between serine/ asparagine in FRH1 and Trp in FRH2. The size of CDRH1 in all antibodies, including 6A and 8C mAb, was five aa. The CDRH2 region was surrounded by conserved residues, between glycine in FRH2 and arginine in FRH3. The length of CDRH2 was 16 aa in all selected mAbs except one (1 of 32), which had 13 residues. The CDRH3 was flanked by conserved residues; the start residue was the third amino acid after the last conserved Cys in FRH3, while the end of this gene was just before the Trp in FRH4. The CDRH3 was the most variable region in size compared with CDRH1 and CDRH2 regions. The average length of CDRH3 was 23.34 ± 10.60 aa, and, as shown in , the length range was between 6 and 63 aa and the majority of selected mAbs had 17–26 residues (28 of 32). The size of CDRH3 in 6A and 8C mAbs was 21 and 14 residues, respectively.
Figure 8. VH and CDRH3 size of selected AD8 gp140+ mAbs among isolated mAbs. (A) The length of VH region. Black points show all selected mAbs. Red point represents 6A mAb and blue point 8C mAb. (B) CDRH3 length of selected mAbs. The size of CDRH3 was defined as the number of aa in this region. Blue columns represent the number of all selected B cell clones while the black columns represent the number of residues in CDRH3 of 6A and 8C mAbs.
High frequency of Cys but low number of aromatic residues in VH and CDRH3 regions
No Cys was present in the sequence of CDRH1, whereas the presence of Cys aa in CDRH2 region was restricted to only 5 mAbs with the frequency of 6.25% of the total aa in this gene (1 of 16). 6A and 8C mAbs did not have any Cys in their CDRH2 region. The sequencing analysis revealed that Cys was more frequent in the CDRH3 region compared with CDRH1 and CDRH2 regions. Except for one mAb that had a CDRH3 length of 6 residues, all selected mAbs had 1–4 Cys. Two mAbs with very long CDRH3 of 59 and 63 aa had 6 and 8 Cys, respectively (). The average percentage of Cys was 7.33 ± 4.11 (range of 0–8 Cys). The frequency of Cys in the total residues of CDRH3 of 6A and 8C mAb was 9.52% and 14.28%, respectively.
Figure 9. SHM and Cys and aromatic residues in VH and CDRH3 regions. (A) Number of Cys. (B) The average percentage of Cys in total CDRH3 residues. Black points show all selected mAbs. Red point represents 6A mAb and blue point shows 8C mAb. Green ovals represent the mAbs with the value above the average. (C) Number of aromatic residues. (D) The average percentage of aromatic residues in total CDRH3 residues. (E) Percentage of SHMs in CDRH3 of selected mAbs. (F) Percentage of SHMs in VH of selected mAbs. (G) 6A and 8C VH genes alignment with germline genes.
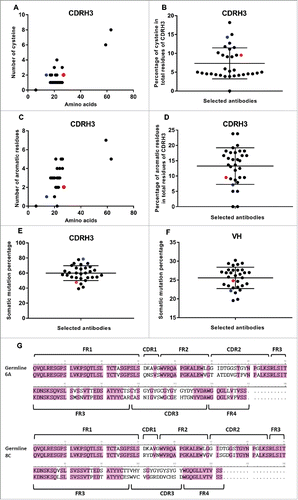
In most of the antibodies (26 of 32), the aromatic residue Tyr (20% frequency) was observed in CDRH1. In addition, there was one Trp in one of the selected mAbs. One Tyr was observed in 8C mAb CDRH1, while 6A mAb sequence did not reveal any aromatic residue in CDRH1. Tyr, and in some cases aromatic residues Trp or Phe, were observed in CDRH2 of selected mAbs (26 of 32). There was one Phe and one Tyr in 6A (6.25% of all CDRH2 region aa) and 8C (6.25% of all CDRH2 region aa) mAb, respectively. The majority of selected mAbs (20 of 26) had one aromatic residue in their CDRH2 regions, while there was one mAb with 3 aromatic residues and 5 mAb with 2 aromatic residues. The average frequency of aromatic residues was 6.53 ± 4.49 of all CDRH2 region aa. As shown in , the frequency of aromatic residues such as Trp, Tyr and Phe in CDRH3 region was between 0 and 7 in selected mAbs. Two mAbs had no aromatic residues in their CDRH3 regions. The majority of selected mAbs had 2–5 aromatic residues. Two mAbs with the very long CDRH3 regions had 7 and 5 aromatic residues, respectively. The average frequency of aromatic residues in total residues of CDRH3 was 13.26 ± 5.97. In 6A and 8C mAbs, the percentage of aromatic residues was 9.52% and 7.14%, respectively ().
The number of Cys in light variable regions of all selected mAbs was only two, and no diversity could be detected. Tryptophan frequency in most of the antibodies was only one, with the exception of only a few antibodies showing two or three tryptophan. 6A and 8C mAbs have only one tryptophan (data not shown).
Average somatic hypermutation degree in VH regions of 6A and 8C mAbs
The bovine lambda chain has a limited role in antigen binding recognition,Citation57 so the degree of SHM was not analyzed in the selected mAbs. The analysis of SHMs was performed according to the alignment of CDRH genes with the available germline genes of Holstein Friesian cowsCitation58 The CDRH1 and CDRH2 genes were aligned with germline IGHVH boVH1 gene, whereas CDRH3 gene was aligned with the best matched germline genes of IGHV boVH1, IGHD (1–8) and IGHJJ1.Citation47 The average of SHM in CDRH1 was 69.37% ± 22.13% (range: 0–100%). The SHM rate in both 6A and 8C mAbs was 80%. The SHM average of CDRH2 was 42.96 ± 11.08 in alignment with best matched germline gene of IGHV boVH1. The SHM rate in CDRH2 of 6A and 8C mAbs was 62.5% and 37.5%, respectively. The CDRH3 region of each mAb was aligned with germline genes of IGHV boVH1, IGHD (1–8) and IGHJ1. The average SHMs degree was 59.34 ± 10.16. The percentage of SHMs in 6A and 8C mAb was 47.62 and 78.57, respectively (). Among the germline genes, 56.25% of mAbs (18 of 32) were similar to IGHD5 gene, 15.62% of mAbs (5 of 32) were similar to IGHD3 gene, 15.62% of mAbs (5 of 32) were similar to IGHD7 gene, 6.25% of mAbs (2 of 32) were similar to IGHD2 gene, 3.12% of mAbs (1 of 32) were similar to IGHD4 gene, and 3.12% of mAbs (1 of 32) were similar to IGHD8 gene. 8C mAb showed the highest SHM degree among all selected mAbs (). 6A and 8C germline genes were IGHD7 and IGHD8, respectively. The average degree of SHM of entire VH gene was 25.46 ± 2.83. 6A mAb alignment with germline genes revealed 24.81% SHM, which was in the average range. 8C mAb alignment with its germline genes, showed 22.13% SHM, which was below the average (). The alignment of 6A and 8C VH genes with their germline genes are displayed in .
Trp and Cys residue play critical role in gp140 binding of 6A and 8C mAbs
The mutated mAbs were assessed for AD8 gp140 Env-binding by ELISA. The end-point concentration of wild type 6A mAb was 8 ng/ml, which was impacted negatively through alanine (Ala) substitution (). The mutation Y115A exhibited the least effect on antigen-binding activity (end-point concentration of 40 ng/ml), whereas the substitution of Trp (W106A) and His (H111A) reduced the anti-gp140 binding activity by ∼125 fold (end-point concentration of 1000ng/ml compared with wild type end-point concentration of 8 ng/ml). The mutation of Cys (C108) to Ala showed the highest negative influence by hindering the interaction between 6A mAb and soluble gp140 (end-point concentration above 1000 ng/ml) ().
Table 1. Effect of mutations on structural change and Env-binding.
