ABSTRACT
The increased number of bispecific antibodies (BsAb) under therapeutic development has resulted in a need for mouse surrogate BsAbs. Here, we describe a one-step method for generating highly pure mouse BsAbs suitable for in vitro and in vivo studies. We identify two mutations in the mouse IgG2a and IgG2b Fc region: one that eliminates protein A binding and one that enhances protein A binding by 8-fold. We show that BsAbs harboring these mutations can be purified from the residual parental monoclonal antibodies in one step using protein A affinity chromatography. The structural basis for the effects of these mutations was analyzed by X-ray crystallography. While the mutation that disrupted protein A binding also inhibited FcRn interaction, a bispecific mutant in which one subunit retained the ability to bind protein A could still interact with FcRn. Pharmacokinetic analysis of the serum half-lives of the mutants showed that the mutant BsAb had a serum half-life comparable to a wild-type Ab. The results describe a rapid method for generating panels of mouse BsAbs that could be used in mouse studies.
Introduction
Therapeutic biologics programs are increasingly turning to bispecific antibodies (BsAbs), which are Abs containing two distinct antigen binding (Fab) regions. Indeed, many such bispecific therapeutics for dual-antigen targeting, cell redirection efforts, and immune checkpoint modulation are currently in clinical trials, Citation1-3 although high-throughput production of highly pure BsAbs presents challenges. The development of human IgG-based BsAbs to be generated on a manufacturing scale was initially limited by the difficulty of separating the desired BsAb from undesired antibody species that could be present in production mixtures.Citation4 For example, BsAb can be generated by co-transfection of two heavy chains (HC) and two light chains (LC), resulting in a small amount (< 10%) of desired BsAb among a majority of contaminating species, but this approach is not feasible for large-scale or high-throughput efforts. Although novel Fv-based molecules and non-IgG based scaffolds can alleviate HC-HC and HC-LC pairing issues, Citation5 IgG-based bispecific molecules are often attractive on the basis of their longer serum half-lives, lower likelihood of having anti-drug responses, and their amenability to engineering to modulate Fc effector functions such as antibody-dependent cell-mediated cytotoxicity (ADCC) or antibody-dependent cellular phagocytosis (ADCP).
Thus, substantial effort has been put into optimizing pairing between HC-HC and between HC-LC. To generate BsAbs, the HC molecules require heterodimerization while the LC molecules must pair with the correct cognate HC. Some examples of successful strategies to influence HC-HC pairing include knobs-into-holes (KIH), Citation6 electrostatic steering, Citation7 and controlled Fab arm exchange (cFAE).Citation8-10 These methods all involve introduction of complementary mutations into the CH3 domain to favor formation of heterodimerization of Fc regions. To generate BsAb by co-transfection of HC plasmids requires additional methods to ensure correct HC-LC pairing.Citation4,11,12 The cFAE method ensures proper HC-LC pairing by using separately expressed and purified parental mAbs containing the complementary HC mutations F405L or K409R. The parental mAbs) that are mixed and incubated under mild reducing conditions and subsequent oxidation. The reaction generally results in greater than 90% heterodimer formation of human IgG1 BsAb, Citation8 and was later shown to be effective when separately expressed mAbs are subjected to cFAE directly from culture supernatants.Citation13
To better validate the therapeutic efficacy of IgG biologics, drug discovery programs frequently use mouse IgG Abs as surrogates for in vivo studies, particularly mouse IgG2a and mouse IgG2b since they display greater Fcγ receptor-mediated effector function compared with mouse IgG1 and mouse IgG3 Abs.Citation14 Mouse IgG2a and IgG2b Abs can be generated by cFAE, although efficient BsAb formation requires two additional mutations (T/V370K, F405L and K409R, R411T) to favor HC pairing.Citation15 Mouse BsAbs have also been generated by other methods, including KIH technology.Citation16,17 While these methods can lead to relatively efficient BsAb formation, removal of contaminating parental mAb and Ab fragments is still required, especially in cases where parental Ab can have a confounding effect on experimental results. Additionally, in cases where parental mAbs are co-transfected, expression levels of each polypeptide chain results in significant populations of undesired contaminating species. Removal of these species, which can have similar biophysical characteristics as the desired BsAb, can be time consuming. Thus, methods for high-throughput purification of mouse BsAb heterodimers are of critical importance.
Large scale and high-throughput purification of bispecific IgG molecules requires robust, preferably single-step purification methods that use commercially available resins, such as those based on staphylococcal protein A. Stability-enhancing mutations introduced into a domain of protein A led to a synthetic fragment termed the Z-domain.Citation18 Tandem Z-domains have been engineered into commercial Fc affinity resins that are resistant to high pH treatment and which bind only the Fc region.Citation19,20 Abs are purified using Z-domain affinity resin by binding at neutral pH and eluting in acidic pH buffer. The pH value at which IgG can be eluted from protein A resin depends on its affinity for protein A. In fact, differences in affinities for protein A have been observed between IgG isotypes, and these affinity differences result in different pH values at which they can be eluted from immobilized protein A.Citation21,22
Modulation of the protein A binding characteristics of Abs can significantly decrease serum half-life since both protein A and the neonatal Fc receptor (FcRn) share a binding site on the Fc.Citation23 FcRn is largely responsible for maintaining the long serum half-life of IgG Citation24-29 via its ability to bind with high affinity to IgG at acidic pH (< 6.5) in the recycling endosome and to dissociate at neutral pH, releasing the IgG back into the serum and thus saving IgG from lysosomal degradation.Citation30,31 For example, mutation of isoleucine at position 253 (I253), which is conserved across human and mouse IgG molecules, to alanine almost entirely abolishes binding to FcRn.Citation32,33
We sought to develop a higher-throughput method to purify BsAb from contaminating parental mAbs and Ab fragments. We hypothesized that two different mouse IgG variants that bound Z-domain with different affinities could be separated by protein A-affinity chromatography at different pH values, allowing one-step generation of BsAb, analogous to a previous effort using human IgG1 Abs.Citation34 Here, we describe two sets of mutations in mouse IgG2a and IgG2b, one which abolished protein A binding and one which, remarkably, enhanced protein A binding. BsAbs in which each heavy chain harbored one of these mutations were separated from the residual parental mAbs by step elution from protein A resin at different pH values. We analyzed the effects of these mutation sets on Fc receptor interaction in vitro and on serum half-life in vivo. The structural basis for the effects of these mutations was determined by X-ray crystallography. Combined, the results described a method for one-step purification of > 95% pure mouse BsAb having serum half-lives comparable to wild-type mouse Abs.
