ABSTRACT
Glycosylation is an important attribute of biopharmaceutical products to monitor from development through production. However, glycosylation analysis has traditionally been a time-consuming process with long sample preparation protocols and manual interpretation of the data. To address the challenges associated with glycan analysis, we developed a streamlined analytical solution that covers the entire process from sample preparation to data analysis. In this communication, we describe the complete analytical solution that begins with a simplified and fast N-linked glycan sample preparation protocol that can be completed in less than 1 hr. The sample preparation includes labelling with RapiFluor-MS tag to improve both fluorescence (FLR) and mass spectral (MS) sensitivities. Following HILIC-UPLC/FLR/MS analyses, the data are processed and a library search based on glucose units has been included to expedite the task of structural assignment. We then applied this total analytical solution to characterize the glycosylation of the NIST Reference Material mAb 8761. For this glycoprotein, we confidently identified 35 N-linked glycans and all three major classes, high mannose, complex, and hybrid, were present. The majority of the glycans were neutral and fucosylated; glycans featuring N-glycolylneuraminic acid and those with two galactoses connected via an α1,3-linkage were also identified.
Abbreviations
| AA | = | 2-aminoanthanilic acid |
| 2-AB | = | 2-aminobenzamide |
| ABS | = | Arthrobacter ureafaciens sialidase |
| ACN | = | Acetonitrile |
| ADCC | = | antibody-dependent cell-mediated cytotoxicity |
| BKF | = | bovine kidney fucosidase |
| BTG | = | bovine testis β-galactosidase |
| C | = | Celsius |
| CID | = | Collision-induced Dissociation |
| DDA | = | Data-directed Analysis |
| DIA | = | Data-independent acquisition |
| DMF | = | Dimethylformamide |
| ESI | = | Electrospray ionization |
| FA | = | Formic Acid |
| FLR | = | Fluorescence |
| Gal | = | Galactose |
| gal-α1,3-gal | = | Two galactoses linked via an α1-3 bond |
| GlcNAc | = | N-acetylglucosamine |
| GU | = | Glucose Unit |
| GUH | = | β-N-acetylhexosaminidase |
| HILIC | = | Hydrophilic Interaction Liquid Chromatography |
| hr | = | Hour |
| IgG | = | Immunoglobulin G |
| kV | = | kilovolt |
| M5 | = | Mannose-5 glycan |
| mAb | = | monoclonal antibody |
| MD | = | Mass Detector |
| min | = | Minute |
| mg | = | milligram |
| mM | = | millimolar |
| MS | = | Mass Spectrometry |
| MSE | = | A mode of data independent acquisition |
| NAN1 | = | recombinant sialidase |
| NHS | = | N-hydroxyuccinimidyl |
| ppm | = | parts-per-million |
| QTOF | = | Quadrupole Time-of-Flight |
| RFMS | = | RapiFluor-MS |
| RM | = | Reference Material |
| SDS | = | sodium dodecyl sulfate |
| SPE | = | Solid-phase extraction |
| μg | = | microgram |
| μL | = | microliter |
| UPLC | = | Ultra-performance Liquid Chromatography |
| UV | = | Ultraviolet |
| V | = | Volt |
Introduction
Over recent decades, substantial effort has been devoted to the development of monoclonal antibodies (mAbs) and other proteins, including enzyme replacements, fusion proteins, and clotting factors, as therapeutic agents. Many of these products are glycosylated, and the glycans attached to the protein influence its physicochemical characteristics. Thus, detailed analyses of glycans are routinely performed on biopharmaceutical products throughout their development and production to ensure proper function, efficacy, and safety, as well as for quality control and release purposes. Glycosylation is, therefore, one of the critical quality attributes that must be rigorously characterized and monitored. Among the glycosylation attributes that are monitored are the overall levels of the high mannose glycans, in particular, mannose-5 (M5), which may alter the pharmacokinetic properties of a mAb drug by increasing its serum clearance rate and increasing its antibody-dependent cell-mediated cytotoxicity (ADCC).Citation1 Other glycan attributes routinely monitored include non-human carbohydrate attributes that are associated with the production of therapeutic proteins in non-human cell lines, such as N-glycolylneuraminic acid and two galactoses connected through an α1,3 linkage (gal-α1,3-gal). Both of these epitopes can lead to immunogenic responses.Citation2-3 Additionally, the level of core fucosylation is frequently assessed because a decreased amount of this monosaccharide increases the ADCCCitation4-5 and can enhance the efficacy of the drug. Monitoring and characterizing glycosylation is also important to companies developing biosimilar therapeutics. Comparison of the glycan profiles of the biosimilar and reference molecule is one of the ways that biosimilarity can be demonstrated.Citation6-7
Recent advances in liquid chromatographic (LC) and mass spectrometric (MS) techniques have proven to be very effective for detailed characterizations of the glycans attached to biotherapeutic proteins.Citation8-9 However, glycans are challenging to analyze by LC-MS because they are largely invisible in their native states to optical detection methods and are plagued by low MS ionization efficiencies. Thus, to enable sensitive analytical measurements, glycans are frequently derivatized. Historically in the biopharmaceutical industry, 2-aminobenzamide (2-AB) or 2-aminoanthranilic acid (2-AA) have been conjugated to the glycan structure via a multi-hour reductive amidation reaction to allow for fluorescence (FLR) detection.Citation10 Similarly, glycans have been derivatized with 4-aminobenzoic acid 2-(diethylamino)ethyl esterCitation11 to enhance their electrospray ionization (ESI) efficiency by incorporating a basic functionality. With this label, the resulting derivatives were reported to have a 5,000-fold improvement in their MS signals over their native counterparts. However, while generally effective at enhancing the analytical performance, these types of methods require a significant time investment due to long enzymatic release times and complicated sample preparation protocols.