Figure 10. ELISA assay of mutated 6A and 8C mAbs. Ala mutagenesis of aromatic residues, His and Cys in heavy CDRs of 6A and 8C mAbs. (A-B) Soluble gp140-specific binding of wild type and mutated 6A and 8C mAbs against AD8 Env. Cut-off signal was 2 times above the background. Values represent the mean of three replicates.
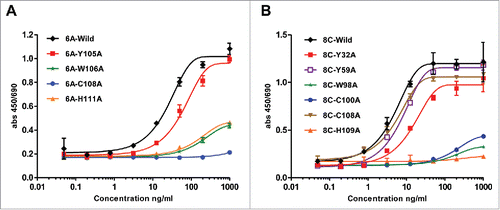
Regarding mAb 8C, the wild type had an end-point concentration of 0.32 ng/ml and His mutation (H109A) did not have any significant effect on antigen binding of the mAb (). The Tyr mutations to Ala reduced the antigen binding 5 times in comparison with that to wild type antibody (Y32A, Y59A end-point concentration of 1.6 ng/ml). Mutation of Cys residues (C100A or C108A) dramatically reduced the antigen-antibody interaction by 625-fold. The major end-point concentration was determined to be 1000 ng/ml for W98A, indicating a markedly impeded antigen-binding activity (3125 times), and that the principal role of this residue is direct antigen binding or maintaining the required structure for antigen-antibody structure ().
Modeling of wild type heavy bovine V genes
The homology-modeling revealed a protruding CDRH3 for 6A VH chain with Trp and Cys on the tip, predicting a potentially important role in antigen binding ( and ). The structural modeling was performed to predict the position of aa in CDRHs that participate in potential strong hydrogen binding, such as aromatic residues (Trp and Tyr that also form deeply recessed hydrophobic interactions with globular proteins), and His. The heavy CDRs of 6A and 8C also contained Cys aa that may form disulfide bonds that constrain tertiary protein structure (). Although the modeling of CDRH3 is challenging, the result of ELISA for mutated antibodies is consistent with the result we obtained from modeling.
Figure 11. 3D structure homology-modeling of wild type and mutated 6A and 8C bovine V genes. (A) 6A wild type and mutated mAbs. (B) 8C wild type and mutated mAbs. The 8C and 6A VH regions are shown in gray and the heavy CDRs are as follow: CDRH1: green, CDRH2: red and CDRH3: blue. The mutated residues are shown as black and with arrow in each mutated mAbs. the VH region was modeled using a fully automated protein structure homology-modeling server (SWISS-MODEL, http://swissmodel.expasy.org). The server used bovine ultra-long CDRH3 as template (Protein data bank code: 4k3d.1.A for 6A mAb and 4k3e.2.A for 8C mAb). The figures were generated using PyMOL program.Citation88
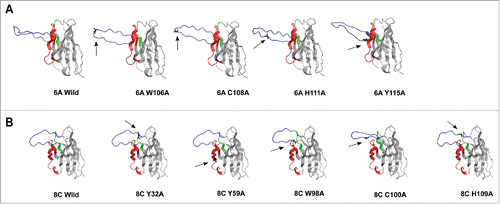
In addition, the Swiss-model homology tool used almost 10,000 templates to create the best-matched 3D structure. The final templates that the tool selected were X-ray crystallography of 4k3d.1.A bovine antibody for 6A mAb and 4k3e.2.A bovine antibody for 8C mAb. The Global Model Quality Estimation was 0.84 and 0.83 for 8C and 6A mAb, respectively. Since this score is usually reported between 0 and 1, the values close to 1 demonstrate high reliability on the modeled structures. Sequence identity also gives a confidence range. If the identity of the modeled structure compared with the template is 70% or over, the structure is reliable. The sequence identity of 6A to its template was 70%, while this value was 76.23% for 8C mAb.
To investigate the effect of Ala substitution of Trp, His and Cys on the 3D structure of CDRH3, the SWISS-MODEL serverCitation59-62 was used to model the entire V regions with the selected mutations. The predicted structure changed in 3D modeling is summarized in . All Ala substitutions in 6A mAb were performed in CDRH3, while in 8C mAb, Y32A and Y59A mutations were performed in CDRH1 and CDRH2, respectively. As shown in and , replacement of Trp (both mAbs) and His (only 6A mAb) changes the structure of CDRH3, which can be the potential cause of a reduction in antigen-binding activity. Exchange of C100 and C108 residues to Ala in 8C and 6A mAbs, respectively, showed reduction in Env-binding, which could be explained by structural change in modeled CDRH3 regions ( and ).
Discussion
Numerous attempts have been made to develop an effective HIV vaccine, but only modest progress has been made in stimulating broad neutralizing antibodies in humans. While most studies examine serum antibody to assess vaccine efficacy, the appearance of antigen-specific memory B cells in PBMC is a critical outcome supporting immunization efficiency. ELISPOT assays estimate the number of total and HIV-specific memory B cells circulating in blood or other tissues of vaccinated animals/individuals and HIV infected patients. In HIV-infected individuals, a range of 0.1–7% HIV-specific memory B cells within the total memory B cell pool has been reported previously.Citation63,64 Immunization of macaques with YU-2 gp140 resulted in a high percentage of HIV-specific memory B cells. Sundling and colleagues estimated that YU-2 gp140 trimer-specific memory B cells were 10–20% of total memory B cells of PBMC in immunized macaques.Citation65,66 Here, we report that the frequency of AD8 gp140-specific memory B cells was in agreement with the AD8 gp140 antibody titer in serum samples. The third year immunization resulted in a consistent level of 5.26–6.38% AD8 Env memory B cells within the total memory B cell pool. While the ELISPOT assay only estimates the frequency of HIV-specific memory B cells, recent improvements in instrumentation for fluorescent cell detection and isolation (flow cytometry assay) have facilitated the isolation of single HIV-specific B cells suitable for Ig gene cloning and mAb production.Citation67-72 In our study, the percentage of HIV-specific memory B cells identified by flow cytometry (0.87%) was considerably lower than the result achieved by ELISPOT. However, this variation could just reflect a difference between assays, where the memory B cell ELISPOT assay includes an incubation step of several days that could potentially amplify clones and relies on secreted antibody rather than surface expression of immunoglobulin. It is most likely the difference between ELISPOT and flow cytometry reflects differences in sensitivity.
Construction of chimeric or humanized mAbs offers insights into the molecular evolution of the antibody response, and the preparation of potentially useful materials. Extensive engineering of the antibodies toward humanization is usually required to prevent the generation of a human-anti-mAb response in patients in which these mAbs will be potentially used therapeutically. Previous studies showed that humanized murine-derived antibodies can retain antigen binding and neutralization activity.Citation73,74 Our results showed that the anti-HIV bovine V regions can be fully functional for HIV AD8 gp140 and SOSIP gp140 Env-binding when joined with the human-IgG1 C region. These results provide a starting point to investigate humanization of bovine mAb for potential in-vivo applications in future. MAb chimerization, which substitutes the bovine Fc for a human Fc region, could be advantageous in removing deleterious functions associated with the bovine Fc. Replacing bovine Fc with known human Fc functions may, on the other hand, be a disadvantage if there were any novel benefit provided by the bovine Fc region in generating the potent antiviral activity reported in the whole bovine antibody mixture. Further studies are required to understand any enhanced anti-HIV-1 activity provided by the bovine antibody Fc region, while limiting the immunogenicity to humans from the bovine IgG Fc.
