ABSTRACT
Mesothelin is a glycosylphosphatidylinositol (GPI)-anchored membrane protein that shows promise as a target for antibody-directed cancer therapy. High levels of soluble forms of the antigen represent a barrier to directing therapy to cellular targets. The ability to develop antibodies that can selectively discriminate between membrane-bound and soluble conformations of a specific protein, and thus target only the membrane-associated antigen, is a substantive issue. We show that use of a tolerance protocol provides a route to such discrimination. Mice were tolerized with soluble mesothelin and a second round of immunizations was performed using mesothelin transfected P815 cells. RNA extracted from splenocytes was used in phage display to obtain mesothelin-specific antigen-binding fragments (Fabs) that were subsequently screened by flow cytometry and ELISA. This approach generated 147 different Fabs in 34 VH-CDR3 families. Utilizing competition assays with soluble protein and mesothelin-containing serum obtained from metastatic cancer patients, 10 of these 34 VH-CDR3 families were found to bind exclusively to the membrane-associated form of mesothelin. Epitope mapping performed for the 1H7 clone showed that it does not recognize GPI anchor. VH-CDR3 sequence analysis of all Fabs showed significant differences between Fabs selective for the membrane-associated form of the antigen and those that recognize both membrane bound and soluble forms. This work demonstrates the potential to generate an antibody specific to the membrane-bound form of mesothelin. 1H7 offers potential for therapeutic application against mesothelin-bearing tumors, which would be largely unaffected by the presence of the soluble antigen.
Introduction
Mesothelin is a glycosylphosphatidylinositol (GPI)-linked glycoprotein synthesized as a 69 kDa precursor and proteolytically processed into a 30 kDa NH2-terminal secreted form (formerly referred to as megakaryocyte potentiating factor) and a 40 kDa membrane-bound mesothelin.Citation1 Mesothelin is present at relatively low levels in mesothelial cells of the pleura, peritoneum and pericardium of healthy individuals, but is highly expressed in several different cancers, including mesotheliomas, stomach cancer, squamous cell carcinomas, prostate cancer, pancreatic cancer, lung cancer, and ovarian cancer.Citation2-6 In particular, it has been reported that most serous carcinomas of the ovary and adenocarcinomas of the pancreas express high levels of mesothelin.Citation7 In addition, high levels of mesothelin have been detected in greater than 55% of lung cancers and greater than 70% of ovarian cancers.Citation5,8 The limited expression of mesothelin on normal cells makes it a viable target for tumor immunotherapy. Administration of antibodies against mesothelin has been proposed as a strategy for mesothelioma as well as lung, ovarian and pancreatic cancer.
Although mesothelin is considered a membrane-bound protein, soluble forms of mesothelin have also been reported in patients' serum and in the stroma of tumors, including malignant mesothelioma, ovarian cancers or highly metastatic cancers. Soluble mesothelin has been reported to result from both alternative splicing and protease cleavage.Citation9,10 Tumor necrosis factor-α converting enzyme (TACE), a member of the MMP/ADAM family, has been identified as a mesothelin sheddase.Citation11
Soluble receptors can be released in the tumoral microenvironment, especially when expressed on cancer cells, and act as a decoy for therapeutic antibodies. It has previously been shown that a GPI anchor can affect protein conformation, indeed the presence of a GPI anchor can result in sufficient conformational changes in the protein to be specifically targeted by antibodies, in contrast to their cleaved, non-GPI, forms.Citation12-14
The use of monoclonal antibodies (mAbs) against membrane targets has been shown to increase the survival of patients having several cancers.Citation15 Several antibody-based molecules that target mesothelin have been developed. These include the chimeric anti-mesothelin mAb MORAb-009,Citation16 immunotoxins SS1P and RG7787,Citation17,Citation18 and the antibody-drug conjugates BAY94–9343,Citation19 and BMS986148.Citation20 Recently, a human single-domain antibody (SD1-hFc) has also been described. SD1-hFc is reported to elicit a potent anti-tumor activity by generating a complement-dependent cytotoxicity (CDC) reaction targeting an epitope in mesothelin close to the cancer cell surface.Citation21 Although these mesothelin-targeting agents showed a therapeutic effect against some mesothelin-expressing tumors, they were not effective in lower doses. One possible explanation for the lack of efficacy might be the presence of soluble mesothelin in the blood as well as in the extracellular space of tumors, which could reduce the efficacy of the therapeutic mAb by competition.Citation22 A recent study demonstrated that reduced shedding of surface mesothelin as a result of treatment with paclitaxel (Taxol) improves the efficacy of mesothelin-targeting recombinant immunotoxins.Citation23 Similarly, mutation of the protein responsible for mesothelin cleavage such that it is not shed results in improved efficacy of the immunotoxin SS1P.Citation23
Here, we propose a new approach to overcome the effect of shed (soluble) mesothelin on antibody-based therapies. Our goal was to identify an antibody that exclusively targets the whole cell membrane-associated form of mesothelin without interfering with the soluble form of the protein. Identifying antibodies that specifically recognize membrane-bound mesothelin could have a significant effect on patients' outcome. For this purpose, we used a new method to generate antibodies comprising: 1) immunizing an animal with a soluble form of the antigen, 2) administering an agent that selectively kills rapidly dividing cells,Citation24 3) immunizing the animal with a membrane-bound form of the antigen, and 4) screening for an antibody that can bind to the second antigen without binding the first one.
Phage display technology was used to produce antibody fragments because of known advantages in specific epitope targeting. It has been shown to be a promising technology to produce specific antibodies against targets on the cell surface.Citation25 Phage display libraries have also been reported as a viable alternative to hybridoma technology for development of antibodies with desired antigen-binding characteristics.Citation26 Indeed, occluded epitopes on the cell surface may be difficult to access by full-size immunoglobulin G (IgG) antibodies produced by hybridoma technology. These epitopes can be targeted by fragments of antibodies such as antibody-binding fragments (Fabs) and single-chain variable fragments (scFvs).
As reported here, from a total of 480 Fab clones obtained by phage display technology, we identified 147 clones as mesothelin specific, among which 116 clones bound to the native form of mesothelin on Hela cells. Following further analysis of these 116 antibodies, we isolated the clone 1H7, which targets selectively the membrane-bound form of mesothelin without binding shed forms of this antigen.