Results
Rationale for mutations and their effects on protein A binding
We measured the affinity of IgG molecules for protein A by isothermal titration calorimetry (ITC) using a previously described synthetic three-helix Z-domain derived from protein A (Supplementary Fig. 1a).Citation18 As a control, the affinity (KD) of human IgG1 for Z-domain was measured as 23 nM (, Supplementary Fig. 1b), which was similar to previously reported measurements.Citation19,35-37 Mouse IgG2a and IgG2b bound Z-domain with weaker affinity, having KD values of 0.2 and 3.6 μM, respectively (, , Supplementary Fig. 1c and ). Since mutation of the highly conserved I253 was known to disrupt protein A binding, Citation32,33 we introduced I253D to generate a weak-binding parental Ab. Mutation of isoleucine 253 to aspartate (I253D) in mouse IgG2a or IgG2b abolished binding to Z-domain (, Supplementary Fig. 1e). Because of the weaker binding of mouse IgG2a and IgG2b relative to human IgG1, we sought to identify mutations that would enhance binding to protein A, leading to a parental Ab that would elute only at low pH. IgG binds Z-domain using three loops in the Fc region, two from the CH2 and one from CH3 domain.Citation38 By sequence comparison of mouse and human IgG Fc regions, we identified P307 and Q309 in mouse IgG2a and IgG2b, which are conserved in mouse IgG2a, IgG2b, and IgG3 but differ in human IgG1, as potentially contributing to the difference in affinity (Supplementary Fig. 2a). Human IgG1, which binds Z-domain much more tightly, contains threonine 307 (T307) and leucine (L309), which contribute to high affinity protein A binding.
Figure 1. Binding of IgG2b to Z-domain and differential affinity purification. (A) Isotherms resulting from the binding of Z-domain to mouse IgG2b variants. Top panels show power input vs mole ratio of injectant (Z-domain) to titrant (IgG). Bottom panels plot the corresponding binding enthalpy. (B) Purification chromatograms of mouse IgG2b variants by 3-step pH gradient using mAbSelect Sure (GE Healthcare) affinity resin. The left y-axis shows absorbance at 280 nm and the right y-axis shows buffer pH vs elution volume. One mg each of mouse IgG2b was loaded in each experiment. (C) The elution fractions from each elution step: pH 7.2, 4.0, and 3.4 for the mixed sample containing both parental Abs and the bispecific Ab (orange trace in panel b) were analyzed by hydrophobic interaction chromatography (HIC). Dashed lines indicate the elution volume of the two parental mouse Abs and the mouse BsAb. Note that the elution volume of the mouse BsAb was intermediate between that of the parental mouse Abs.
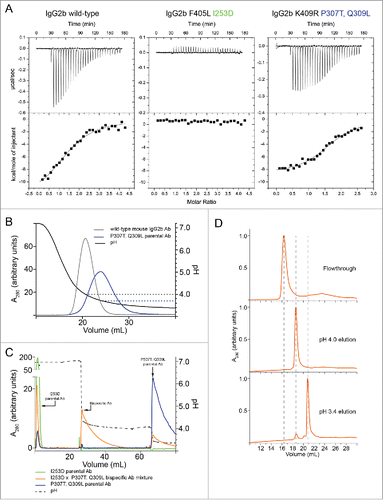
Table 1. Binding constants and stoichiometry of Z-domain binding to IgG variants.
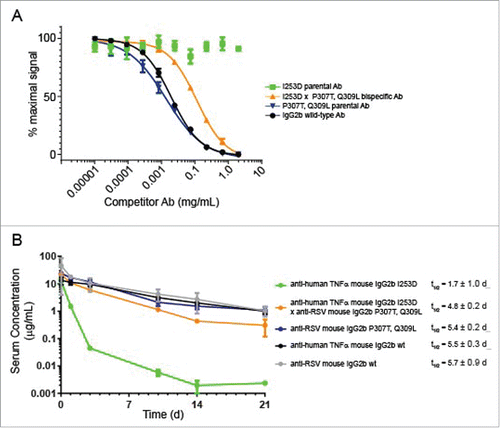
Figure 2. Analysis of FcRn interaction and PK analysis. (A) Competition binding of mouse IgG2b variants with wild-type IgG2b for FcRn using AlphaScreen assay. The graph displays % maximum signal plotted vs concentration of competitor. Mouse IgG2b wild-type compares the ability of the Ab to compete with itself as a control. (B) Pharmacokinetic analysis of mouse IgG2b variants in C57BL/6 mice. The graph displays the concentration of each Ab vs time after injection. Each point represented the mean ± standard error of 6 animals per group. Antibody specificities are indicated on the plot.
The mutations proline 307 to threonine (P307T) and glutamine 309 to leucine (Q309L) increased the affinity of mouse IgG2b for Z-domain by ∼8-fold relative to wild-type mouse IgG2b and provided a modest affinity increase for mouse IgG2a (, Supplementary Fig. 2b, ). The difference in affinity of the mutants for Z-domain resulted in elution from immobilized protein A resin at a lower pH than a wild-type mouse Ab at the same ionic strength (). We hypothesized that if the difference in elution pH for mouse Abs possessing the I253D or the P307T, Q309L mutations was large, then a mouse BsAb, wherein each heavy chain subunit harbored one of the mutation sets would elute at an intermediate pH value. Thus, the mouse BsAb could be purified from contaminating parental mouse mAbs using a pH step-gradient.
Purification of mutant bispecific antibodies
We tested whether differences in affinity would result in different pH values at which the mouse IgG2a and IgG2b variants could be eluted from protein A resin. Note that the affinity resin (mAbSelect SuRe, GE Healthcare), which comprised tandem Z-domain moieties, has a higher affinity for the Fc than native protein A and does not bind the Fab region.Citation20 The binding between mouse IgG and Z-domain peptide was confirmed to be solely within the Fc since Z-domain bound to the Fc (containing the P307T, Q309L mutations) alone with an affinity similar to that of the full-length mouse mAb (, Supplementary Fig. 2c). Wild-type mouse IgG2b eluted at pH 4.4 from the resin (). As expected, the mouse IgG2b I253D mutant did not bind the resin, and eluted in the flowthrough at pH 7.2, while the P307T, Q309L mutant eluted only at pH 3.4. The large difference in elution pH between the two mutants suggested that a mouse BsAb, with each heavy chain subunit harboring one of these two mutation sets, would elute at an intermediate pH, allowing its purification from the mutant parental mouse mAbs.
To test this hypothesis, we first prepared a mouse IgG2b BsAb using the cFAE method.Citation8 To test whether the BsAb could be efficiently purified by differential protein A elution, the BsAb solution was mixed with the two parental mAbs at an equimolar ratio. This mixture of parental and bispecific mouse Abs were resolved by a multiple pH step elution (). The composition of each elution step was analyzed by hydrophobic interaction chromatography (HIC), which showed that the I253D Abs could be separated from the P307T, Q309L Abs based on the different hydrophobicity of their Fab regions (). The flowthrough was found to contain the I253D parental mAb, while the pH 4.0 eluent contained only BsAb, and the pH 3.4 eluent contained almost exclusively (> 95%) parental mAb harboring the P307T, Q309L mutation.