To increase sample preparation throughput and improve the analytical performance of glycans, a new labelling reagent has been developed.Citation12 This molecule, commercially known as RapiFluor-MS™ (RFMS), contains three key components: an N-hydroxysuccinimidyl (NHS)-carbamate with rapid labelling kinetics of amino functionalities, a quinolinyl core that enables highly sensitive FLR measurements, and a tertiary amine with a high gas-phase proton affinity that results in sensitive MS measurements in the positive MS mode of operation. It is noteworthy that this molecule was designed to improve measurement sensitivity for both FLR (10x over 2-AB) and MS (150x over 2AB) detectors.Citation12 The ability of this label to allow very sensitive analytical measurements of biotherapeutic glycans was demonstrated in a comparison of the glycan profiles of innovator and biosimilar infliximab.Citation8 In this study, the majority of the glycans observed to have the greatest differences in their respective abundance levels between the two molecules were present at very low levels. The improved MS sensitivity due to labelling with RFMS was critical to obtain high-quality MS data needed for confident identification and confirmation. Further, the 1:1 labelling chemistry of glycans with RFMS allows precise relative quantitation using FLR data when using highly efficient hydrophilic interaction chromatography (HILIC)-based ultra-performance liquid chromatography (UPLC) separations.Citation13 The measurement precision was also important to discern differences in abundances of low level glycans.
Beyond the experimental challenges associated with glycan analysis, data interpretation and structural identification remain difficult and are perhaps the greatest bottlenecks for a streamlined glycan analysis. Traditionally, this task most commonly requires manual data interpretation, although several software tools have been developed and have proven to be very useful.Citation14-16 Among these, is Glycobase,Citation17 an online collection of libraries for glycans derivatized with commonly used tags, including 2AB, 2AA, APTS for capillary electrophoresis, and now RFMS. Here, the retention times for each glycan have been converted to glucose units (GU)Citation18 for each tag. As the chemical nature of the tag influences retentivity, each tag will have a unique GU value for a particular glycan. This approach has been used to successfully minimize chromatographic variability often observed across different instruments and columns. The method has provided reliable glycan structural assignments, but Glycobase does not contain any mass information. Another library with 170 unique entries has been developed, and glycan identification is based on accurate mass and chromatographic retention time when porous graphitic carbon was used as the stationary phase medium.Citation14 In the library described here, we combine the strengths of both of these approaches: retention times that have been converted to GU values with subsequent assignment verification using accurate mass data (less than 5 ppm), which we have found to be very beneficial to correctly assign a glycan to a given FLR peak during characterization studies of proteins with unknown glycan profiles. While it may be possible in some cases to base assignments by GU for well-characterized glycoproteins, the library presented in this communication contains 168 glycans, and several instances exist where glycans with significantly different structures have very similar GU values and some of these examples are important to the biopharma industry. For example, using only GU values, it would be difficult at best to correctly distinguish M5 (GU=6.17) from A2[6]G(4)1 (GU 6.18) and A1G(4)1Ga(3)1 (GU=6.50) from A4 (GU=6.50). The approach used in this work to successfully distinguish M5 from A2[6]G(4)1 and A1G(4)1Ga(3)1 from A4 used accurate mass data.
Here, we present a streamlined end-to-end analytical platform used for the characterization of N-linked glycans derived from the NIST mAb Reference Material 8671. In this platform, a simplified sample preparation protocol where N-linked glycans were prepared in less than one hour and included glycan release, labelling with RFMS, and sample purificationCitation12 was used. The analytes were then subjected to HILIC-UPLC/FLR/MS investigations and the resulting analytical data were searched against a GU library of RFMS-labelled glycans for structural assignment.