Results
Production of mesothelin targeting Fabs
To identify new antibodies that selectively target the whole cell membrane-bound form of mesothelin, we hypothesized that there was a conformational difference between the membrane-bound and soluble form of the common components of mesothelin, which results in different conformational epitopes. A novel subtractive immunization was thus used to generate antibodies that only recognize epitopes selectively derived from membrane-bound proteins. To prevent mice from producing antibodies against soluble mesothelin antigens, mice were tolerized by immunizing them with the full-length recombinant mesothelin protein, and then treated with cyclophosphamide to kill the activated lymphocytes. To generate antibodies targeting membrane-bound mesothelin, these mice were subsequently immunized with mesothelin-transfected P815 cells (P815-Meso). After 4 cycles of immunizations, mice were killed, total RNA was extracted from splenocytes, and 5 Fab libraries were created by using phage display technology (). To avoid crosslinking with antibodies recognizing other antigens on P815 cells, phage libraries were first selected, against mesothelin-expressing Chinese hamster ovary (CHO) cells (CHO-Meso). Then, the second round of panning was performed against P815-Meso cells (). Non-transfected CHO and P815 cells were also used as a control for panning to assess specific phage enrichment (data not shown). After 2 rounds of panning for each library, Fab-containing periplasmic extractions of 96 single colony cultures were collected and screened on CHO-Meso cells. Overall, 147 Fabs binding to mesothelin were identified. A study pathway was designed to first eliminate only the molecules that bound the transfected form, and then to identify only membrane-form associated mesothelin binders by using competition tests with soluble mesothelin ().
Figure 1. Generation of mouse anti-mesothelin Fabs. 5 mice were first tolerized with soluble form of mesothelin, then immunized with P815-Meso cells. After 4 cycles of immunization mice were killed and spleens were resected. Total RNA was extracted from splenocytes and cDNA was produced. Vh-Ch1 and Vk-Ck sequences were amplified, cutted and inserted to pCB3 phage vector. (A). 2 rounds of panning were performed on libraries with CHO-Meso (45%) vs CHO-WTcells (Round I) and P815-Meso (90%) vs P815 wt cells (Round II) respectively (B). Number of positive FABs screened on CHO-Meso and Hela cells for each mouse phage display library (C). Study pathway to identify non-competing Fabs which bind only membrane associated form of mesothelin (D).
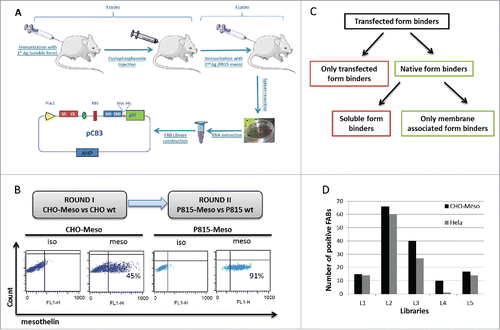
To identify antibodies binding specifically to mesothelin on cancer cell membranes (referred to as native form of mesothelin), we re-screened all positive Fabs able to recognize Hela, a cervix adenocarcinoma cell line that expresses naturally high levels of mesothelin. Only 116 of the 147 Fabs were identified as native form specific binders on Hela cells (). Then, all 147 Fabs were sequenced and their VH-CDR3 regions were determined for further analysis.
Regrouping Fabs in families following their VH-CDR3 similarities
VH-CDR3 sequences of antibodies have already been reported as the most important region for epitope recognition.Citation27-29 On this basis, VH-CDR3 regions of all Fabs were aligned and associated in 34 families according to their amino acid sequence similarities. Fabs recognizing Hela cells were distributed in only 20 of 34 VH-CDR3 families. The screening data of Fab staining on CHO-Meso and Hela cells revealed that all clones of each family showed uniform staining on cells, suggesting that all of them recognized the same epitope (Fig. S1). On the basis of these data, only one clone of each family was selected for further testing.
Eliminating soluble mesothelin binding Fabs
After the identification of native form binders, the next screening was designed to eliminate Fabs that recognize soluble mesothelin. To compare their ability to bind either only membrane-bound or both membrane-bound and soluble forms of mesothelin, the binding of Fabs to Hela cells was tested in competition with recombinant mesothelin protein. Fab concentration was adjusted by end point dilution assay. To establish the end point dilution, 2-fold dilution series for each Fab were performed. These solutions were tested on Hela cells to identify the limited concentration (end point dilution) for which the Fab signal decreases in flow cytometry analysis (Fig. S2). To identify the Fabs that discriminate between membrane-bound and soluble antigen, each Fab was incubated at its end point dilution for 30 minutes with 0.8 µg of soluble mesothelin, before assessing the binding capacity on Hela cells. From the flow cytometry analysis, only 10 VH-CDR3 families were identified as partial or completely discriminating Fabs. Representative staining on Hela cells of Fabs 1H7, 3C2 and 3C1 is shown in . 1H7 and 3C2 Fabs bound 69% and 71% on Hela cells, respectively. Incubation with 0.8 µg of mesothelin before staining did not alter the binding capacity of any of the selected Fabs, while the staining capacity of 3C1 decreased significantly. To confirm the results obtained by flow cytometry, a biotinylated mesothelin protein was attached to a streptavidin-coated plate and used for the screening of discriminating and non-discriminating Fabs using direct enzyme-linked immunosorbent assay (ELISA). Only non-discriminating Fabs, 3C1 for example, could bind to mesothelin. In contrast, Fabs that do not bind soluble mesothelin were not able to detect mesothelin in ELISA assay ().
Figure 2. Competition tests with soluble recombinant mesothelin protein. Example of non-competing (3C2 and 1H7) and competing (3C1) Fabs with staining on Hela cells with or without presence of 0.8 µg soluble mesothelin protein. (A). ELISA assay to detect the binding capacity of Fabs (3C1, 3C2 and 1H7) to soluble recombinant mesothelin protein (B).
These experiments confirmed that the methodology used for immunization allows us to identify discriminating Fab families. Next, we sought to investigate if the soluble mesothelin-related peptides (SMRP) present in physiologic fluids, specifically those present in mesothelin-expressing cancer patients reduces the proportion of discriminating Fabs binding the antigen presented on the cell surface.
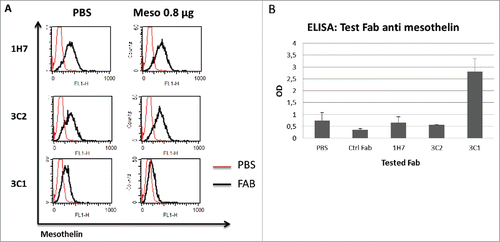
Competition tests with SMRP in patients' serum and cell supernatant
High levels of SMRP is a prognostic factor for mesothelioma, ovarian and pancreatic cancers.Citation30-34 The presence of SMRP can also alter the binding capacity of mesothelin-targeting therapeutic agents.Citation22,23 On the basis of these data, we used the serum of 21 patients with metastatic colon and pancreatic cancers in competition assays with selected Fabs. Serum concentration of mesothelin from the 21 patients and from 2 healthy volunteers (A and B) was first determined by ELISA assay (). Mesothelin concentration was shown to be significantly higher in all of the 21 patients compared with healthy donors. According to their mesothelin concentration, serum samples were pooled in 2 groups: high- (> 20 ng/ml) and low-serum (< 20 ng/ml) samples, respectively. Hela cell-derived supernatant containing 35 ng/ml of mesothelin was also used as another source of SMRP (). Competition assays were performed with high- or low-serum samples and with Hela supernatant. The samples were pre-incubated with selected Fabs for 30 minutes, and then added to Hela cells before flow cytometry analysis. Five families were revealed as discriminating Fabs against physiologic mesothelin-containing fluids. shows that the presence of high- and low-serum samples or Hela cell supernatant containing 35 ng/ml of mesothelin did not significantly alter the binding capacity of representative discriminating Fabs (e.g., 1H7 and 3C2). In contrast, the binding ability of a competing Fab (e.g., 3C1) was shown to decrease in the presence of high- or low- serum samples, and with concentrated Hela supernatant as well (24 vs. 9, 11 and 12 percent, espectively; cf. ).