We introduced the mutations into a mouse IgG2a framework, and analyzed them in the same way. Mutation of T370K further improved yields of mouse BsAb during cFAE when combined with K409R, so this mutation was included for the mouse IgG2a studies (data not shown). As expected, mouse IgG2a harboring the I253D mutation did not bind Z-domain and thus flowed through the affinity column during a wash step (Supplementary Fig. 3a). Surprisingly, although mouse IgG2a P307T, Q309L bound Z-domain more tightly than the corresponding mouse IgG2b variant at pH 7.2 (KD ∼ 126 nM vs 468 nM), this mouse IgG2a variant eluted at higher pH values (4.4 vs 3.4). This was also true of the wild-type mouse molecules (). However, the introduction of the P307T, Q309L mutations provided a modest improvement in affinity and decreased the elution pH of mouse IgG2a Abs, allowing these molecules to be eluted at lower pH values. An equimolar mixture of an I253D parental mouse mAb, a P307T, Q309L parental mouse mAb, and the resultant mouse BsAb (∼ 33% purity of each species), was separated using a 3-step pH gradient (pH 7.2, 4.9, and 3.4), allowing the mouse BsAb to be purified to ∼ 82% purity (Supplementary Fig. 3), based on HIC analysis.
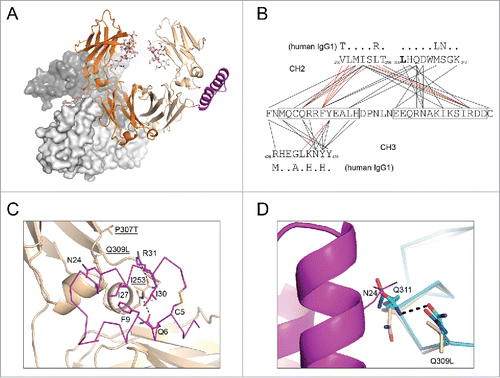
Figure 3. Structural basis of altered protein A binding by IgG2b variants. (A) Fc and Z34C are shown in cartoon with I253D-containing chain colored orange; P307T, Q309L-containing chain colored wheat; Z34C colored magenta. N297 linked glycans are shown in stick (light grey). The crystallographically related Fc dimer that blocks the equivalent Z34C binding site on the I235D chain is shown in light and dark grey surface. (B) Contact map for Z34C bound to heterodimeric Fc. Lines drawn between Z34C sequence (middle) and sequences from mouse IgG2b CH2 (top) and CH3 (bottom) domains indicate interatomic contacts (5 Å cutoff). Red lines indicate contacts that involve only Fc main chain atoms. For comparison, the human IgG1 sequence is shown above and below CH2 and CH3 domain sequences, respectively, with dots indicating sequence identity. (C) P307T, Q309L -containing chain of heterodimeric Fc shown in cartoon and colored wheat. Residues P307T, Q309L, and I253 was shown in stick (labeled, underlined). Z34C (magenta) was shown in ribbon with select residues shown in stick, and those within 5 Å of Fc residues I253 and Q309L were labeled. (d) CH2 domains (ribbon) from chain A (blue) and chain B (cyan) of PBD 2RGS are shown aligned that of the P307T, Q309L chain of mouse IgG2b from the present work. The side chain of N24 (line) from Z34C (pink cartoon) as well as the side chains of residues at positions 309 and 311 (stick) of the CH2 domains are also shown.
We also tested the purification of a muIgG2b BsAb formed in culture supernatants. A mixture of culture supernatants containing approximately equimolar amounts of parental mAbs with either I253D or P307T, Q309L mutations was prepared and subjected to in-supernatant cFAE as described.Citation13 The mixture was then purified by protein A-affinity chromatography using a step gradient, and the fractions were analyzed by HIC (Supplementary Fig. 4). The intermediate pH elution contained > 95% pure mouse BsAb while the low pH elution (pH 3.4) contained < 3% residual BsAb, having ∼ 97% P307T, Q309L parental Ab. This result showed that the differential protein A purification method was also amenable to purification from in-supernatant cFAE-derived material.
Analysis of Fc receptor interaction and serum half-life
To assay the extent to which the mutations affected FcRn binding, we used two in vitro methods. In an AlphaScreen competition-based assay (Perkin Elmer), the binding of a labeled mouse IgG to labeled mouse FcRn results in a luminescence signal. The binding was competed using unlabeled wild-type Ab or different mutant Abs, resulting in a dose-dependent decrease in signal (). We also measured the relative on- and off-rates of Fc receptor binding by surface bilayer interferometry (Supplementary Fig. 5). At pH 6.0, the mutant mouse IgG2b parental Ab harboring P307T, Q309L on both heavy chain subunits competed as well as wild-type mouse IgG2b for FcRn, indicating that this mutation had no effect on FcRn binding (concentration at half-maximal signal of ∼ 0.03 mg/mL) (). Conversely, the mutant containing I253D on both heavy chain subunits could not compete for FcRn binding up to 2 mg/mL, consistent with previous reports.Citation33 Interestingly, the mouse BsAb in which one heavy chain subunit contained I253D while the other heavy chain subunit contained P307T, Q309L could compete for FcRn with a half-maximal signal at 0.1 mg/mL. This suggested that while this molecule was impaired for FcRn binding, the activity could be rescued to some extent by having a single functional Fc subunit.
Using surface bilayer interferometry, we confirmed the effects of the mutations on the abilities of the IgG2b variants to bind FcRn (Supplementary Fig. 5a). The results, consistent with those from AlphaScreen, showed the IgG2b I253D parental Ab displayed almost no binding to FcRn. This was also true for IgG2a I253D parental Ab. For IgG2b, the P307T, Q309L parental Ab bound FcRn equally as well as wild-type IgG2b, whereas the analogous mutant in IgG2a displayed somewhat tighter binding to FcRn, having a relative kon ∼ 2.7-fold faster compared to wild-type IgG2a. FcRn binds to the CH2-CH3 interface of the Fc, whereas other Fcγ receptors bind to the upper CH2 region, Citation39 suggesting that the mutations would have little effect on binding. To confirm this, we analyzed the abilities of the mutants to bind mouse FcγRI, FcγRIIb, FcγRIII, and FcγRIV. All the mutants displayed on- and off-rates of binding within two-fold of the wild-type antibodies, except in the case of IgG2b binding to FcγRI. In this case, the I253D appeared to confer weak binding to the receptor whereas wild-type IgG2b had very low binding to FcγRI. The mechanism by which the I253D point mutation conferred slightly enhanced binding to FcγRI was unknown, since this mutation had no apparent effect on the ability of IgG2a to bind FcγRI. Overall, the mutations had little effect on Fcγ receptor interaction, suggesting that they would have similar effector functions in vivo.