In this study, among 32 expressed mAbs, we did not detect any neutralizing mAb. We also used AD8 gp140 Env to isolate HIV-specific memory B cells and to assess the specific Env binding activity of cloned mAbs. Characterization of 6A and 8C mAbs showed that they could also bind to AD8 SOS-IP gp140. However, these two antibodies did not show any neutralizing activity against AD8 and MN pseudoviruses. Failure in isolating neutralizing antibodies could potentially be due to the structure of soluble AD8 gp140, which is more open than the natural Env structure and may not exhibit some neutralizing epitopes that are accessible on native form of Env or may expose more non-neutralizing epitopes.Citation75 In addition, it is also possible that the probe was aggregated over time and the proportion of correctly structured trimeric gp140 was relatively low, which led to the low number of HIV-specific memory B cell selection. Another explanation could be that the probe structure was conserved, but biotinylation and conjugation of streptavidin PE was not fully successful. We also predict that the Env proteins that were used for the vaccination and B cell FACS selection were not 100% pure and may have been contaminated with some human-derived proteins (from cell culture), which led to selection of some B cells against human proteins. The Env protein used in the ELISA assay to determine the anti-HIV antibodies was from a different production batch, and it may not have contained the human-derived protein contaminations which resulted in the low “hit rate.” Finally, the expressed antibody screening/ selection strategy in which the probe is immobilized on a surface of ELISA plate could have been at fault. It is possible that immobilization could alter the conformation of some neutralizing epitopes or cause them to be less accessible for antibody binding, leading to isolation of mainly non-neutralizing antibodies in the screening step.
Though CD4bs is one of the neutralizing epitopes for BrNAbs, the lack of neutralizing activity for 6A and 8C mAbs despite competition with sCD4 could be due to the expression conditions during in vitro culture in human cells. For instance, it has been shown that strong glycosylation of the antibodies in CHO-K1 cells compared with the hybridoma parental cells can preserve the neutralizing activity of BrNAbs.Citation76 With the high abundance of aromatic aa in the bovine V-region, tyrosine sulfation is another post-translation mechanism that may contribute to neutralizing activity. For instance, some CD4-inducible antibodies, including 47e, 412d, CM51, and E51, are modified by sulfation.Citation77 Furthermore, expression of antibodies on the surface of susceptible cells (membrane-bound antibody) can help to conserve their existing neutralization. Recently, it was found that, although TG15 does not exhibit any neutralizing activity when expressed as a soluble protein,Citation78,79 it can inhibit various clades of HIV-1 when expressed on the surface of HIV-1 susceptible cells. In our study, we investigated the neutralization activity of the antibodies in the standard neutralization assays using TZM-bl and CF23th cells. However, probably more relevant subsequent neutralization assays are need to investigate the inhibition of virus replication in PBMCs or purified CD4 T cell cultures. In addition, it is questionable whether the 6A and 8C mAbs' neutralization is impeded by V3-loop glycans Citation80 and further studies on neutralization and binding of these mAbs against glycan-deficient viruses could address this question.
Though SOSIP AD8 gp140 mimics the epitopes on the native Env spike,Citation81,82 the binding of 6A and 8C mAbs to SOSIP AD8 gp140 in the absence of any detectable neutralization activity might demonstrate that these mAbs bind to non-neutralizing epitopes that are available not only on soluble trimeric gp140, but also on strain-matched SOSIP gp140. Competition with sCD4, but not b12 and VRC01 BrNAbs, showed these mAbs may target epitopes within the CD4bs that do not overlap with b12 and VRC01 epitopes. Another explanation could be that 3D structural changes in Env following sCD4 binding result in less accessibility of mAbs to their epitopes. However, further studies such as X- ray crystallography may reveal the precise site of binding.
Sequencing analysis revealed that the best matched IGHD germline genesCitation58 for CDRH3 (IGHD7 for 6A mAb and IGHD8 for 8C mAb) have a moderate size (13–20) compared with other germline genes.Citation58 In addition, the percentage of SHM of VH genes in 6A and 8C mAbs was close to the average across all mAbs prepared here, demonstrating that the bovine antibodies targeting HIV gp140 Env do not necessarily need above average rates of SHM. However, future studies on additional HIV Env-binding mAbs are needed to reveal if average somatically mutated and moderate sized CDRH3 from cows are optimal for HIV neutralization like in the human species, and whether the bovine antibodies need to undergo extensive affinity-maturation for broadly neutralization activity.
Previous studies of BrNAbs demonstrated that the presence of Cys and aromatic residues such as Trp and Tyr occurs rarely in human anti-HIV antibodies, though they may be critically involved in the antigen binding or neutralization activity.Citation51-53 On the other hand, it was shown that Cys residues are more frequent in bovine ultra-long CDRH3 than short or moderate-sized CDRH3.Citation45 So, we aimed to examine the potential role of Cys and aromatic residues in 6A and 8C mAbs. In this study, substitution of Cys aa in CDRH3 region had a strong negative effect and eliminated antigen binding activity. This is indicative of the important contribution of this residue in the interaction of the antibody with the antigen. Mutation of Trp in CDRH3 region, and to some extent His residues, which are the strongest amino acid participants in hydrogen bonding, significantly decreased the antigen-antibody interaction. This result suggests that both the later residues play a crucial role in the epitope binding. However, mutation of the Tyr aa that are a weaker participant in hydrogen bond formation exhibited undetectable or very slight effect in the antigen binding activity. The potential function of specific CDRH3 residues in the antigen binding can be explained in different ways: direct binding to the epitope, involvement in direct association of residues of both molecules and providing the required intramolecular 3D structure for the epitope detection. We used structural modeling to show that Trp, His and Cys residues may play a role maintaining the functional 3D structure of CDRH3 necessary for antigen binding activity (). This study only investigates the role of aromatic, polar His and Cys residues in CDRs, whereas un-investigated residues may be involved in the two molecules interactions. To determine the precise contribution of different residues in CDRs, X-ray crystallography of the antigen combining site/epitope interface would be more informative.
Although the use of soluble trimeric gp140 resulted in a broad polyclonal neutralizing response, vaccination of the cows with SOSIP gp140, which shares close structure with the virion-bound functional form of Env, might target BrNAbs germline B cells receptors (BCRs) more efficiently than soluble trimeric gp140. Using SOSIP proteins and virus-like particles for B cell selection might increase the chance of isolating B cells with BCRs binding to the native epitopes on HIV Env. Furthermore, including a high-throughput method such as microneutralization after flow cytometry selection of B cells may facilitate a functional investigation of secreted antibodies produced in very low amount from one cell. This could be a powerful strategy to clone and express only variable genes of BrNAbs rather than those only binding to HIV Env. Overall, the unusual features of the bovine immunoglobulin diversity mechanism suggest that bovine mAbs may be produced to antigenic epitopes that are very difficult for other species to engage. Potential avenues to move forward with bovine antibodies are encouraging and humanization of bovine BrNAbs as a long-acting antiviral therapy in HIV infected individuals may serve as an adjunct to the existing daily oral antiviral regimens.
Material and methods
Immunization of cow and anti- HIV AD8 gp140 antibody titer in serum samples
Oligomeric clade B AD8 gp140 of Env (AD8 clone of ADA) truncated at the membrane proximal external region was created as described previously.Citation14 Briefly, 100 μg of purified AD8 gp140 in adjuvant (Montanide ISO 206; Seppic, France) was injected intramuscularly into the flank. Over a four year period (48.5 months), the cow achieved three pregnancies and was boosted with 3 – 4 doses of gp140 during each pregnancy.Citation15 Serum samples were collected at different time points and anti-HIV AD8 gp140 antibody titres were measured by ELISA.Citation14
Isolation of PBMC
Blood samples from immunized and non-immunized cows were taken in citrate-treated tubes (from the last pregnancy: 46–48.5 months) and PBMCs were isolated by the use of density gradient centrifugation. The isolation was performed as described previously Citation83 with some minor modifications. Briefly, 30 ml of diluted blood (1:1 (v/v) in Dulbecco's Phosphate-Buffered Saline (DPBS, ThermoFisher scientific) was layered over 20 ml of Ficol paque™ plus (GE Healthcare) according to the manufacturer's instructions. Then, the tube was spun at 1600 ×g for 40 min at room temperature (RT) and the isolated PBMC from the interface were removed. The cells were washed with cold DPBS/2mM Ethylenediaminetetraacetic acid (EDTA) and centrifuged at 500 ×g for 10 min at 4˚C. The final cell pellet was resuspended in 3ml of 0.83% ammonium chloride for 5 min on ice to lyse red blood cells. Subsequently, the cells were washed with cold DPBS and frozen in heat-inactivated 90% horse serum (Sigma Aldrich) and 10% dimethylsulfoxide (DMSO, Merck).