Figure 3. Competition tests with patients' sera and concentrated Hela supernatant. Mesothelin concentration in patients' serum determined by ELISA assay (1 to 21). 2 safe donors (A and B) and PBS was used as a negative control for mesothelin expression and ELISA specificity, respectively (A). Mesothelin concentration in 10X concentrated Hela supernatant with ELISA assay. Hela cell lysate was used as a mesothelin containing control. (B). Examples of staining on Hela cells with non-competing Fabs (1H7 and 3C2) and competing FAB (3C1) in presence of 10X concentrated Hela supernatant and patients' sera high and low (C).
Mesothelin is anchored to cell membranes with GPI. GPI anchors with attached proteins are released from cells either in complexes with membrane lipids or exosomes.Citation35 For this reason, we investigated whether mesothelin resided in tumor-released exosomes. Exosomes were isolated from Hela cell supernatant by ultracentrifugation and mesothelin was quantified by ELISA assay (Fig. S3A). Mesothelin expression was confirmed by confocal microscopy showing co-localization of the protein with the exosome specific marker CD63 (Fig. S3B). The binding capacity of Fabs was tested in competition with exosomes. In those experiments, mesothelin-expressing exosomes failed to prevent Fab binding to Hela cells (Fig. S3C).
Altogether, these experiments demonstrate that the discriminating Fabs can selectively target mesothelin expressed on the surface of cancer cells without being subject to the antagonist effects of SMRP present in physiologic fluids.
1H7-hFc specifically binds mesothelin-expressing cancer cells
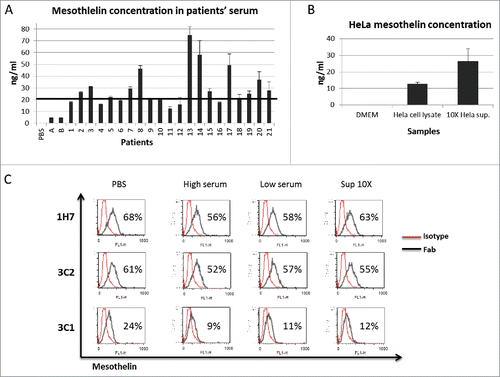
We next sought to determine if our discriminating antibodies target mesothelin-expressing cancer cells. For this purpose, we chose the Fab 1H7 as a discriminating antibody, for chimerization with human IgG1. The binding capacity of 1H7-hFc was then assessed on different cancer cell lines. We observed that 1H7-hFc bound Hela and CHO-Meso, a cell line over-expressing human mesothelin, but not CHO cells. 1H7-hFc strongly bound breast cancer cell line MDA-MB231, but moderately recognized colon cancer cells HT29, while non-small cell lung cancer cells A549 were weakly bound by this clone. No binding could be observed for cervix cancer cells C33A, which is a mesothelin negative cell line (). To assess if 1H7-hFc could recognize primary cell lines, we tested 3 different pancreatic cancer cells BesPac-C7, BesPac-C8 and BesPac-C12, obtained in our laboratory from ascites of pancreatic cancer patients. Flow cytometry analysis showed that 1H7-hFc bound strongly to BesPac-C7 and BesPac-C12, and weakly to BesPac-C8 cells. Mesothelin expression on these cells was confirmed by qRT-PCR and western blotting experiments (data not shown). These results demonstrated that 1H7-hFc selectively recognizes native mesothelin on different types of cancer cells.
Figure 4. Test on other cancer cell lines. FACS analysis of staining by 1H7-hFc on different cancer cells. CHO-Meso vs CHO-WT cells were used as a specificity control for 1H7-hFc.
To show the monospecificity of our anti-mesothelin antibody 1H7, we performed an immunoprecipitation (IP) assay in mild conditions on Hela and CHO-Meso cells and migrated the product in acrylamide gel. Staining of the gel showed a band at 40 kDa that strongly corresponds to mesothelin. We also analyzed this IP product using 1H7-hFc in western blotting with non-reduced and non-heated conditions and obtained bands at 40 and 69 kDa that corresponds mature and non-mature mesothelin, respectively (Fig. S4).
1H7 does not recognize GPI anchor
The experiments presented above demonstrate that 1H7 is selective for the membrane-associated conformation of the mesothelin protein and does not recognize other conformations such as the recombinant form or that contained within the physiologic fluids from mesothelin-expressing cancer patients. A new set of experiments was designed to investigate the role of the presence of GPI in epitope recognition by 1H7-Fc. To show the specificity of 1H7-hFc to mesothelin, Hela cells were treated with 1U and 10U of GPI-specific phospholipase C (PI-PLC), which cleaves the phosphor-glycerol bond and leaves protein from cell membrane with the GPI anchor attached.Citation11 Depletion of mesothelin expression on cell membrane was confirmed by cytometry analysis (),
Figure 5. 1h7-hFc does not bind to GPI. Mesothelin staining by 1H7-hFc on Hela cells after treatment with 1 and 10 U/ml of PtdIns-PLC for 1 hour (A). FACS analysis of PC3 and LN-Cap cells by CD59 and 1H7-hFc staining (B). RMFI on Hela cells by staining differents concentrations (25, 50 and 100 nM) of FLAER (C). FACS analysis of staining on Hela cells by 1H7-hFc with or without the presence of 50 nm of FLAER (D).
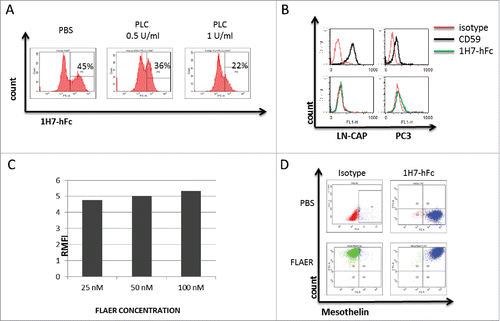
It remains possible that 1H7 is recognizing some component of the GPI anchor or a shared epitope between the GPI anchor and the protein. To decipher the role of the GPI-anchor in the epitope recognition of 1H7-hFc, we assessed whether 1H7 might be a non-specific antibody to GPI. To eliminate this hypothesis, PC3 and LN-CAP cells, which do not express mesothelin but highly express another GPI-anchored protein CD59, were stained by 1H7-hFc. The cells were highly positive for CD59 but not for 1H7-hFc, which confirmed that 1H7 does not recognize the GPI anchor ().