We then examined how in vitro binding to FcRn would manifest with respect to the serum half-life of mouse IgG2b variants. A pharmacokinetic (PK) study was performed to evaluate the effect of the two sets of mutations (I253D or P307T, Q309L) on the serum half-lives of mouse IgG2b and IgG2a Abs harboring the mutation on both heavy chains and on a BsAb harboring one set of mutations on each heavy chain subunit. C57Bl/6 mice were injected intravenously with a single dose of the antibody, and serum levels of each Ab were measured by ELISA over 21 d (, , Supplementary Fig. 3C). After ∼ 1 h, the clearance rates of all Abs were approximately linear. The I253D parental Ab was cleared from serum rapidly– having a serum half-life of approximately 1.7 d or 0.5 d for the IgG2b or IgG2a isotype, respectively. In the case of the IgG2b isotype, the Ab harboring the P307T, Q309L mutations had a serum half-life of 5.4 days, which was similar to that of the two wild-type parental Abs, which had serum half-lives of 5.5 or 5.7 days (). The BsAb harboring the I253D mutation on one arm and the P307T, Q309L mutations on the other arm, had a half-life of 4.8 days, which was intermediate between the half-lives of its two parental Abs, but was closer to a wild-type half-life than it was to the I253D parental Ab. Analogous to IgG2b Abs, the IgG2a BsAb had a half-life value (t/12 = 8.8 d) that was intermediate between that of the two parental Abs (∼ 14.5 or 9.3 d), but closer to the wild-type Ab (Supplementary ). This result was consistent with previous observations that antibodies require only a single Fc subunit competent for FcRn binding to retain a long serum half-life.Citation40 Consistently, the BsAbs had maximum drug concentration or Cmax and area under the curve (AUC) values similar to their wild-type parental Abs (). Together, these results show that mouse IgG2a and IgG2b BsAbs harboring the I253D x P307T, Q309L mutations are suitable for in vivo studies, since they have half-live values comparable to that of wild-type Abs. However, they are cleared slightly faster, likely due to having only a single Fc subunit capable of binding FcRn.
Table 2. Pharmacokinetics properties of IgG variants.
Crystal structure of Z34C bound to heterodimeric mouse IgG2b Fc
In an effort to determine the structural basis by which the I253D and P307T, Q309L mutations affected binding to protein A, we determined the crystal structure to 2.7 Å resolution of a heterodimeric Fc harboring the two mutation sets on opposing chains in complex with Z34C, Citation37 a peptide analog of the Z-domain. (, Supplementary ). While Z34C lacked the third helix contained in the canonical Z-domain, it bound mouse IgG2b with similar affinity (KD = 2.6 μM vs 3.6 μM) (Supplementary Fig. 6a). Although the peptide was added in greater than two-fold excess of Fc dimer, the asymmetric unit contained a single copy of the heterodimeric Fc with Z34C bound to only to the chain harboring the P307T, Q309L mutations. The equivalent binding site on the opposite chain was blocked by a symmetry-related Fc molecule (). The electron density maps confirmed that the subunit bound by Z34C contained the P307T and Q309L mutations (Supplementary Fig. 6b), consistent with the enhanced affinity of these mutants for protein A. The electron densities for the side chain for the I253D mutation as well as the Z34C peptide were also clearly resolved (Supplementary Fig. 6c, d).
The mode of binding of Z34C at the CH2-CH3 interface of the P307T, Q309L-containing chain was similar to that observed for its interaction with human IgG1 Fc (PDB 1L6X; Supplementary Fig. 7). Interactions involving the C-terminal helix of Z34C were limited to the CH2 domain, whereas the N-terminal helix interacted with both CH2 and CH3 domains (). The side chain of P307T adopted a different rotamer than what was observed in the structure of the human IgG1 complex. In the case of the latter, the side chain of T307 formed a hydrogen bond with the side chain of N286 from the CH2 domain, which was a threonine in mouse IgG2b (Supplementary Fig. 7b). Neither T307 in the human IgG1 structure nor P307T in the present mouse IgG2b structure were within contact distance of bound Z34C peptide. In the case of the latter, the distance of closest approach was 5.7 Å between P307T atom Cγ2 and the Nη2 atom of R31 in Z34C. Similarly, interactions of Q309L with Z34C were limited, with only the Cδ1 atom of Q309L falling within 5 Å of the sidechain of Z34C residue N24 (). In the P307T, Q309L chain, I253 was positioned on a hydrophobic surface of the Z34C peptide packing between the phenyl ring of F9 and the hydrophobic portion of the R31 sidechain.
P307T and Q309L mutations were not found to result in any large backbone perturbations relative to wild-type mouse IgG2b (PDB 2RGS; Supplementary Fig. 8). Alignment of extended main chain atoms of the CH2 domain from the P307T, Q309L-containing chain with the CH2 domain from chain A in PDB 2RGS resulted in a positional root mean square deviation (r.m.s.d.) over the same atoms of 0.8 Å. Based on this alignment, the short stretch of sequence from T305 to Q311 encompassing the sites of mutation had an r.m.s.d. of 0.5 Å. The most notable change in structure in the vicinity of the bound peptide relative to wild type mouse IgG2b Fc region (PDB 2RGS) was an altered side chain conformation for Q311 that was required to avoid a clash with Z34C residue N24 (). The I253D mutation on the opposing Fc chain was also found to result in no significant change in backbone conformation relative to wild-type mouse IgG2b (PDB 2RGS; Supplementary Fig. 8). Alignment of extended main chain atoms of the I253D-containing CH2 domain with the CH2 domain from chain A in PDB 2RGS resulted in an r.m.s.d. of 0.94 Å, and based on this alignment, for the stretch of sequence from residue V250 to residue T256, an extended main chain r.m.s.d. of 0.8 Å is calculated.
Discussion
Multispecific Abs are an increasingly important part of the biological therapeutic repertoire, but they require more effort than mAbs to produce with high yield and purity. The presence of contaminating Ab fragments or parental Abs in differing concentrations can result in irreproducible functional responses that are required for hit selection and validation. The results of this work led to a method that can significantly accelerate the production of highly pure mouse BsAbs. We identified two sets of mutations in the CH2 domain (I253D and P307T, Q309L) that can be incorporated into a set of parental mouse IgG2a or IgG2b molecules for cFAE to allow the formation of mouse BsAb that could be simultaneously purified, re-oxidized, and purified using differential protein A purification from mixed supernatants of parental mAb expression cultures. Conceptually, similar methods exist for human IgG1 BsAbs, wherein one parental Ab contains the H435R, Y436F mutations in the CH3 domain that completely abolish protein A binding.Citation34,41 Thus, the formation of a BsAb using the aforementioned construct with the wild-type human IgG1 Ab that binds to protein A with high affinity requires no additional mutations to allow efficient purification of the BsAb. Nonetheless, we have developed a method specifically for mouse IgG2a and IgG2b BsAbs that allows higher throughput purification of the BsAb with little effects on Fc receptor interactions and that should be generally applicable to any mouse Ab subtypes.
One caveat to this method was that the BsAb must be eluted at a pH value just below that at which the molecule was retained on the resin. As a result, greater elution volumes were required to obtain high yields. Nevertheless, the resulting highly pure mouse BsAb molecules retained a serum half-life comparable to wild-type mouse Abs, suggesting that the mutant mouse BsAbs prepared in this way were amenable for in vivo studies.