Elispot
PBMCs of immunized cow 7004 (47.5 weeks) and non-immunized cow were thawed at 37°C followed by a wash step using culture media containing Iscove's Modified Dulbecco's Medium with L-glutamine and 25mM Hepes (Gibco) containing penicillin (10units/mL), streptomycin (10μg/mL), 1×non-essfeiential amino acids (ThermoFisher scientific), 1mM sodium pyruvate (ThermoFisher scientific) and 15% heat-inactivated horse serum (Sigma Aldrich, Cat.No. H1138). The cells were centrifuged at 500×g 10 min and resuspended in culture media again. To stimulate PBMCs, 106 PBMC/ml were cultured in culture media containing 20U/ml recombinant human IL-2 (Ray-Biotech, Cat. No. 213–10206), 1µg/ml (1/1000 dilution) Pokeweed (PWM, Sigma Aldrich, Cat. No. L8777), 20 ng/ml recombinant bovine IL-10 (Kingfisher Biotech, Cat. No. KINFRP0379B-005) and 1 µg/ml anti-bovine CD40 (IL-A156, Serotec, Cat. No. SEMCA2431GA) for 6 d.
The frequency of total and antigen-specific bovine memory B cells was determined as described previouslyCitation84 with some minor modifications. MultiScreen ™-HA ELISPOT plates (Millipore, Watford, UK) were coated overnight at 4°C with 1/1000 mouse anti-bovine IgG (clone BG-18, Sigma, Cat. No. B6901) in carbonate buffer pH 9.6 (0.2M Na2CO3, 0.2M NaHCO3) for total memory B cells detection or 2.5 µg/ml AD8 gp140 Env protein for HIV-specific memory B cells. Afterward, the plate was washed 5 times with PBS and blocked with culture media for 2 hours at 37°C. The plate was washed with PBS and subsequently the stimulated cells were cultured and incubated overnight at 37°C in a 5% CO2 incubator. The following day, the plate was washed and incubated with 1/1000 sheep anti-bovine IgG (HRP) conjugated (AbD Serotec, Cat. No. SEAA123P) for 5 hours at RT. Finally, the plate was washed again and the reaction was developed using 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Mabtech) and stopped by washing in water. The blue spots were counted using AID ELISPOT reader.
Flow cytometry
PBMCs from the vaccinated cow were incubated with LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (ThermoFisher scientific) for 20 min at 4°C, then the cells were stained using 1/200 anti-bovine CD21-FITC (AbD Serotec, Cat. No. MCA1424F) for 30 min at 4°C. The CD21+ cells were sorted and subsequently the cells were stained with 5 ug/ml Streptavidin PE- conjugated AD8 gp140 (prepared as mentioned in protocol S1) for 30–50 min at 4°C. Separately, anti-bovine IgG (Sigma, Cat. No. B6901) was conjugated to pacific-blue fluorophore using mouse IgG1 labeling kit (ThermoFisher scientific). After a washing step, anti-bovine IgG (final dilution 1/300) was used to stain all PBMC samples for 30 min at 4°C. Finally, the data was collected on a LSR Fortessa flow cytometer using FACSDiva software (BD Biosciences).
Amplification of bovine Ig V gene and chimeric/bovine mAb cloning and production
HIV-specific B cells were single-cell sorted by flow cytometry into 96-well PCR plates containing RNase inhibitors and stored at −80°C. Then, cDNA was synthesized from RNA for each single cell using SuperScript® III reverse transcriptase (Invitrogen) and stored at −20°C until further analysis.Citation85 The Ig heavy gamma (IgHγ) and light lambda (IgLλ) V genes were amplified independently in nested PCR. Each reaction was performed in 50 µl total volume containing 200 µM dNTPs, 200 nM from each external primer, 1.5 mM MgCl2 (Promega), 1.2U Go taq HotStart® Taq DNA polymerase (Promega), PCR buffer and 3µl cDNA (or first round PCR product for second round PCR). The PCR reaction primers and conditions are listed in Table S1. The external primers target the signal peptide gene upstream of Ig V genes (IgHγ Fw1 or IgLλ Fw1) and the Ig C gene downstream of Ig V genes (IgHγ Rv1 or IgLλ Rv1), while internal primers (IgHγ EcoRI Fw2/ IgHγ NheI Rv2 or IgLλ EcoRI Fw2 / IgLλ AvrII Rv2) bind to conserved regions amplifying the entire V genes. To verify the accuracy and efficiency of the chimeric antibody construction, VRC01 V genes were used as positive control. VRC01 heavy and VL genes were amplified from CMVR VRC01 H and CMVR VRC01 L plasmids (NIH AIDS Reagent Program) (Table S1). The amplified IgV fragments were cloned into the expression vectors of pFUSEss-CHIg-hG1 and pFUSE2ss-CLIg-hL2 (Invivogen), which express human constant heavy (gamma) and light (lambda) genes, according to the manufacturer's instruction. Sequencing analysis confirmed the accuracy of cloning. The fully bovine expression DNA plasmids were constructed as mentioned in protocol S2.
Chimeric/fully bovine antibody production
Then, the pair heavy/light expression DNA plasmids (Heavy: light ratio of 2:3) were transfected into human embryonic kidney 293T (HEK293T) cells using Lyovec transfection reagent (Invivogen) according to the manufacturer's instructions. The supernatants were analyzed by western blotting for the expression of the mAbs and by ELISA assay to investigate the anti-HIV binding activity.
Verification of antibody expression
Antibody-containing supernatants were subjected to 12.5% reducing SDS gel and blotted onto nitrocellulose membrane. Membranes were blocked in 5% skim milk for 1 hour at RT and then incubated with 1/10000 ZyMAX goat anti-human IgG (H+L) – HRP conjugated (ThermoFisher scientific, Cat. No. A18811) or 1/1000 sheep anti-bovine IgG (HRP) conjugated (AbD Serotec, Cat. No. SEAA123P) for 2 hours (RT). The membranes were washed with PBS- 0.1% Tween 20 after each step. Finally, Supersignal west pico chemiluminescent substrate (ThermoFisher Scientific) was applied to the membrane for 5 min and the bands were visualized using MF-ChemiBis 3.2 imaging system (DNR).
ELISA assay
The assay was performed to screen anti-HIV antibodies and assess the antigen binding activity of each chimeric mAb. In brief, a 96-well polystyrene flat-bottom plate was coated with purified trimeric gp140 (AD8, YU-2, MW and PSC 89) or with monomeric gp120 in coating buffer (pH 9.8; 2 mM Tris, 10 mM NaCl) followed by blocking with 5% skim milk. Then, the chimeric antibodies were added and detected using ZyMAX goat anti-human IgG (H+L) – HRP conjugated (ThermoFisher scientific). Color reaction was developed using TMB substrate and the absorbance was measured at 450 nm against a reference of 690 nm.
Purification of chimeric/fully bovine antibodies
The anti-HIV gp140 mAbs were produced in large scale. Cell debris were removed by centrifugation at 500 × g for 10 min followed by collection of supernatant and storage with the addition of 0.02% sodium azide at 4°C. The purification of antibodies was performed using protein G (prt G) agarose Fast flow (Millipore) according to manufacturer's instruction. Briefly, 150 ml supernatant of each antibody was loaded onto a column containing 1 ml of 100% prt G agarose and eluted with 50 mM glycine pH 2.7. The eluted antibodies were immediately neutralized using 2 M Tris pH 10. Finally, the antibodies were buffer exchanged to PBS and concentrated by Amicon Ultra 30kDa filter (Millipore). Purified IgGs were filter sterilized and the concentration was calculated according to OD 280 nm measurement.