Second, we hypothesized that 1H7-hFc might bind a shared epitope between the GPI anchor and mesothelin. This hypothesis was also eliminated by using a FITC coupled pre-aerolysin protein, FLAER, which binds specifically to GPI.Citation36 We determined the saturation concentration of FLAER by staining Hela cells. Then, cells were incubated with 25, 50 and 100 nM of FLAER and analyzed by flow cytometry. Relative median fluorescent intensities (RMFI) were 4.8, 5 and 5.2 for each concentration, respectively. A saturation concentration of 50 nM was selected (), and Hela cells were incubated with 50 nM of FLAER one hour before staining with 1H7-hFc. Flow cytometry analysis showed that the binding properties of 1H7 were not altered by the presence of FLAER (). These results suggested that GPI is not involved directly in the epitope recognition by 1H7.
Together, these experiments indicate that GPI has no direct role in the recognition of cancer cells by 1H7 and suggest that 1H7 recognizes a conformational epitope generated by molecular interactions between mesothelin and the GPI anchor.
VH-CDR3 sequence comparison
Due to the differences in epitope recognition between discriminating and non-discriminating Fabs, we looked to see if there were consistent differences in their VH-CDR3 sequences. Regardless of epitope recognition, the VH-CDR3 length was 10 to 16 amino acids for all Fabs. According to IMGT unique numbering and color menu for amino acids,Citation37 all 105–117 positions of VH-CDR3 sequences were identified and amino acid frequencies were calculated for all positions of the CDR3 region (). At position 105, both the discriminating and non-discriminating sequences frequently encoded alanine, but 30% of discriminating sequences encoded threonine, inversely to leucine (10 %) and serine (10%) in non-discriminating sequences. At position 106, arginine was used frequently for both sequences. At positions 107 and 110, glycine was more frequent (50% and 60%, respectively) in discriminating sequences. Positively charged lysine, hydrophobic leucine, isoleucine and glycine were found more in discriminating sequences compared non-discriminating sequences at position 108 and 109. At position 113, only tryptophan (10% for both) and serine (20% and 10%, respectively) were coded commonly for both discriminating and non-discriminating sequences, but aspartic acid (10%), methionine (10%), phenylalanine (10%), leucine (30%), and isoleucine (10%) were only present in discriminating sequences. At position 114 in discriminating sequences, proline (10%) and serine (10%) were observed, but not in non-discriminating sequences. The presence of tyrosine, phenylalanine, and glycine were common for both discriminating and non-discriminating sequences. No significant difference of frequencies of amino acids was noted at positions 115, 116 and 117.
Figure 6. VH-CDR3 sequence analysis of competing and non-competing Fabs. The frequency of individual amino acids at the specific 11 positions of competing (n = 10) (A) and non-competing binders'(n = 10) (B) VH CDR3 sequences. The color menu for amino acids is according to IMGT. CDR3 positions are shown according to the IMGT unique numbering. Only membrane associated form binders (non-competing Fabs) in panel A, soluble form binders (competing Fabs) in panel B.
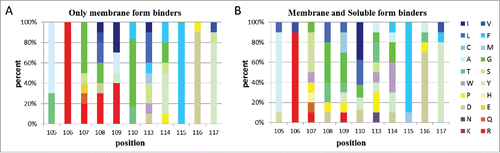
Comparison of VH-CDR3 sequences of discriminating Fabs versus non-discriminating Fabs showed that the frequencies of amino acids at positions 107 to 114 were significantly different in discriminating Fabs. Further research is warranted to confirm our data and determine the role of these amino acids in epitope recognition.
Discussion
This work describes the generation of a novel anti-mesothelin antibody (1H7) resulting from a subtractive immunization protocol inducing tolerance to the soluble antigen followed by the construction of mouse phage display libraries. 1H7 binds preferentially to mesothelin-expressing cells and does not compete with cleaved or recombinant mesothelin. It binds an epitope that appears to exist only when the protein is on the cell surface, and disappears in aqueous solutions.
Our results demonstrated the possibility of obtaining antibodies that could discriminate the membrane bound and soluble form of antigen by using a novel method of immunization. Although most of the obtained antibodies were specific to the membrane-bound form, we have also obtained some soluble form binders. This could be the result of ineffective tolerization, which could be improved by using higher doses of cyclophosphamide or more cycles of the immunization by soluble antigen and tolerance induction. Another choice to improve the effectiveness of our method could be to incubate phages with soluble antigen and then pursuit the selection on cells in the panning step of antibody selection.
Mesothelin has already been described in the literature as a good target for cancer therapy.Citation38 Several agents, including immunotoxins, chimeric mAbs, antibody-drug conjugates, anti-mesothelin tumor vaccines and anti-mesothelin chimeric antigen receptors (CARs) are in various stages of development for the treatment of patients with mesothelin-expressing tumors.Citation39-41 A chimeric antibody (MORAb-009) and an immunotoxin (SS1P) constructed from the same murine SS1 scFv targeting mesothelin have already been developed and are currently being evaluated in clinical trials treatments of mesothelioma and pancreatic cancers. The clinical use of SS1P is limited by its propensity to induce neutralizing antibodies. To date, 2 different approaches have been used to improve the efficiency of SS1P: 1) depletion of lymphocytes by chemical agents to diminish immune response to the bacterial toxin component,Citation42 and 2) use of mutated variants to improve the activity and decrease the immunogenicity of SS1P.Citation17 A further limitation for clinical use of SS1P is the high concentration of mesothelin in patients' serum and interstitial space of tumors.Citation43,44 Zhang et al. highlighted that the administration of SS1P combined with Taxol decreased the concentration of shed mesothelin.Citation22 They demonstrated that the synergy of SS1P with Taxol was due to the Taxol-induced fall in shed antigen levels. Here, we suggest that conjugating the antibody 1H7 with a less immunogenic toxin than SS1P might substantially enhance the efficacy of treatment by both reducing clearance of the conjugate and avoiding the issue of the therapeutic antibody binding to shed antigen.
The implication of the GPI anchor in conformational epitope recognition has been investigated in several studies. Bradley et al. showed that, by using de-lipidation of Thy-1 by different enzymes and constructing a recombinant Thy-1 without a GPI anchor, the recognition of Thy-1 by the mAbs K117, 5E10, and AS02 in western blots is abolished or greatly diminished.Citation45 However Thy-1 was detected in western blotting after exclusive partitioning into the insoluble phase. MAbs that bind to GPI-anchored proteins such as VSG or gp23 on the surface of parasites do not react with the same proteins when they are in soluble form.Citation46-48 This suggests the existence of different conformational epitopes in soluble and membrane-associated forms of GPI-anchored proteins that play a critical role in antibody binding. Changes in GPI-dependent antibody reactivity have also been observed in mammalian proteins; for example, human CD59 antibodies whose reactivity decreases after GPI-specific phospholipase D (GPI-PLD) treatment. This phenomenon has also been observed for human CD52 and Thy-1 proteins.Citation49,50 The work described above shows that membrane-associated mesothelin can be targeted specifically by mAb fragments that do not recognize the soluble form.