Mouse surrogate models play a critical role in research and development. As a result of their different affinities for Fcγ receptors, mouse IgG isotypes and subtypes vary significantly in their abilities to modulate effector functions such as ADCC and ADCP. Mouse IgG2a and IgG2b with active Fc regions are commonly used for surrogate studies requiring mouse immune cell activity. The mutations introduced here were unlikely to affect these functions because interactions with Fcγ receptors occurred mainly through the regions of the CH2 domains closest to the hinge region.
While the PK results correlated with in vitro FcRn binding results, the extent to which the effects of the I253D mutation could be rescued by pairing with a P307T, Q309L heavy chain subunit was unexpected. In particular, the presence of a single FcRn-binding subunit was sufficient to increase the serum half-life from ∼1 to 5 days. However, serum, half-life can also be influenced by Fab identity, Citation42, 43 and indeed the Fab contained in the P307T, Q309L parental may have conferred a modestly extended half-life for IgG2a Abs (). However, in this example, the effect of Fab identity was less significant than the effects of direct binding site mutations, supporting the suggestion that these bispecific mutants are suitable for in vivo studies.
We determined the crystal structure of a heterodimeric mouse IgG2b mutant Fc harboring the I253D mutation in one chain and P307T and Q309L mutations in the other in complex with the protein A analog peptide Z34C. Ten of 15 residues having side chains involved in contacts with the Fc were identical between Z34C and the Z-domain construct used in ITC studies.Citation35,44 For two of the five positions that differed from Z-domain, only a single contact involving an atom of the side chain and the Fc was observed. The Cβ atom of Z34C residue C5 was positioned 4.6 Å from the Cγ2 atom of Fc residue I253, and the Cγ2 of Z34C residue I30 was positioned 4.8 Å from the Cγ1 of the same Fc residue. The side chain of Z34C residue R7, an asparagine in Z-domain, made contacts with a single residue (N434) in the CH3 domain. Contacts involving the side chain of M3, a lysine in Z-domain, were similarly limited to N434 and Y436 in the CH3 domain. The fifth position of difference, R31 in Z34C, was conserved as a lysine in the Z-domain. The residues observed to contribute most significantly to the interaction between Z34C and Fc are identical in Z-domain, consistent with the similar affinity with which Z34C and Z-domain bound to mouse IgG2b. Since crystals of IgG2b in complex with Z-domain diffracted to only 3.9 Å (data not shown), the effects of the I253D and P307T/Q309L sets of mutations were considered in light of the structure of the Fc-Z34C complex.
The structure suggested how the described mutations could alter binding to Z-domain. As previously stated, I253 present in the P307T, Q309L chain of the heterodimeric mIgG2b Fc was observed to pack between the phenyl ring of F9 and the hydrophobic portion of the R31 sidechain. I253 adopted an identical conformation and similarly engaged Z34C in the structure of the human IgG1 complex (Supplementary Fig. 6c). Therefore, the introduction of a negative charge upon mutating this residue to aspartate resulted in reduced affinity by perturbing this hydrophobic interaction. However, the mechanism by which the P307T and Q309L mutations enhanced the affinity for Z-domain could be more complex. These mutations did not significantly perturb the backbone conformation of the CH2 domain, and direct interactions with Z34C were limited. Furthermore, upon structural alignment of the CH2 domain of wild-type mouse IgG2b to the P307T, Q309L chain of the present structure, not only should the conformation of Q309 present in structure 2RGS be sterically permitted, but it would also be well-positioned to engage in a hydrogen bond with the side chain of Z34C residue N24. Rather this result could be explained by the observed conformational difference for the Q311 side chain between the wild-type and mutant structures. In one of two chains present in 2RGS, the side chains of Q309 and Q311 were positioned at a distance suitable for a hydrogen bond; in the second chain, χ angles of Q311 were only slightly altered (). In the structure of the mutant Fc bound to Z34C, the side chain of Q311 was flipped, a change necessary to avoid a clash with Z34C residue N24 (). Although unlikely to account fully for the near order of magnitude enhancement in affinity for Z-domain of the P307T, Q309L mouse IgG2b mutant relative to wild-type, it was possible that the Q309L mutation facilitated the conformational adjustment of Q311 necessary for Z34C binding by weakening the interaction between the two residues. Also, P307T, Q309L, or both mutations could exert a subtle effect on backbone flexibility that allowed the EF loop to more easily accommodate binding to peptide, an effect that would not be discernible in a static crystal structure of the Fc region bound to the peptide.
Further work will be required to expand this strategy into other species and IgG isotypes and subtypes since the affinity of the wild-type parental mouse Ab for protein A plays a large part in the ease with which a mutant can be separated based on this technique. However, these considerable efforts may be worthwhile due to the large number of BsAbs in development and the routine use of protein A-based resins for their affinity purification. Likewise, it is possible to use such mutations to prepare bispecific or multispecific Fc fusion molecules.
Materials and methods
Proteins
Mouse IgG2a and IgG2b variants were generated from an anti-human tumor necrosis factor (TNF) mAb (F405L heavy chain subunit molecules) or an anti-respiratory syncytial virus (RSV) mAb (K409R heavy chain subunit molecules). EU numbering of IgG was used throughout this report. Mouse IgG2a and IgG2b heavy chain plasmids were co-transfected with light chain plasmids at a 1:3 molar ratio. Mutations were generated by gene synthesis (Genewiz, Inc.). Proteins were expressed in Expi293 cells (Invitrogen, Inc.) according to the manufacturer's protocol and purified by affinity chromatography using protein G or Ni-NTA resin. BsAbs were produced using the controlled Fab-arm exchange (cFAE) method, according to previously described methods.Citation8 Proteins were confirmed by SDS-PAGE and size-exclusion chromatography for purity. The extent of bispecific formation was monitored by analytical HIC using a butyl-NPR column (Tosoh Bioscience LLC). Proteins were eluted using a gradient of 20 mM sodium phosphate pH 6.0 containing 1.5 M to 0 M (NH4)2SO4 to quantify the relative population of each species. The Z-domain peptide from staphylococcal protein A having the sequence: VDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAEAKKLNDAQAPK and the Z34C peptide having the sequence: FNMQCQRRFYEALHDPNLNEEQRNAKIKSIRDDC with a disulfide bridge formed between the side chains of C5 and C34 were synthesized (New England Peptide, Inc.) and verified by LC-MS. The concentration of Z-domain was determined using the extinction coefficient at 280 nm of 1490 M−1 cm−1. All proteins were stored at 4 °C in phosphate-buffered saline pH 7.2 (PBS), unless noted. Z34C used for crystallography was reconstituted from lyophilized powder in 50 mM NaCl, 20 mM HEPES, pH 7.5 and stored at −20°C.