Assessment of mAbs binding to denatured AD8 gp140 Env
To investigate if the anti-HIV gp140 mAbs bind to linear epitopes on gp140 in reducing conditions, 300 ng of AD8 gp140 was run on 8% SDS gel, then transferred onto the nitrocellulose membrane. Afterward, the membrane was blocked with 5% skim milk and incubated with chimeric mAb overnight at 4°C. The membrane was washed with PBST, then the attached antibodies were detected using 1/10000 ZyMAX goat anti-human IgG (H+L) – HRP conjugated (ThermoFisher scientific) for 2 hours at RT. The membrane was washed, substrate was added and the bands were visualized as described above.
Assessment of mAbs binding to different form of AD8 Env
An IP assay was performed to investigate the binding of anti-AD8 Env mAb to soluble AD8 trimeric gp140, AD8 SOSIP gp140 (stabilized, soluble cleaved trimeric form of Env protein), cleavage competent gp140 and chimeric gp140 Env proteins (SIV gp41-AD8 gp120; SIV gp120-AD8 gp41). An IP assay with soluble gp140 was performed to verify the binding of anti-Env mAbs to non-immobilized gp140 Env. SOSIP gp140 was also used in the IP assay because it has the closest structure to the native Env. The cleavage competent gp140 converts mostly to monomeric gp120 and gp41, and an IP assay using this form of Env reveals whether the mAbs are able to bind monomeric gp120 or gp41. To identify the binding site of mAbs on AD8 gp140 Env, chimeric gp140 Env, which is composed of AD8 human-derived gp120 and SIV macaque-derived gp41 (SIV gp41-AD8 gp120) or vice versa (SIV gp120-AD8 gp41), was used.
The IP assay was performed as following: 40 µl 50% prt G agarose was incubated with 10 µg/reaction (50 µg/ml) of chimeric mAbs or 100 ng/reaction (0.5 µg/ml) VRC01/ 2F5 (as positive control) in lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% IGEPAL and protease inhibitor (Roche Diagnostics) for 2 hours at 4°C to allow the Fc region binding to prt G. Then, the beads were washed with lysis buffer to remove the unbound antibodies. The beads coupled to the mAbs were incubated with Env proteins overnight. The concentration of the proteins was 25 µg/ml for soluble AD8 trimeric gp140 and AD8 SOSIP gp140 or 100–200 µl of harvested transfection supernatant for chimeric gp140 Env proteins. After several washings with lysis buffer, the beads were boiled with 25 µl of 5X SDS buffer (containing 5% mercaptoethanol) for 10 minutes, vortexed and spun for 10 minutes at 12,000× g. Ten microliters of eluted proteins from the IP assay were run on reducing SDS gel and then blotted onto the nitrocellulose membrane. After blocking with 5% skim milk, the membrane was washed with PBST and incubated with 1/1000 anti-gp120 DV-012 sheep polyclonal antibody (NIH reagent) or 1/3000 anti-gp120 D7324 sheep polyclonal antibody (Aalto Bio Reagents, Cat. No. D7324) to detect gp120 and 1 µg/ml 2F5 (NIH reagent) antibody to detect gp41 at 4°C overnight. The membrane was washed and incubated with 1/1000 rabbit anti-sheep IgG (H+L) HRP (ThermoFisher scientific, Cat. No. 31480) or 1/1000 goat anti-human IgG (H+L) HRP for 2 hours at RT. After the final wash, substrate was added and the bands were visualized.
Competition ELISA with BrNAbs and sCD4
To investigate the epitopes of AD8 Env-binding mAbs, a competition ELISA was performed using the BrNAbs b12 and VRC01 (CD4bs), 2G12 (mannose-dependent epitope on gp120), PG9 and PG16 (N glycan linked V1-V2 region), PGT121, PGT126 and PGT135 (N glycan linked V3 region) and 2F5 and 4E10 (MPER region). The plate was coated and blocked as described previously using AD8 gp140 Env protein. Then, serial dilutions of fully bovine mAb antibodies were added and the plate was incubated for 2 hours at RT. After washing, 1 µg/ml BrNAbs was added to each well and incubated for 2 hours at RT. The plate was washed and the binding of BrNAbs was detected using goat anti-human IgG (H+L) HRP (1/1000). Color reaction was developed using TMB substrate. For sCD4 competition ELISA, the plate was coated with 1 µg/ml soluble AD8 gp140 then blocked with 5% skim milk. Then, the wells were incubated with 20 µg/ml sCD4 and serial concentration of chimeric mAbs (200 ng/ml in the first well) for 3 hours at RT. The binding of chimeric mAbs was detected using 1/1000 goat anti-human IgG (H+L) HRP. Color reaction was developed using TMB substrate.
Neutralization assay
The neutralization assay of TZM-bl and CF2th/CD4/CCR5/CXCR4 canine cells was performed as described previously. HIV pseudoviruses were produced by co-transfection of full-length Env expression plasmids (AD8 (pCMV-AD8; prepared from pAD8 provided by M. Martin, National Institute of Allergy and Infectious Diseases); MN (pSVIII-MN; provided by J. Sodroski, Division of AIDS, Harvard Medical School); SF162 (pCAGGS-SF162; provided by L. Stamatatos, Fred Hutchinson Cancer Research Center and C. Cheng-Mayer, Aaron Diamond AIDS Research Center); MuLV) and an EGFP-expressing proviral reporter plasmid (pNL-4.3ΔenvNef-EGFP). Pseudovirus-containing supernatants were harvested after 48–72 hours. Ten µl of virus (10% infectivity) was incubated with 20 µl of mAbs at different concentrations for 1 hour at 37°C. After the incubation time, 70 µl of CF2th cells (2×104) was added to each well followed by spinoculation at 1200xg for 2 hours (RT). Residual virus and mAbs were removed and 200 µl fresh media was added. The infectivity of the pseudoviruses in the presence of mAbs was analyzed by flow cytometry (LSRII).Citation86 To perform TZM-bl neutralization assay, 200 TCID50 of each pseudovirus was incubated with different concentration of mAbs (in 100 µl volume) for 1 hour at 37°C. Afterward, 100 µl TZM-bl cells (104) was added to each well and the plate was incubated for 48 hours at 37°C. The infectivity of pseudoviruses was calculated according to luciferase relative light unit readout using Fluostar plate reader (BMG Labtech).Citation87
Structure- based modeling of VH region
To define the location of aromatic, His and Cys residues in both 6A and 8C antibodies, and the structure changes following Ala substitution, the VH region was modeled using a fully automated protein structure homology-modeling server (SWISS-MODEL, http://swissmodel.expasy.org). The server used bovine ultra-long CDRH3 as template (Protein data bank code: 4k3d.1.A for 6A mAb and 4k3e.2.A for 8C mAb).
Ala substitution mutagenesis
To investigate the potential role of aromatic residues and Cys in VH regions, some mutations were introduced into the CDRH regions of 6A and 8C by PCR using Pfu Ultra II Fusion HotStart DNA Polymerase (Promega) or Phusion High-Fidelity PCR DNA Polymerase (Promega) according to the manufacturer's instructions. The PCR reaction was performed as following: 95°C 5 mins, 30 cycles of 95°C 30 s, 48°C /50°C 30s, 72°C 8 mins and 72°C 15 mins. The sequence of primers is listed in Table S1. For each primer, the complementary sequence was used as the reverse pair primer. The expression of the antibodies was performed as described previously. The supernatants were harvested and the antigen binding of mAbs was investigated in ELISA assay.