Therapeutic antibody-based anti-cancer approaches rely on the overexpression of the target antigen on cancer cells relative to the expression on normal tissues. The selectivity relies on this differential expression being sufficient to allow a killing action on the target cells without substantial damage to normal tissues. Our discriminating Fab thus might be a potential candidate for conversion of its single chain to clinically relevant-molecules: 1) conjugated with chemotherapy drug,Citation51 2) linked with an immunotoxin,Citation52 or 3) formatted as an IgG with effector functions such as antibody-dependent cell-mediated cytotoxicity.Citation53 In addition, studies on genetically modified T cells with CARs, including anti-mesothelin CAR T cells, report significant efficacy in different malignancies.Citation54,55 This membrane-bound antigen specificity could be of interest for generating anti-mesothelin CAR T cells using one of these discriminating Fabs.
Materials and methods
Patient samples
Blood samples from metastatic colon and pancreatic cancer patients were obtained from the oncology department of University Hospital of Besancon, France. Healthy donor blood samples were provided by the blood bank of Bourgogne-Franche-Comte.
Cell culture
CHO, P815, C33A, HT29, A549, PC3, LN-CAP and Hela cells were purchased from ATCC. CHO, C33A, HT29, A549, and Hela cells were cultured in DMEM-Glutamax (GIBCO), PC3, LN-CAP and P815 in RPMI 1640 supplemented with 10% of heat-inactivated fetal bovine serum ((FBS-HI)-Gibco), and 1% of penicillin-streptomycin (10,000 U/mL) (GIBCO). BesPac-C7, BesPac-C8, and BesPac-C12, were purified in our laboratory from ascites of pancreatic cancer patients donated as above and cultured in DMEM-Glutamax supplemented with 10% of FBS-HI. All cells were cultured in T175 Flasks at 37°C in a 5% CO2 incubator and sub-cultured every 2 d when cells confluence reached 80–90% of the cell flask. P815 and CHO cells were transfected with mesothelin-expressing PCMV6-XL4 vector (Invitrogen) to obtain CHO-Mesothelin and P815-Mesothelin cells.
Immunization of Mice
Four weeks old female BALB/c mice were first immunized intraperitoneally with 1 µg of full-length mesothelin recombinant protein (RayBiotech, ref: 230–00043) and, for the next 2 days, each day 1 mg of cyclophosphamide was injected intraperitoneally to each mouse. This procedure was performed 4 times at intervals of 2 weeks. Four days after the last cyclophosphamide injection, mice were immunized intraperitoneally (3 times at intervals of 2 weeks) with 10 million P815 cells transfected with mesothelin-expressing vector. A further 2 weeks later, an intravenous injection was performed with 1 million P815-Meso cells. Four days after the last injection, the mice were killed and the spleens were collected.
RNA extraction and cDNA construction
All spleens were separately mechanically degraded and splenocytes were collected. RNA was extracted using Qiagen RNeasy® Midi Kit according to manufacturer's instructions. 40 µg of RNA was used to make the cDNA by using SuperScript III First Strand Synthesis System by Invitrogen (cat#18080–051).
Sub-library construction
PCR reactions were performed with primers to amplify specifically the VH-Ch1 and Vk-Ck coding regions of lymphocytes. The PCR amplifications were performed using the Expand High Fidelity System by Roche (cat# 4738268001) in 100 µl reaction volume using 6 µl of non-purified cDNA as template and 500 nM of each combination of primers. After the migration and purification of PCR products, second PCR reactions were performed, this time with restriction enzyme sites containing primers. After the second PCR, all products for VH-Ch1 and Vk-Ck were pooled and digested with SfiI/NotI and ApaLI/AscI, respectively. Phage-mid pCB3 vector was digested with the same enzymes and ligation was performed separately to obtain 2 sub-libraries, VH-CH1 and Vk-Ck. After the transformation of E. coli strain TG1 by electroporation, sub-libraries with the size of greater than 1E+08 colonies were created.
Library construction
After overnight culture, midi-prep was performed for each of the sub-libraries. VH-Ch1 regions were cloned on the Vk-Ck sub-libraries after digestion by SfiI/NotI enzymes. Transformation was performed with electroporation on E. coli strain TG1 and libraries with sizes of greater than 1E+08 colonies were obtained.
Panning of library
Phages were incubated with 5E+06 cells in solution for one hour at 4°C and then washed several times with 10% FBS/phosphate-buffered saline (PBS). Bound phages were retrieved using trypsin (1 mg/ml) at room temperature. Protease activity was immediately neutralized by applying 16 mM protease inhibitor AEBSF. On the first round of screening the CHO-Mesothelin cells were used to select the phage libraries; on the second round the P815-Mesothelin cells were used to select the first round rescued phage.
Preparation of periplasmic extracts containing soluble Fabs
Soluble Fabs were produced from individual E. coli clones as described.Citation56 Fab expression was induced by adding 1 mM of isopropyl 1-thio-β-D-galactopyranoside. Periplasmic extracts (P.E.s) containing soluble Fab were extracted by a freeze/thaw cycle; bacteria were pelleted and frozen at −80°C for 2 hours and then thawed at room temperature and resuspended in 110 μl of PBS. The P.E. containing the soluble Fab was collected after 30 min shaking at room temperature and centrifugation.
Flow cytometry and screening
Twenty µl of extracted Fabs were tested for binding to CHO-Mesothelin and to CHO-WT cells using a Flow cytometer BD Accuri™ C6 system. The binding of the soluble Fabs to CHO-Mesothelin and to CHO-WT cells (1E+05 cells) was detected with an anti c-myc antibody (Myc 9E10 Diaclone cat#855.313.020 and a goat anti-mouse IgG-FITC (Cappel MP cat#55526). Fab binding levels were determined by the percentage of positive cells signal and mean intensity levels in the FL1 channel detector. Each monoclonal Fab sample was also tested on non-expressing cells, CHO-WT, as a control. The positive clones were then tested in the presence (0.8 µg/well) and absence of soluble recombinant human mesothelin (Raybiotech, cat#230–00043–50). Selected clones were also tested with the same method on Hela cells to determine the natural form binders.
Human mesothelin fluorescein-conjugated antibody (ref: FAB32652F) from Bio-techne was used for positive control staining on mesothelin-expressing cells. Anti-CD59 antibody was purchased from Diaclone.
FLAER were purchased from Cedarlane Labs and used according to the manufacturer's instructions.
Sequencing of antibody genes
Agar stabs were prepared from the panel of positive clones, by inoculating with 2 μl of stock culture flat-bottom 96-well plates containing LB Agar supplemented with ampicillin at 100 μg/ml and 2% glucose, according to LGC Genomics recommendations. The variable heavy chain was sequenced with primer pelb3 and the variable light chain was sequenced with primer M13R. The amino acid sequences of the Fab variable domains obtained from LGC genomics were extracted using CLC Workbench or SIMPLE antibody extractor software. The Fabs were grouped in families based on the VH-CDR3 amino acid sequence homology.