Protein A affinity chromatography
To determine the approximate pH at which each IgG molecule eluted from protein A resin, 1 mg of each sample was loaded onto a mAbSelect SuRe column, composed of immobilized tandem protein A-derived Z-domains (GE Healthcare Life Sciences 11-0034-93) in PBS. Proteins were first eluted using a linear gradient from PBS pH 7.2 to 50 mM citrate pH 3.0 over 30 column volumes. For purification, proteins were eluted in a two-step gradient using 50 mM citrate buffer pH 4.0 and 3.4 for IgG2b and pH 4.9 and 3.4 for IgG2a. Elution fractions were assayed by HIC
Isothermal titration calorimetry
ITC experiments were performed at 25 °C using a MicroCal VP-ITC calorimeter. Both titrant and injectant were dialyzed in PBS. All titrations were performed analogously, having 20 μM IgG in the cell and 200 μM of Z-domain in the syringe. Each 10 μL injection of Z-domain peptide spanned 20 s, with 5 min spacing between injections. Data were analyzed using Origin software using a 1:1 binding model.
AlphaScreen
Abs were biotinylated using the SureLINK Chromophoric Biotin Labeling kit (KPL Inc.), according to the manufacturer's protocol. His-tagged mouse FcRn was purchased from Sino Biological. Assays were performed in PBS pH 6.0, supplemented with 0.05% (w/v) bovine serum albumin (BSA) and 0.01% (v/v) Tween-20. Note that while BSA can also bind FcRn, this concentration was ∼ 0.0005 mg/mL, which was low compared to the concentration of IgG, and binding between BSA and FcRn is non-competitive with IgG.Citation45 Biotinylated wild-type mouse IgG2b at 1 μg/mL was bound to streptavidin-conjugated donor beads, and His-tagged FcRn at 0.2 μg/mL was bound to nickel-conjugated acceptor beads. Competitor Abs were prepared at 2 mg/mL and were serially diluted by 3-fold for each point. Luminescence between 520–620 nm was recorded using an EnVision plate reader (Perkin Elmer). Data were analyzed using Prism 6.01 software (GraphPad Software, Inc.) and fitted using a 4-parameter competition model, as described previously.Citation46
Pharmacokinetics studies
Abs were targeted against either anti-respiratory syncytial virus (RSV) F-glycoprotein (P307T, Q309L mutants and parental Abs), or anti-human TNF (I253D mutants and parental Abs) so that there would be no target-mediated internalization, which would confound the PK analysis. The test Abs were injected into C57BL/6 mice intravenously via tail vein at a dose of 10 mg/kg with 6 animals per group. Two sets of animals were used for each test Ab and time points were taken at 30 min, 1 d, 7 d, 21 d and at 1 h, 3 d, 14 d, and 21 d. Serial retro-orbital bleeds were obtained from CO2-anesthesized mice at the indicated time points and terminal bleeds were taken by cardiac puncture. After 30 min at room temperature, blood samples were centrifuged at 3,000 X g for 15 min and serum collected for analyses. The PK study was approved by the Institutional Animal Care and Use Committee at Janssen Research & Development, LLC. For detection of the mouse test Abs in mouse sera, an electrochemiluminescent immunoassay was used. Streptavidin Gold multi-array 96-well plates (Meso Scale Discovery) were coated for 2 h on a shaker with 50 µL/well of 2 µg/mL of recombinant TNF (R&D systems) for anti-TNF Abs or recombinant RSV-F glycoprotein (Sino Biological) for anti-RSV Abs. Plates were then washed with Tris-buffered saline supplemented with 0.05% (v/v) Tween-20. Serum samples and standards were diluted in 1 X PBS supplemented with 2% (w/v) BSA, 0.5% (v/v) Tween-20, added to plates and incubated for 2 hours on a shaker at room temperature. Bound Ab was detected using a ruthenium-labeled goat anti-mouse IgG, Fc specific (Mesoscale Discovery cat. # R32AC) at 2 µg/mL for 2 h on a shaker. Plates were washed, 200 µL Read Buffer T (Meso Scale Discovery) was added per well, and plates were read on the MSD Sector Imager 6000. Serum concentrations of the Abs were determined from a standard curve using a 5-parameter non-linear regression program in Prism 7 software. For the estimation of terminal Ab half-life, a non-compartmental model was used and calculated as: t1/2 = 0.693 / slope.Citation47 The terminal half-life value for an Ab was determined by taking the average of the t1/2 values calculated for each animal within the test group.
Crystallization and structure determination
Z34C peptide was added to recombinant heterodimeric Fc at a 2.2:1 (Fc dimer) molar ratio. Following a 5 h incubation at room temperature, the sample was concentrated to 13.7 mg/mL in buffer containing 50 mM NaCl, 20 mM Tris, pH 8.0. Crystallization experiments employing the sitting drop vapor diffusion method were performed at room temperature using a Mosquito liquid handling robot (TTP Labtech, Ltd). 300 nL drops comprising protein stock and reservoir solution mixed at a 1:1 (v/v) ratio were set up in Corning 3550 plates. Diffraction quality crystals were grown at 20°C and were obtained with a reservoir solution comprising 17% (w/v) PEG 3350, 0.2 M LiCl, 0.1 M Tris, pH 8.0. Crystals were cryo-protected with reservoir solution supplemented with 20% (v/v) glycerol and flash frozen in liquid nitrogen prior to data collection. Diffraction data were collected at the Advanced Photon Source IMCA-CAT beamline 17-ID equipped with a Dectris Pilatus 6M pixel array detector and processed to 2.7 Å with the program XDS.Citation48 Initial phases were determined by molecular replacement with the program Phaser Citation49 using a crystal structure of mouse IgG2b Fc (PDB ID 2RGS Citation50) as a search model. The structure of Z34C from PDB 1L6X Citation38 was subsequently manually docked into available 2Fo-Fc and Fo-Fc electron density followed by additional rounds of rebuilding and refinement using the programs Coot Citation51 and Phenix, Citation52 respectively. Data collection and final refinement statistics are given in . Analysis of contacts was performed using the program NCONT as implemented in the CCP4 suite of crystallographic programs Citation53 using a cutoff distance of 5 Å. For comparison to other structures, alignments with performed with either the program LSQMANCitation54 or PyMol (www.schrodinger.com/pymol). Figures were generated with PyMol.
Table 3. Crystal data, X-ray data, and refinement statistics.
Accession codes
The atomic coordinates and structure factors have been deposited in the Protein Data Bank with accession code 5UBX.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Suppl_mat.docx
Download MS Word (1.7 MB)Acknowledgments
We thank Dr. Bernie Scallon (Janssen R&D) for helpful discussions during this work. The authors also acknowledge the staff of the Industrial Macromolecular Crystallography Association Collaborative Access Team (IMCA-CAT) beamline 17-ID at the Advanced Photon Source (APS) of Argonne National Laboratory. We thank Jan Reichert, Aran Labrijn, Janine Schuurman, and Paul Parren for helpful comments to this manuscript.