Ethics statement
A four-year-old female Holstein Friesian cattle (Bos taurus) who had successfully delivered a calf was subsequently enrolled into our previously reported one-year long vaccination and calving study.Citation14 The animal, called 7004, was then enrolled into an ongoing study that was performed on an open-grazing experimental farm for cattle under a Scientific Premises License from the Bureau of Animal Welfare, in the state of Victoria (POCTAA(3)003 A04). Long-term repeated vaccinations were conducted with approval from the Immuron Animal Ethics Committee constituted according to the government of Victoria Bureau of Animal Welfare [approval POCTAA (3)003; A04].
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.zip
Download Zip (1.3 MB)Acknowledgment
We thank Ian Gill and Noel Russell for Veterinary care of the cows and Victor Wong and Gerhardt Rank for technical assistance.
References
- Gunthard HF, Saag MS, Benson CA, del Rio C, Eron JJ, Gallant JE, Hoy JF, Mugavero MJ, Sax PE, Thompson MA, et al. Antiretroviral Drugs for Treatment and Prevention of HIV Infection in Adults 2016 Recommendations of the International Antiviral Society-USA Panel. JAMA 2016; 316(2):191-210; PMID:27404187; http://dx.doi.org/10.1001/jama.2016.8900
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science 2013; 341(6151):1199-204; PMID:24031012; http://dx.doi.org/10.1126/science.1241144
- Watkins JD, Siddappa NB, Lakhashe SK, Humbert M, Sholukh A, Hemashettar G, Wong YL, Yoon JK, Wang W, Novembre FJ, et al. An anti-HIV-1 V3 loop antibody fully protects cross-clade and elicits T-cell immunity in macaques mucosally challenged with an R5 clade C SHIV. PLoS One 2011; 6(3):e18207; PMID:21483815; http://dx.doi.org/10.1371/journal.pone.0018207
- Mascola JR, Montefiori DC. The role of antibodies in HIV vaccines. Annu Rev Immunol 2010; 28:413-44; PMID:20192810; http://dx.doi.org/10.1146/annurev-immunol-030409-101256
- Kramer VG, Siddappa NB, Ruprecht RM. Passive immunization as tool to identify protective HIV-1 Env epitopes. Curr HIV Res 2007; 5(6):642-55; PMID:18045119; http://dx.doi.org/10.2174/157016207782418506
- Casadevall A, Dadachova E, Pirofski LA. Passive antibody therapy for infectious diseases. Nat Rev Microbiol 2004; 2(9):695-703; PMID:15372080; http://dx.doi.org/10.1038/nrmicro974
- Melchers M, Bontjer I, Tong T, Chung NP, Klasse PJ, Eggink D, Montefiori DC, Gentile M, Cerutti A, Olson WC, et al. Targeting HIV-1 envelope glycoprotein trimers to B cells by using APRIL improves antibody responses. J Virol 2012; 86(5):2488-500; PMID:22205734; http://dx.doi.org/10.1128/JVI.06259-11
- Beddows S, Franti M, Dey AK, Kirschner M, Iyer SP, Fisch DC, Ketas T, Yuste E, Desrosiers RC, Klasse PJ, et al. A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 2007; 360(2):329-40; PMID:17126869; http://dx.doi.org/10.1016/j.virol.2006.10.032
- Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 2009; 361(23):2209-20; PMID:19843557; http://dx.doi.org/10.1056/NEJMoa0908492
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT. HIV vaccine design and the neutralizing antibody problem. Nat Immunol 2004; 5(3):233-6; PMID:14985706; http://dx.doi.org/10.1038/ni0304-233
- Kovacs JM, Nkolola JP, Peng H, Cheung A, Perry J, Miller CA, Seaman MS, Barouch DH, Chen B. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc Natl Acad Sci U S A 2012; 109(30):12111-6; PMID:22773820; http://dx.doi.org/10.1073/pnas.1204533109
- Kang YK, Andjelic S, Binley JM, Crooks ET, Franti M, Iyer SP, Donovan GP, Dey AK, Zhu P, Roux KH, et al. Structural and immunogenicity studies of a cleaved, stabilized envelope trimer derived from subtype A HIV-1. Vaccine 2009; 27(37):5120-32; PMID:19567243; http://dx.doi.org/10.1016/j.vaccine.2009.06.037
- McCoy LE, Quigley AF, Strokappe NM, Bulmer-Thomas B, Seaman MS, Mortier D, Rutten L, Chander N, Edwards CJ, Ketteler R, et al. Potent and broad neutralization of HIV-1 by a llama antibody elicited by immunization. J Exp Med 2012; 209(6):1091-103; PMID:22641382; http://dx.doi.org/10.1084/jem.20112655
- Kramski M, Center RJ, Wheatley AK, Jacobson JC, Alexander MR, Rawlin G, Purcell DF. Hyperimmune bovine colostrum as a low-cost, large-scale source of antibodies with broad neutralizing activity for HIV-1 envelope with potential use in microbicides. Antimicrob Agents Chemother 2012; 56(8):4310-9; PMID:22664963; http://dx.doi.org/10.1128/AAC.00453-12
- Heydarchi B, Center RJ, Gonelli C, Muller B, Mackenzie C, Khoury G, Lichtfuss M, Rawlin G, Purcell DF. Repeated Vaccination of Cows with HIV Env gp140 during Subsequent Pregnancies Elicits and Sustains an Enduring Strong Env-Binding and Neutralising Antibody Response. PLoS One 2016; 11(6):e0157353; PMID:27300145; http://dx.doi.org/10.1371/journal.pone.0157353
- Ohno S, Mori N, Matsunaga T. Antigen-binding specificities of antibodies are primarily determined by seven residues of VH. Proc Natl Acad Sci U S A 1985; 82(9):2945-9; PMID:3921967; http://dx.doi.org/10.1073/pnas.82.9.2945
- Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu Rev Immunol 2000; 18:495-527; PMID:10837067; http://dx.doi.org/10.1146/annurev.immunol.18.1.495
- Smider V, Chu G. The end-joining reaction in V(D)J recombination. Semin Immunol 1997; 9(3):189-97; PMID:9200330; http://dx.doi.org/10.1006/smim.1997.0070
- Tonegawa S. Somatic generation of antibody diversity. Nature 1983; 302(5909):575-81; PMID:6300689; http://dx.doi.org/10.1038/302575a0
- Lopez O, Perez C, Wylie D. A single VH family and long CDR3s are the targets for hypermutation in bovine immunoglobulin heavy chains. Immunol Rev 1998; 162:55-66; PMID:9602352; http://dx.doi.org/10.1111/j.1600-065X.1998.tb01429.x
- Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 2007; 76:1-22; PMID:17328676; http://dx.doi.org/10.1146/annurev.biochem.76.061705.090740
- Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 2011; 208(1):181-93; PMID:21220454; http://dx.doi.org/10.1084/jem.20101352
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009; 458(7238):636-40; PMID:19287373; http://dx.doi.org/10.1038/nature07930
- Tiller T, Tsuiji M, Yurasov S, Velinzon K, Nussenzweig MC, Wardemann H. Autoreactivity in human IgG+ memory B cells. Immunity 2007;26(2):205-13; PMID:17306569; http://dx.doi.org/10.1016/j.immuni.2007.01.009
- Prabakaran P, Zhu Z, Chen W, Gong R, Feng Y, Streaker E, Dimitrov DS. Origin, diversity, and maturation of human antiviral antibodies analyzed by high-throughput sequencing. Front Microbiol 2012; 3:277; PMID:22876240; http://dx.doi.org/10.3389/fmicb.2012.00277
- Mouquet H, Scheid JF, Zoller MJ, Krogsgaard M, Ott RG, Shukair S, Artyomov MN, Pietzsch J, Connors M, Pereyra F, et al. Polyreactivity increases the apparent affinity of anti-HIV antibodies by heteroligation. Nature 2010; 467(7315):591-5; PMID:20882016; http://dx.doi.org/10.1038/nature09385