Production and purification of chimeric monoclonal antibodies
1H7 was chimerized at RD Biotech (Besançon, France) with human IgG1-expressing mammalian vector and transfected to CHO cells. Cell culture supernatants were collected and antibody concentration was determined by RD Biotech.
ELISA
Binding of selected clones to soluble mesothelin was also assayed by ELISA. Wells of Maxisorp plates (Nunc, cat#456537) were coated with 100 ng of full-length recombinant mesothelin protein in 100 µl of PBS and incubated overnight at 4 °C. The wells were saturated with 200 µl of 5% bovine serum albumin solution for 2 hours. 100 µl of PBS containing 20 µl of extracted Fabs was applied to each well and incubated for 1 hour at room temperature. After washing several times with 1X washing buffer, bound Fabs were detected with biotinylated anti c-myc antibody and revealed with HRP coupled streptavidine for 30 minutes. After washing several times with 1X washing buffer, 100 µl of HRP substrate TMB was added to each well. Color development was stopped after 5 minutes by adding 100 µl of 2N H2SO4, and the OD readings were taken at 450 nm.
To determine mesothelin quantity in samples an ELISA assay kit from Biolegend (cat: 438604) was used. ELISA was performed following manufacturer's instructions.
Immunoprecipitation and western blotting
Immunoprecipitation (IP) of cell lysates from Hela and CHO-Meso cells was performed by using Dynabeads Protein G. At first, to preclear the cell lysate, 200 μl of Dynabeads Protein G was washed 3 times with PBS tween (0.025%) and incubated with cell lysate at 4°C overnight, and then the beads were removed by centrifugation 5 minutes at 20000 g and supernatant was recovered. In parallel, 30 µl of Dynabeads Protein G was also washed with Tris-buffered saline (TBS) tween (0.025%) and incubated with 5 µg of 1H7-hFc at 22°C for 2 hours. Then, the beads were precipitated and added to the supernatant recovered after the preclearing step and incubated at 4°C for 4 hours. Immune complex was precipitated by magnetic capturing, and washed 4 times with 600 µl of TBS tween (0.025%). The precipitated immune-complex was dissociated by heating to 70°C for 5 minutes and recuperated in 50 µl of Laemmli 1X buffer without adding β-mercaptoethanol. Five µl of the IP product was used for SDS gel migration and coloration and also for western blotting experiments.
Cellular proteins were extracted in Laemmli 1X buffer without adding β-mercaptoethanol and not heated to 95°C. They were separated by electrophoresis (SDS-PAGE) in a polyacrylamide gel in the presence of sodium dodecyl sulfate (SDS) according to their molecular weight, and stained by InstantBlue (Sigma) following manufacturer's instructions.
After the migration in a polyacrylamide gel, all proteins were transferred to a polyvinylidene difluoride film for staining by a specific antibody (1H7-hFc) and then detection by secondary anti-human antibody coupled to peroxidase was performed. 15 µg of lysed cells (Input) was also used as a control.
Exosome extraction
Exosomes were extracted from Hela cell culture. Cell culture was centrifuged at 300 x g for 10 mins to precipitate the cells, 2000 x g for 10 mins to pellet dead cells, 3000 x g for 30 mins to pellet cell debris and supernatant was collected. Then, ultracentrifugation was performed at 100000 x g for 70 mins to precipitate exosomes. Supplementary wash step with PBS 1X at 100000 x g for 70 mins was also performed to remove the contaminant proteins.
Exosomes were detected by confocal microscopy with CD63 staining and mesothelin concentration was determined by ELISA assay.
Disclosure of potential conflicts of interest
The authors declare that they have not competing interests.
Supplemental_Data.pptx
Download MS Power Point (811.3 KB)Acknowledgments
The authors would like to thank Doctor Andy Clark and Guadalupe Tizon for English writing assistance and also Dr Yann Godet for his advice on the article design.
References
- Yamaguchi N, Hattori K, Oh-eda M, Kojima T, Imai N, Ochi N. A novel cytokine exhibiting megakaryocyte potentiating activity from a human pancreatic tumor cell line HPC-Y5. J Biol Chem 1994; 269:805-08; PMID:8288629
- Hassan R, Bera T, Pastan I. Mesothelin: a new target for immunotherapy. Clin Cancer Res Off J Am Assoc Cancer Res 2004; 10:3937-42; PMID:15217923; http://dx.doi.org/10.1158/1078-0432.CCR-03-0801
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin compared with paclitaxel and cisplatin in patients with stage III and stage IV ovarian cancer. N Engl J Med 1996; 334:1-6; PMID:7494563; http://dx.doi.org/10.1056/NEJM199601043340101
- Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ, et al. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE). Clin Cancer Res Off J Am Assoc Cancer Res 2001; 7:3862-68; PMID:11751476
- Hassan R, Kreitman RJ, Pastan I, Willingham MC. Localization of mesothelin in epithelial ovarian cancer. Appl Immunohistochem Mol Morphol AIMM Off Publ Soc Appl Immunohistochem 2005; 13:243-47; PMID:16082249; http://dx.doi.org/10.1097/01.pai.00000141545.36485.d6
- Li M, Bharadwaj U, Zhang R, Zhang S, Mu H, Fisher WE, Brunicardi FC, Chen C, Yao Q. Mesothelin is a malignant factor and therapeutic vaccine target for pancreatic cancer. Mol Cancer Ther 2008; 7:286-96; PMID:18281514; http://dx.doi.org/10.1158/1535-7163.MCT-07-0483
- Yen MJ, Hsu C-Y, Mao T-L, Wu T-C, Roden R, Wang T-L, Shih I-M. Diffuse mesothelin expression correlates with prolonged patient survival in ovarian serous carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res 2006; 12:827-31; PMID:16467095; http://dx.doi.org/10.1158/1078-0432.CCR-05-1397
- Ho M, Bera TK, Willingham MC, Onda M, Hassan R, FitzGerald D, Pastan I. Mesothelin expression in human lung cancer. Clin Cancer Res Off J Am Assoc Cancer Res 2007; 13:1571-75; PMID:17332303; http://dx.doi.org/10.1158/1078-0432.CCR-06-2161