References
- Jachimowicz RD, Borchmann S, Rothe A. Multi-specific antibodies for cancer immunotherapy. BioDrugs: Clinical Immunotherapeutics, Biopharmaceuticals And Gene Therapy. 2014;28:331-343. doi:10.1007/s40259-014-0091-4. PMID:24638872
- Byrne H, Conroy PJ, Whisstock JC, O'Kennedy RJ. A tale of two specificities: bispecific antibodies for therapeutic and diagnostic applications. Trends in biotechnology. 2013;31:621-632. doi:10.1016/j.tibtech.2013.08.007. PMID:24094861
- Thakur A, Lum LG. “NextGen” Biologics: Bispecific Antibodies and Emerging Clinical Results. Expert opinion on biological therapy. 2016;16:675-688. doi:10.1517/14712598.2016.1150996. PMID:26848610
- Spiess C, Zhai Q, Carter PJ. Alternative molecular formats and therapeutic applications for bispecific antibodies. Molecular immunology. 2015;67:95-106. doi:10.1016/j.molimm.2015.01.003. PMID:25637431
- Weidle UH, Kontermann RE, Brinkmann U. Tumor-antigen-binding bispecific antibodies for cancer treatment. Seminars in oncology. 2014;41:653-660. doi:10.1053/j.seminoncol.2014.08.004. PMID:25440609
- Ridgway JB, Presta, LG, Carter, P. ‘Knobs-into-holes’ engineering of antibody CH3 domains for heavy chain heterodimerization. Protein Eng. 1996;9:617-621. doi:10.1093/protein/9.7.617. PMID:8844834
- Gunasekaran K, Pentony M, Shen M, Garrett L, Forte C, Woodward A, Ng SB, Born T, Retter M, Manchulenko K, et al. Enhancing antibody Fc heterodimer formation through electrostatic steering effects: applications to bispecific molecules and monovalent IgG. J Biol Chem. 2010;285:19637-19646. doi:10.1074/jbc.M110.117382
- Labrijn AF, Meesters JI, de Goeij BE, van den Bremer ET, Neijssen J, van Kampen MD, Strumane K, Verploegen S, Kundu A, Gramer MJ, et al. Efficient generation of stable bispecific IgG1 by controlled Fab-arm exchange. Proc Natl Acad Sci U S A. 2013;110:5145-5150. doi:10.1073/pnas.1220145110. PMID:23479652
- Gramer MJ, van den Bremer ET, van Kampen MD, Kundu A, Kopfmann P, Etter E, Stinehelfer D, Long J, Lannom T, Noordergraaf EH, et al. Production of stable bispecific IgG1 by controlled Fab-arm exchange: scalability from bench to large-scale manufacturing by application of standard approaches. MAbs. 2013;5:962-973. doi:10.4161/mabs.26233. PMID:23995617
- Strop P, Ho WH, Boustany LM, Abdiche YN, Lindquist KC, Farias SE, Rickert M, Appah CT, Pascua E, Radcliffe T, et al. Generating bispecific human IgG1 and IgG2 antibodies from any antibody pair. J Mol Biol. 2012;420:204-219. doi:10.1016/j.jmb.2012.04.020. PMID:22543237
- Schaefer W, Regula JT, Bähner M, Schanzer J, Croasdale R, Dürr H, Gassner C, Georges G, Kettenberger H, Imhof-Jung S, et al. Immunoglobulin domain crossover as a generic approach for the production of bispecific IgG antibodies. Proc Natl Acad Sci U S A. 2011;108:11187-11192. doi:10.1073/pnas.1019002108
- Lewis SM, Wu X, Pustilnik A, Sereno A, Huang F, Rick HL, Guntas G, Leaver-Fay A, Smith EM, Ho C, et al. Generation of bispecific IgG antibodies by structure-based design of an orthogonal Fab interface. Nature biotechnology. 2014;32:191-198. doi:10.1038/nbt.2797
- Paul S, Connor J, Nesspor T, Haytko P, Boakye K, Chiu ML, Jiang H. An efficient process of generating bispecific antibodies via controlled Fab-arm exchange using culture supernatants. Protein Expr Purif. 2016;121:133-140. doi:10.1016/j.pep.2016.01.014. PMID:26826313
- Bruhns, P. Properties of mouse and human IgG receptors and their contribution to disease models. Blood. 2012;119:5640-5649. doi:10.1182/blood-2012-01-380121
- Labrijn AF, Meesters JI, Bunce M, Armstrong AA, Somani S, Nesspor TC, Chiu ML, Altintaş I, Verploegen S, Schuurman J, et al. Efficient Generation of Bispecific Murine Antibodies for Pre-Clinical Investigations in Syngeneic Rodent Models. Sci Rep. 2017;7:2476. doi:10.1038/s41598-017-02823-9
- Couch JA, Yu YJ, Zhang Y, Tarrant JM, Fuji RN, Meilandt WJ, Solanoy H, Tong RK, Hoyte K, Luk W, et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood-brain barrier. Sci Transl Med. 2013;5(183ra157):181-112. PMID:23636093
- Lo M, Kim HS, Tong RK, Bainbridge TW, Vernes JM, Zhang Y, Lin YL, Chung S, Dennis MS, Zuchero YJ, et al. Effector-attenuating Substitutions That Maintain Antibody Stability and Reduce Toxicity in Mice. J Biol Chem. 2017;292:3900-3908. doi:10.1074/jbc.M116.767749
- Nilsson B, Moks T, Jansson B, Abrahmsén L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlén M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107-113. doi:10.1093/protein/1.2.107. PMID:3507693
- Linhult M, Gülich S, Gräslund T, Simon A, Karlsson M, Sjöberg A, Nord K, Hober S. Improving the tolerance of a protein a analogue to repeated alkaline exposures using a bypass mutagenesis approach. Proteins. 2004;55:407-416. doi:10.1002/prot.10616. PMID:15048831
- Jansson B, Uhlen M, Nygren, PA. All individual domains of staphylococcal protein A show Fab binding. FEMS Immunol Med Microbiol. 1998;20:69-78. doi:10.1016/S0928-8244(97)00108-9. PMID:9514577
- Boot JH, Geerts, ME, Aarden LA. Monoclonal anti-peroxidase isotype switch variants. Applications in studies of protein A binding and characterization of rat monoclonal antibodies. J Immunol Methods. 1987;103:69-77
- Ey PL, Prowse SJ, Jenkin CR. Isolation of pure IgG1, IgG2a and IgG2b immunoglobulins from mouse serum using protein A-sepharose. Immunochemistry 1978;15:429-436. doi:10.1016/0161-5890(78)90070-6. PMID:30693
- DeLano WL, Ultsch MH, de Vos AM, Wells JA. Convergent solutions to binding at a protein-protein interface. Science 2000;287:1279-1283. doi:10.1126/science.287.5456.1279. PMID:10678837
- Hinton PR, Xiong JM, Johlfs MG, Tang MT, Keller S, Tsurushita N. An engineered human IgG1 antibody with longer serum half-life. J Immunol. 2006;176:346-356. doi:10.4049/jimmunol.176.1.346. PMID:16365427