- Zhou T, Xu L, Dey B, Hessell AJ, Van Ryk D, Xiang SH, Yang X, Zhang MY, Zwick MB, Arthos J, et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 2007; 445(7129):732-7; PMID:17301785; http://dx.doi.org/10.1038/nature05580
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 1994; 266(5187):1024-7; PMID:7973652; http://dx.doi.org/10.1126/science.7973652
- Pancera M, McLellan JS, Wu X, Zhu J, Changela A, Schmidt SD, Yang Y, Zhou T, Phogat S, Mascola JR, et al. Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1. J Virol 2010; 84(16):8098-110; PMID:20538861; http://dx.doi.org/10.1128/JVI.00966-10
- Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J Mol Biol 2003; 325(2):337-54; PMID:12488099; http://dx.doi.org/10.1016/S0022-2836(02)01222-6
- Scherer EM, Leaman DP, Zwick MB, McMichael AJ, Burton DR. Aromatic residues at the edge of the antibody combining site facilitate viral glycoprotein recognition through membrane interactions. Proc Natl Acad Sci U S A 2010; 107(4):1529-34; PMID:20080706; http://dx.doi.org/10.1073/pnas.0909680107
- Ofek G, McKee K, Yang Y, Yang ZY, Skinner J, Guenaga FJ, Wyatt R, Zwick MB, Nabel GJ, Mascola JR, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol 2010; 84(6):2955-62; PMID:20042512; http://dx.doi.org/10.1128/JVI.02257-09
- Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A 2009; 106(48):20234-9; PMID:19906992; http://dx.doi.org/10.1073/pnas.0908713106
- Julien JP, Huarte N, Maeso R, Taneva SG, Cunningham A, Nieva JL, Pai EF. Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J Virol 2010; 84(9):4136-47; PMID:20147404; http://dx.doi.org/10.1128/JVI.02357-09
- Ofek G, Tang M, Sambor A, Katinger H, Mascola JR, Wyatt R, Kwong PD. Structure and mechanistic analysis of the anti-human immunodeficiency virus type 1 antibody 2F5 in complex with its gp41 epitope. J Virol 2004; 78(19):10724-37; PMID:15367639; http://dx.doi.org/10.1128/JVI.78.19.10724-10737.2004
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 1994; 10(4):359-69; PMID:7520721; http://dx.doi.org/10.1089/aid.1994.10.359
- Purtscher M, Trkola A, Gruber G, Buchacher A, Predl R, Steindl F, Tauer C, Berger R, Barrett N, Jungbauer A, et al. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses 1994; 10(12):1651-8; PMID:7888224; http://dx.doi.org/10.1089/aid.1994.10.1651
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 2013; 20(7):796-803; PMID:23708606; http://dx.doi.org/10.1038/nsmb.2594
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 2012; 37(3):412-25; PMID:22999947; http://dx.doi.org/10.1016/j.immuni.2012.08.012
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 2012; 109(47):E3268-77; PMID:23115339; http://dx.doi.org/10.1073/pnas.1217207109
- Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc Natl Acad Sci U S A 2010; 107(25):11483-8; PMID:20534513; http://dx.doi.org/10.1073/pnas.1004600107
- Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, Schroeder HW, Jr., Kirkham PM. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. J Mol Biol 2003; 334(4):733-49; PMID:14636599; http://dx.doi.org/10.1016/j.jmb.2003.10.007
- Wu TT, Johnson G, Kabat EA. Length distribution of CDRH3 in antibodies. Proteins 1993; 16(1):1-7; PMID:8497480; http://dx.doi.org/10.1002/prot.340160102
- Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW, Jr. Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. J Immunol 2005; 174(12):7773-80; PMID:15944280; http://dx.doi.org/10.4049/jimmunol.174.12.7773
- Wang F, Ekiert DC, Ahmad I, Yu W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, et al. Reshaping antibody diversity. Cell 2013; 153(6):1379-93; PMID:23746848; http://dx.doi.org/10.1016/j.cell.2013.04.049
- Zhao Y, Jackson SM, Aitken R. The bovine antibody repertoire. Dev Comp Immunol 2006; 30(1-2):175-86; PMID:16054212; http://dx.doi.org/10.1016/j.dci.2005.06.012
- Sinclair MC, Gilchrist J, Aitken R. Bovine IgG repertoire is dominated by a single diversified VH gene family. J Immunol 1997; 159(8):3883-9; PMID:9378976
- Saini SS, Allore B, Jacobs RM, Kaushik A. Exceptionally long CDR3H region with multiple cysteine residues in functional bovine IgM antibodies. Eur J Immunol 1999; 29(8):2420-6; PMID:10458755; http://dx.doi.org/10.1002/(SICI)1521-4141(199908)29:08%3c2420::AID-IMMU2420%3e3.0.CO;2-A
- Kaushik AK, Kehrli ME, Jr., Kurtz A, Ng S, Koti M, Shojaei F, Saini SS. Somatic hypermutations and isotype restricted exceptionally long CDR3H contribute to antibody diversification in cattle. Vet Immunol Immunopathol 2009; 127(1-2):106-13; PMID:19012969; http://dx.doi.org/10.1016/j.vetimm.2008.09.024
- Sather DN, Armann J, Ching LK, Mavrantoni A, Sellhorn G, Caldwell Z, Yu X, Wood B, Self S, Kalams S, et al. Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 2009; 83(2):757-69; PMID:18987148; http://dx.doi.org/10.1128/JVI.02036-08
- Ofek G, Zirkle B, Yang Y, Zhu Z, McKee K, Zhang B, Chuang GY, Georgiev IS, O'Dell S, Doria-Rose N, et al. Structural basis for HIV-1 neutralization by 2F5-like antibodies m66 and m66.6. J Virol 2014; 88(5):2426-41; PMID:24335316; http://dx.doi.org/10.1128/JVI.02837-13
- West AP, Jr., Diskin R, Nussenzweig MC, Bjorkman PJ. Structural basis for germ-line gene usage of a potent class of antibodies targeting the CD4-binding site of HIV-1 gp120. Proc Natl Acad Sci U S A 2012; 109(30):E2083-90; PMID:22745174; http://dx.doi.org/10.1073/pnas.1208984109
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 2014; 509(7498):55-62; PMID:24590074; http://dx.doi.org/10.1038/nature13036
- Kramski M, Lichtfuss GF, Navis M, Isitman G, Wren L, Rawlin G, Center RJ, Jaworowski A, Kent SJ, Purcell DFJ. Anti-HIV-1 antibody-dependent cellular cytotoxicity mediated by hyperimmune bovine colostrum IgG. Euro J Immunol 2012; 42(10):2771-81; PMID:22730083; http://dx.doi.org/10.1002/eji.201242469
- Zhao Y, Kacskovics I, Rabbani H, Hammarström L. Physical Mapping of the Bovine Immunoglobulin Heavy Chain Constant Region Gene Locus. J Biol Chem 2003; 278:35024-32; PMID:12829708; http://dx.doi.org/10.1074/jbc.M301337200
- Ekman A, Niku N, Liljavirta J, Iivanainen1 A. Bos taurus genome sequence reveals the assortment of immunoglobulin and surrogate light chain genes in domestic cattle. BMC Immunol 2009; 10(22); PMID:19405939; http://dx.doi.org/10.1186/1471-2172-10-22
- Queen C, Schneider WP, Selick HE, Payne PW, Landolfi NF, Duncan JF, Avdalovic NM, Levitt M, Junghans RP, Waldmann TA. A Humanized Antibody That Binds to the Interleukin-2 Receptor. Proc Natl Acad Sci U S A 1989; 86(24):10029-10033; PMID:2513570; http://dx.doi.org/10.1073/pnas.86.24.10029
- KOTI M, Kataevab G, Kaushika A. Novel atypical nucleotide insertions specifically at VH–DH junction generate exceptionally long CDR3H in cattle antibodies. Mol Immunol 2010; 47:2119-28; PMID:20435350; http://dx.doi.org/10.1016/j.molimm.2010.02.014
- Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M, Bordoli L, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res 2014; 42(Web Server issue):W252-8; PMID:24782522; http://dx.doi.org/10.1093/nar/gku340
- Guex N, Peitsch MC, Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 2009; 30(Suppl 1):S162-73; PMID:19517507; http://dx.doi.org/10.1002/elps.200900140
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 2009; 37(Database issue):D387-92; PMID:18931379; http://dx.doi.org/10.1093/nar/gkn750
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006; 22(2):195-201; PMID:16301204; http://dx.doi.org/10.1093/bioinformatics/bti770
- Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, et al. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 2009; 83(1):188-99; PMID:18922865; http://dx.doi.org/10.1128/JVI.01583-08
- Fondere JM, Huguet MF, Yssel H, Baillat V, Reynes J, van de Perre P, Vendrell JP. Detection of peripheral HIV-1-specific memory B cells in patients untreated or receiving highly active antiretroviral therapy. AIDS 2003; 17(16):2323-30; PMID:14571183; http://dx.doi.org/10.1097/00002030-200311070-00006
- Sundling C, Martinez P, Soldemo M, Spangberg M, Bengtsson KL, Stertman L, Forsell MN, Karlsson Hedestam GB. Immunization of macaques with soluble HIV type 1 and influenza virus envelope glycoproteins results in a similarly rapid contraction of peripheral B-cell responses after boosting. J Infect Dis 2013; 207(3):426-31; PMID:23162135; http://dx.doi.org/10.1093/infdis/jis696
- Sundling C, Forsell MN, O'Dell S, Feng Y, Chakrabarti B, Rao SS, Lore K, Mascola JR, Wyatt RT, Douagi I, et al. Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J Exp Med 2010; 207(9):2003-17; PMID:20679401; http://dx.doi.org/10.1084/jem.20100025
- Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp CM, Louder MK, Longo NS, Poulsen C, Guenaga J, et al. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J Virol 2012; 86(20):11231-41; PMID:22875963; http://dx.doi.org/10.1128/JVI.01543-12
- Mouquet H, Klein F, Scheid JF, Warncke M, Pietzsch J, Oliveira TY, Velinzon K, Seaman MS, Nussenzweig MC. Memory B cell antibodies to HIV-1 gp140 cloned from individuals infected with clade A and B viruses. PLoS One 2011; 6(9):e24078; PMID:21931643; http://dx.doi.org/10.1371/journal.pone.0024078
- Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods 2009; 343(2):65-7; PMID:19100741; http://dx.doi.org/10.1016/j.jim.2008.11.012
- Sundling C, Li Y, Huynh N, Poulsen C, Wilson R, O'Dell S, Feng Y, Mascola JR, Wyatt RT, Karlsson Hedestam GB. High-resolution definition of vaccine-elicited B cell responses against the HIV primary receptor binding site. Sci Transl Med 2012; 4(142):142ra96; PMID:22786681; http://dx.doi.org/10.1126/scitranslmed.3003752
- Sundling C, Zhang Z, Phad GE, Sheng Z, Wang Y, Mascola JR, Li Y, Wyatt RT, Shapiro L, Karlsson Hedestam GB. Single-cell and deep sequencing of IgG-switched macaque B cells reveal a diverse Ig repertoire following immunization. J Immunol 2014; 192(8):3637-44; PMID:24623130; http://dx.doi.org/10.4049/jimmunol.1303334
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 2010; 329(5993):856-61; PMID:20616233; http://dx.doi.org/10.1126/science.1187659
- Harris JW, Carr FJ, L AK. Recombinant humanized anti-human immunodeficiency virus antibody. Patent CA 2170034 C 2005
- Major JG, Liou RS, Sun LK, Yu LM, Starnes SM, Fung MS, Chang TW, Chang NT. Construction and characterization of chimeric and humanized forms of a broadly neutralizing monoclonal antibody to HIV-1. Hum Antibodies Hybridomas 1994; 5(1-2):9-17; PMID:7858187; http://dx.doi.org/10.3233/HAB-1994-51-202
- Sattentau QJ. Envelope Glycoprotein Trimers as HIV-1 Vaccine Immunogens. Vaccines (Basel) 2013; 1(4):497-512; PMID:26344344; http://dx.doi.org/10.3390/vaccines1040497
- Miranda LR, Duval M, Doherty H, Seaman MS, Posner MR, Cavacini LA. The neutralization properties of a HIV-specific antibody are markedly altered by glycosylation events outside the antigen-binding domain. J Immunol 2007; 178(11):7132-8; PMID:17513762; http://dx.doi.org/10.4049/jimmunol.178.11.7132
- Choe H, Li W, Wright PL, Vasilieva N, Venturi M, Huang CC, Grundner C, Dorfman T, Zwick MB, Wang L, et al. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 2003; 114(2):161-70; PMID:12887918; http://dx.doi.org/10.1016/S0092-8674(03)00508-7
- Zhou P, Goldstein S, Devadas K, Tewari D, Notkins AL. Cells transfected with a non-neutralizing antibody gene are resistant to HIV infection: targeting the endoplasmic reticulum and trans-Golgi network. J Immunol 1998; 160(3):1489-96; PMID:9570571
- Takeda S, Dorfman NA, Robert-Guroff M, Notkins AL, Rando RF. Two-phase approach for the expression of high-affinity human anti-human immunodeficiency virus immunoglobulin Fab domains in Escherichia coli. Hybridoma 1995; 14(1):9-18; PMID:7768538; http://dx.doi.org/10.1089/hyb.1995.14.9
- Pene J, Gauchat JF, Lecart S, Drouet E, Guglielmi P, Boulay V, Delwail A, Foster D, Lecron JC, Yssel H. Cutting edge: IL-21 is a switch factor for the production of IgG(1) and IgG(3) by human B cells. J Immunol 2004; 172(9):5154-7; PMID:15100251; http://dx.doi.org/10.4049/jimmunol.172.9.5154
- Dey AK, David KB, Lu M, Moore JP. Biochemical and biophysical comparison of cleaved and uncleaved soluble, trimeric HIV-1 envelope glycoproteins. Virology 2009; 385(1):275-81; PMID:19135223; http://dx.doi.org/10.1016/j.virol.2008.12.009
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 2013; 9(9):e1003618; PMID:24068931; http://dx.doi.org/10.1371/journal.ppat.1003618
- Ibeagha-Awemu Eveline E, AE. I, Zhao X. The influence of different anticoagulants and sample preparation methods on measurement of mCD14 on bovine monocytes and polymorphonuclear neutrophil leukocytes. BioMed Central 2012; 5:93; PMID:22333045; http://dx.doi.org/10.1186/1756-0500-5-93
- Lefevre EA, Carr BV, Prentice H, Charleston B. A quantitative assessment of primary and secondary immune responses in cattle using a B cell ELISPOT assay. Vet Res 2009; 40(1):3; PMID:18828984; http://dx.doi.org/10.1051/vetres:2008041
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 2008; 329(1–2):112-124; PMID:17996249; http://dx.doi.org/10.1016/j.jim.2007.09.017
- Center RJ, Wheatley AK, Campbell SM, Gaeguta AJ, Peut V, Alcantara S, Siebentritt C, Kent SJ, Purcell DFJ. Induction of HIV-1 subtype B and AE-specific neutralizing antibodies in mice and macaques with DNA prime and recombinant gp140 protein boost regimens. Vaccine 2009; 27(47):6605-12; PMID:19712773; http://dx.doi.org/10.1016/j.vaccine.2009.08.016
- Montefiori DC. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol Biol 2009; 485:395-405; PMID:19020839; http://dx.doi.org/10.1007/978-1-59745-170-3_26
- The PyMOL Molecular Graphics System, Version 1.74 Schrödinger, LLC: New York, NY, USA, 2014. 2014.