- Muminova ZE, Strong TV, Shaw DR. Characterization of human mesothelin transcripts in ovarian and pancreatic cancer. BMC Cancer 2004; 4:19; PMID:15140265; http://dx.doi.org/10.1186/1471-2407-4-19
- Sapede C, Gauvrit A, Barbieux I, Padieu M, Cellerin L, Sagan C, Scherpereel A, Dabouis G, Grégoire M. Aberrant splicing and protease involvement in mesothelin release from epithelioid mesothelioma cells. Cancer Sci 2008; 99:590-94; PMID:18167128; http://dx.doi.org/10.1111/j.1349-7006.2007.00715.x
- Zhang Y, Chertov O, Zhang J, Hassan R, Pastan I. Cytotoxic activity of immunotoxin SS1P is modulated by TACE-dependent mesothelin shedding. Cancer Res 2011; 71:5915-22; PMID:21775520; http://dx.doi.org/10.1158/0008-5472.CAN-11-0466
- Bütikofer P, Malherbe T, Boschung M, Roditi I. GPI-anchored proteins: now you see ‘em, now you don't. FASEB J 2001; 15:545-8; PMID:11156970; http://dx.doi.org/10.1096/fj.00-0415hyp
- Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol 2004; 5:110-20; PMID:15040444; http://dx.doi.org/10.1038/nrm1309
- Paulick MG, Bertozzi CR. The Glycosylphosphatidylinositol Anchor: A Complex Membrane-Anchoring Structure for Proteins. Biochemistry (Mosc) 2008; 47:6991-7000; PMID:18557633; http://dx.doi.org/10.1021/bi8006324
- Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol 2009; 157:220-33; PMID:19459844; http://dx.doi.org/10.1111/j.1476-5381.2009.00190.x
- Ma J, Tang WK, Esser L, Pastan I, Xia D. Recognition of mesothelin by the therapeutic antibody MORAb-009: structural and mechanistic insights. J Biol Chem 2012; 287:33123-31; PMID:22787150; http://dx.doi.org/10.1074/jbc.M112.381756
- Bang S, Nagata S, Onda M, Kreitman RJ, Pastan I. HA22 (R490A) is a recombinant immunotoxin with increased antitumor activity without an increase in animal toxicity. Clin Cancer Res Off J Am Assoc Cancer Res 2005; 11:1545-50; PMID:15746059; http://dx.doi.org/10.1158/1078-0432.CCR-04-1939
- Hollevoet K, Mason-Osann E, Liu X, Imhof-Jung S, Niederfellner G, Pastan I. In vitro and in vivo activity of the low-immunogenic antimesothelin immunotoxin RG7787 in pancreatic cancer. Mol Cancer Ther 2014; 13:2040-49; PMID:24928849; http://dx.doi.org/10.1158/1535-7163.MCT-14-0089-T
- Kelly RJ, Sharon E, Pastan I, Hassan R. Mesothelin targeted agents in clinical trials and in preclinical development. Mol Cancer Ther 2012; 11:517-25; PMID:22351743; http://dx.doi.org/10.1158/1535-7163.MCT-11-0454
- de Goeij BE, Lambert JM. New developments for antibody-drug conjugate-based therapeutic approaches. Curr Opin Immunol 2016; 40:14-23; PMID:26963132; http://dx.doi.org/10.1016/j.coi.2016.02.008
- Tang Z, Feng M, Gao W, Phung Y, Chen W, Chaudhary A, St Croix B, Qian M, Dimitrov DS, Ho M. A human single-domain antibody elicits potent antitumor activity by targeting an epitope in mesothelin close to the cancer cell surface. Mol Cancer Ther 2013; 12:416-26; PMID:23371858; http://dx.doi.org/10.1158/1535-7163.MCT-12-0731
- Zhang Y, Xiang L, Hassan R, Pastan I. Immunotoxin and Taxol synergy results from a decrease in shed mesothelin levels in the extracellular space of tumors. Proc Natl Acad Sci U S A 2007; 104:17099-104; PMID:17940013; http://dx.doi.org/10.1073/pnas.0708101104
- Awuah P, Bera TK, Folivi M, Chertov O, Pastan I. Reduced Shedding of Surface Mesothelin Improves Efficacy of Mesothelin Targeting Recombinant Immunotoxins. Mol Cancer Ther molcanther 2016; 15(7):1648-55; PMID:27196771; http://dx.doi.org/10.1158/1535-7163.MCT-15-0863
- Mapara MY, Pelot M, Zhao G, Swenson K, Pearson D, Sykes M. Induction of stable long-term mixed hematopoietic chimerism following nonmyeloablative conditioning with T cell-depleting antibodies, cyclophosphamide, and thymic irradiation leads to donor-specific in vitro and in vivo tolerance. Biol Blood Marrow Transplant J Am Soc Blood Marrow Transplant 2001; 7:646-55; PMID:11787527; http://dx.doi.org/10.1053/bbmt.2001.v7.pm11787527
- Hairul Bahara NH, Tye GJ, Choong YS, Ong EBB, Ismail A, Lim TS. Phage display antibodies for diagnostic applications. Biol J Int Assoc Biol Stand 2013; 41:209-16; PMID:23647952; http://dx.doi.org/10.1016/j.biologicals.2013.04.001
- Moutel S, Vielemeyer O, Jin H, Divoux S, Benaroch P, Perez F. Fully in vitro selection of recombinant antibodies. Biotechnol J 2009; 4:38-43; PMID:19156724; http://dx.doi.org/10.1002/biot.200800246
- Chothia C, Lesk AM, Tramontano A, Levitt M, Smith-Gill SJ, Air G, Sheriff S, Padlan EA, Davies D, Tulip WR. Conformations of immunoglobulin hypervariable regions. Nature 1989; 342:877-883; PMID:2687698; http://dx.doi.org/10.1038/342877a0
- Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol 1994; 4:857-67; PMID:7536111; http://dx.doi.org/10.1016/0959-440X(94)90267-4
- Xu JL, Davis MM. Diversity in the CDR3 region of V(H) is sufficient for most antibody specificities. Immunity 2000; 13:37-45; PMID:10933393; http://dx.doi.org/10.1016/S1074-7613(00)00006-6
- Canessa PA, Franceschini MC, Ferro P, Battolla E, Dessanti P, Manta C, Sivori M, Pezzi R, Fontana V, Fedeli F, et al. Evaluation of soluble mesothelin-related peptide as a diagnostic marker of malignant pleural mesothelioma effusions: its contribution to cytology. Cancer Invest 2013; 31:43-50; PMID:23249166; http://dx.doi.org/10.3109/07357907.2012.749265
- Cui A, Jin X-G, Zhai K, Tong Z-H, Shi H-Z. Diagnostic values of soluble mesothelin-related peptides for malignant pleural mesothelioma: updated meta-analysis. BMJ Open 2014; 4:e004145; PMID:24566531; http://dx.doi.org/10.1136/bmjopen-2013-004145
- Franko A, Dolzan V, Kovac V, Arneric N, Dodic-Fikfak M. Soluble mesothelin-related peptides levels in patients with malignant mesothelioma. Dis Markers 2012; 32:123-31; PMID:22377706; http://dx.doi.org/10.1155/2012/430689