- Dall'Acqua WF1, Woods RM, Ward ES, Palaszynski SR, Patel NK, Brewah YA, Wu H, Kiener PA, Langermann S. Increasing the affinity of a human IgG1 for the neonatal Fc receptor: biological consequences. J Immunol. 2002;169:5171-5180
- Vaccaro C, Zhou J, Ober RJ, Ward ES. Engineering the Fc region of immunoglobulin G to modulate in vivo antibody levels. Nature Biotech. 2005;23:1283-1288. doi:10.1038/nbt1143
- Stapleton NM, Andersen JT, Stemerding AM, Bjarnarson SP, Verheul RC, Gerritsen J, Zhao Y, Kleijer M, Sandlie I, de Haas M, et al. Competition for FcRn-mediated transport gives rise to short half-life of human IgG3 and offers therapeutic potential. Nat Commun. 2011;2:599. doi:10.1038/ncomms1608
- Yeung YA, Leabman MK, Marvin JS, Qiu J, Adams CW, Lien S, Starovasnik MA, Lowman HB. Engineering human IgG1 affinity to human neonatal Fc receptor: impact of affinity improvement on pharmacokinetics in primates. J Immunol. 2009;182:7663-7671. doi:10.4049/jimmunol.0804182. PMID:19494290
- Hinton PR, Johlfs MG, Xiong JM, Hanestad K, Ong KC, Bullock C, Keller S, Tang MT, Tso JY, Vásquez M, et al. Engineered human IgG antibodies with longer serum half-lives in primates. J Bio Chem. 2004;279:6213-6216. doi:10.1074/jbc.C300470200
- Roopenian DC, Akilesh, S. FcRn: the neonatal Fc receptor comes of age. Nat Rev Immuno. 2007;7:715-725. doi:10.1038/nri2155. PMID:17703228
- Junghans RP. Finally! The Brambell receptor (FcRB). Mediator of transmission of immunity and protection from catabolism for IgG. Immunol Res. 1997;16:29-57. PMID:9048207
- Popov S, Hubbard JG, Kim J, Ober B, Ghetie V, Ward ES. The stoichiometry and affinity of the interaction of murine Fc fragments with the MHC class I-related receptor, FcRn. Mol Immunol. 1996;33:521-530. doi:10.1016/0161-5890(96)00004-1
- Martin WL, West AP, Jr., Gan L, Bjorkman PJ. Crystal structure at 2.8 A of an FcRn/heterodimeric Fc complex: mechanism of pH-dependent binding. Mol Cell. 2001;7:867-877. doi:10.1016/S1097-2765(01)00230-1. PMID:11336709
- Tustian AD, Endicott C, Adams B, Mattila J, Bak H. Development of purification processes for fully human bispecific antibodies based upon modification of protein A binding avidity. MAbs. 2016;8:828-838. doi:10.1080/19420862.2016.1160192. PMID:26963837
- Cedergren L, Andersson R, Jansson B, Uhlen M, Nilsson B. Mutational analysis of the interaction between staphylococcal protein A and human IgG1. Protein Eng. 1993;6:441-448. doi:10.1093/protein/6.4.441. PMID:8332602
- Braisted AC, Wells, JA. Minimizing a binding domain from protein A. Proc Natl Acad Sci U S A. 1996;93:5688-5692. doi:10.1073/pnas.93.12.5688. PMID:8650153
- Starovasnik MA, Braisted AC, Wells JA. Structural mimicry of a native protein by a minimized binding domain. Proc Natl Acad Sci U S A. 1997;94:10080-10085. doi:10.1073/pnas.94.19.10080. PMID:9294166
- Idusogie EE1, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, Meng YG, Mulkerrin MG. Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc. J Immunol. 2000;164:4178-4184
- Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcgamma receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14:94-108. doi:10.1038/nri3666
- Stracke J, Emrich T, Rueger P, Schlothauer T, Kling L, Knaupp A, Hertenberger H, Wolfert A, Spick C, Lau W, et al. A novel approach to investigate the effect of methionine oxidation on pharmacokinetic properties of therapeutic antibodies. MAbs. 2014;6:1229-1242. doi:10.4161/mabs.29601. PMID:25517308
- Smith EJ, Olson K, Haber LJ, Varghese B, Duramad P, Tustian AD, Oyejide A, Kirshner JR, Canova L, Menon J, et al. A novel, native-format bispecific antibody triggering T-cell killing of B-cells is robustly active in mouse tumor models and cynomolgus monkeys. Sci Rep. 2015;5:17943
- Igawa T, Tsunoda H, Tachibana T, Maeda A, Mimoto F, Moriyama C, Nanami M, Sekimori Y, Nabuchi Y, Aso Y, et al. Reduced elimination of IgG antibodies by engineering the variable region. Protein Eng Des Sel: PEDS. 2010;23:385-392. doi:10.1093/protein/gzq009
- Schoch A, Kettenberger H, Mundigl O, Winter G, Engert J, Heinrich J, Emrich T. Charge-mediated influence of the antibody variable domain on FcRn-dependent pharmacokinetics. Proc Natl Acad Sci U S A. 2015;112:5997-6002. doi:10.1073/pnas.1408766112
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 1981;20:2361-2370. doi:10.1021/bi00512a001. PMID:7236608
- Chaudhury C, Brooks CL, Carter DC, Robinson JM, Anderson CL. Albumin binding to FcRn: distinct from the FcRn-IgG interaction. Biochemistry. 2006;45:4983-4990. doi:10.1021/bi052628y. PMID:16605266
- Vafa O, Gilliland GL, Brezski RJ, Strake B, Wilkinson T, Lacy ER, Scallon B, Teplyakov A, Malia TJ, Strohl WR. An engineered Fc variant of an IgG eliminates all immune effector functions via structural perturbations. Methods. 2014;65:114-126. doi:10.1016/j.ymeth.2013.06.035. PMID:23872058
- Gustafson DL, Bradshaw-Pierce in Principles of Anticancer Drug Development. (eds. E. Garrett-Mayer, M. Hidalgo, S.G. Eckhardt & N.J. Clendeninn) (Springer, 2011)
- Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography. 2010;66:125-132
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658-674. doi:10.1107/S0021889807021206. PMID:19461840
- Kolenko P, Dohnálek J, Dusková J, Skálová T, Collard R, Hasek J. New insights into intra- and intermolecular interactions of immunoglobulins: crystal structure of mouse IgG2b-Fc at 2.1-A resolution. Immunology. 2009;126:378-385. doi:10.1111/j.1365-2567.2008.02904.x. PMID:18783468
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486-501. doi:10.1107/S0907444910007493. PMID:20383002
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213-221. doi:10.1107/S0907444909052925
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235-242. doi:10.1107/S0907444910045749
- Kleywegt GJ. Use of non-crystallographic symmetry in protein structure refinement. Acta Crystallogr D Biol Crystallogr. 1996;52:842-857. doi:10.1107/S0907444995016477