- Pass HI, Wali A, Tang N, Ivanova A, Ivanov S, Harbut M, Carbone M, Allard J. Soluble mesothelin-related peptide level elevation in mesothelioma serum and pleural effusions. Ann Thorac Surg 2008; 85:265-72; discussion 272; PMID:18154821; http://dx.doi.org/10.1016/j.athoracsur.2007.07.042
- Wu X, Li D, Liu L, Liu B, Liang H, Yang B. Serum soluble mesothelin-related peptide (SMRP): a potential diagnostic and monitoring marker for epithelial ovarian cancer. Arch Gynecol Obstet 2014; 289:1309-14; PMID:24370956; http://dx.doi.org/10.1007/s00404-013-3128-x
- Rooney IA, Heuser JE, Atkinson JP. GPI-anchored complement regulatory proteins in seminal plasma. An analysis of their physical condition and the mechanisms of their binding to exogenous cells. J Clin Invest 1996; 97:1675-86; PMID:8601633; http://dx.doi.org/10.1172/JCI118594
- van Zanten TS, Cambi A, Koopman M, Joosten B, Figdor CG, Garcia-Parajo MF. Hotspots of GPI-anchored proteins and integrin nanoclusters function as nucleation sites for cell adhesion. Proc Natl Acad Sci U S A 2009; 106:18557-62; PMID:19850864; http://dx.doi.org/10.1073/pnas.0905217106
- Ruiz M. Lefranc MP. Currents in Computational Molecular Biology. Tokyo: Universal Academy Press. Front Sci Ser 2000; 126-27
- Pastan I, Hassan R. Discovery of mesothelin and exploiting it as a target for immunotherapy. Cancer Res 2014; 74:2907-12; PMID:24824231; http://dx.doi.org/10.1158/0008-5472.CAN-14-0337
- Chang M-C, Chen Y-L, Chiang Y-C, Chen T-C, Tang Y-C, Chen C-A, Sun W-Z, Cheng W-F. Mesothelin-specific cell-based vaccine generates antigen-specific immunity and potent antitumor effects by combining with IL-12 immunomodulator. Gene Ther 2016; 23:38-49; PMID:26262583; http://dx.doi.org/10.1038/gt.2015.85
- Kelly RJ, Sharon E, Pastan I, Hassan R. Mesothelin-targeted agents in clinical trials and in preclinical development. Mol Cancer Ther 2012; 11:517-25; PMID:22351743; http://dx.doi.org/10.1158/1535-7163.MCT-11-0454
- O'Hara M, Stashwick C, Haas AR, Tanyi JL. Mesothelin as a target for chimeric antigen receptor-modified T cells as anticancer therapy. Immunotherapy 2016; 8:449-60; PMID:26973126; http://dx.doi.org/10.2217/imt.16.4
- Hassan R, Miller AC, Sharon E, Thomas A, Reynolds JC, Ling A, Kreitman RJ, Miettinen MM, Steinberg SM, Fowler DH, et al. Major cancer regressions in mesothelioma after treatment with an anti-mesothelin immunotoxin and immune suppression. Sci Transl Med 2013; 5:208ra147; PMID:24154601; http://dx.doi.org/10.1126/scitranslmed.3006941
- Weldon JE, Skarzynski M, Therres JA, Ostovitz JR, Zhou H, Kreitman RJ, Pastan I. Designing the furin-cleavable linker in recombinant immunotoxins based on Pseudomonas exotoxin A. Bioconjug Chem 2015; 26:1120-28; PMID:25997032; http://dx.doi.org/10.1021/acs.bioconjchem.5b00190
- Zhang Y, Pastan I. High shed antigen levels within tumors: an additional barrier to immunoconjugate therapy. Clin Cancer Res Off J Am Assoc Cancer Res 2008; 14:7981-86; PMID:19088013; http://dx.doi.org/10.1158/1078-0432.CCR-08-0324
- Bradley JE, Chan JM, Hagood JS. Effect of the GPI anchor of human Thy-1 on antibody recognition and function. Lab Investig J Tech Methods Pathol 2013; 93:365-74; PMID:23358110; http://dx.doi.org/10.1038/labinvest.2012.178
- Cardoso de Almeida ML, Turner MJ. The membrane form of variant surface glycoproteins of Trypanosoma brucei. Nature 1983; 302:349-52; PMID:6188057; http://dx.doi.org/10.1038/302349a0
- Miller EN, Allan LM, Turner MJ. Topological analysis of antigenic determinants on a variant surface glycoprotein of Trypanosoma brucei. Mol Biochem Parasitol 1984; 13:67-81; PMID:6083452; http://dx.doi.org/10.1016/0166-6851(84)90102-6
- Tomavo S, Dubremetz J-F, Schwarz RT. Structural analysis of glycosyl-phosphatidylinositol membrane anchor of the Toxoplasma gondii tachyzoite surface glycoprotein gp23. Biol Cell 1993; 78:155-62; PMID:8241958; http://dx.doi.org/10.1016/0248-4900(93)90126-Y
- Barboni E, Rivero BP, George AJ, Martin SR, Renoup DV, Hounsell EF, Barber PC, Morris RJ. The glycophosphatidylinositol anchor affects the conformation of Thy-1 protein. J Cell Sci 1995; 108 ( Pt 2):487-97; PMID:7539435
- Hale G. Synthetic peptide mimotope of the CAMPATH-1 (CD52) antigen, a small glycosylphosphatidylinositol-anchored glycoprotein. Immunotechnology Int J Immunol Eng 1995; 1:175-87; PMID:9373346; http://dx.doi.org/10.1016/1380-2933(95)00017-8
- Bouchard H, Viskov C, Garcia-Echeverria C. Antibody-drug conjugates—A new wave of cancer drugs. Bioorg Med Chem Lett 2014; 24:5357-63; PMID:25455482; http://dx.doi.org/10.1016/j.bmcl.2014.10.021
- Pastan I, Hassan R, FitzGerald DJ, Kreitman RJ. Immunotoxin therapy of cancer. Nat Rev Cancer 2006; 6:559-65; PMID:16794638; http://dx.doi.org/10.1038/nrc1891
- Seidel UJE, Schlegel P, Lang P. Natural killer cell mediated antibody-dependent cellular cytotoxicity in tumor immunotherapy with therapeutic antibodies. Front Immunol 2013; 4:76; PMID:23543707; http://dx.doi.org/10.3389/fimmu.2013.00076
- Adusumilli PS, Cherkassky L, Villena-Vargas J, Colovos C, Servais E, Plotkin J, Jones DR, Sadelain M. Regional delivery of mesothelin-targeted CAR T cell therapy generates potent and long-lasting CD4-dependent tumor immunity. Sci Transl Med 2014; 6:261ra151; PMID:25378643; http://dx.doi.org/10.1126/scitranslmed.3010162
- Morello A, Sadelain M, Adusumilli PS. Mesothelin-Targeted CARs: Driving T Cells to Solid Tumors. Cancer Discov 2016; 6:133-146; PMID:26503962; http://dx.doi.org/10.1158/2159-8290.CD-15-0583
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol 1991; 222:581-97; PMID:1748994; http://dx.doi.org/10.1016/0022-2836(91)90498-U
- Ng WC, Wong V, Muller B, Rawlin B, Brown EL. Prevention and treatment of influenza with hyperimmune bovine colostrum antibody. PlosOne 2010; e13622; PMID:21049034; http://dx.doi.org/10.1371/journal.pone.0013622