Results
A method to identify glycans that relies on an internal standard for retention time normalization was previously reported.Citation19 Our library follows a similar principal, but it is based on GU values and accurate mass confirmation,Citation18 where the retention time for a given glycan was normalized (externally) against an RFMS-labelled dextran ladder. Expressing retention times as GU values minimizes the uncertainty associated with the chromatographic variability that is commonly observed with different HILIC columns or by using different LC systems. Thus, normalization improves the robustness of the reported retention parameters, GU in this work. To further improve the reliability of the library searches, mass information was used to verify GU structural assignments.
Developing the RFMS glycan GU scientific library
The RFMS GU Scientific Library contains GU values for 168 N-linked glycans derived from nine different glycoproteins: trastuzumab, infliximab, etanercept, NIST mAb, human serum polyclonal IgG, mouse serum polyclonal IgG, erythropoietin, bovine fetuin, and yeast invertase. These glycoproteins were selected to cover the three classes of N-linked glycans: high mannose, complex, and hybrid structures, and to provide extensive coverage of the N-linked glycans that are routinely attached to therapeutic mAbs and other biopharmaceutical products, which cover a broad range of GU values.
The RFMS GU library was developed in multiple stages. The first stage of development was an analysis of the intact N-linked glycan profiles for each glycoprotein and an analysis of their exoglycosidase digests. As these glycoproteins have been previously characterized, this analysis provided preliminary structural assignments using FLR data only. In the second stage, the intact N-linked profiles and their exoglycosidase digests were reanalyzed again by FLR, in addition to a QDa mass detector for mass verification. The QDa ensured that the preliminary structural assignments were correct. The proteins chosen for the library were carefully selected because they have been well characterized and have been used previously for other GU libraries. Therefore, verification of the assignments with average masses was sufficient for these samples. In this work, QDa mass measurements were within ± 0.2 m/z of the expected m/z using average masses. QDa was only employed during the development of the library and was used for verification purposes. This library was designed to aid scientists in rigorously characterizing samples with unknown glycan profiles, and only contains monoisotopic masses and not average masses. Therefore, characterization studies, like the one presented here, should be performed using a high-resolution MS. Once a sample has been rigorously characterized, a QDa can be used for monitoring purposes. The FLR and QDa mass traces for each of these proteins (Supplemental Information Fig. 1) demonstrate significant coverage across the GU range of approximately 3–15. In addition to human-type glycans, some of these glycoproteins also contain glycans with non-human features, including N-glycolylneuraminic acid and glycans with gal-α1,3-gal groups, and several glycans with these features are also included in the library. Additionally, several isomeric glycans differing only in their sialic acid linkages (α2,3- vs α2,6-linked) are also included.
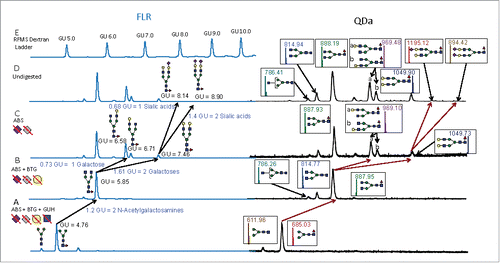
Figure 1. Exoglycosidase digestion array digests of RFMS-labeled N-linked glycans derived from trastuzumab analyzed by HILIC-UPLC/FLR and HILIC-UPLC/FLR/QDa. A) FLR and QDa traces for the digestion with ABS, BTG, and GUH. All glycans were digested down to common core structures (F(6)M3 and M3) with the exoglycosidase enzyme arrays. B) FLR and QDa traces of the ABS and BTG digests to remove sialic acids and galactoses. C) FLR and QDa traces following the removal of sialic acids with ABS. D) The FLR and QDa traces of undigested trastuzumab. E) The FLR chromatogram of the dextran ladder used to determine the GU values for the analytes in Panels A-D. Diagnostic shifts in GU values for the products of each digest revealed the glycan structures. Symbols: blue square- N-acetylglucosamine; green circle- mannose, yellow circle- galactose, red triangle- fucose; grey diamond- N-glycolylneuraminic acid
To generate the library, released and RFMS-labelled N-linked glycans were analyzed by three replicate injections per protein and their associated GU values were calculated by calibrating their retention times against an RFMS-labelled dextran ladder. The reported GU value for a given glycan is the average of all measurements for that glycan. While the glycans derived from each protein were analyzed in triplicate, a single glycan was frequently associated with several different glycoproteins. For example, F(6)A2 is attached to infliximab, etanercept, trastuzumab, human IgG, mouse IgG, and NIST mAb. Therefore, 18 measurements were made for this glycan and the reported GU value for this glycan is the average of all 18 measurements. Likewise, F(6)A2B was only observed on human IgG and the GU was measured three times. The final GU value for this glycan was the average of those three measurements. Using the chromatographic conditions described in the Methods, the GU values were very reproducible and very precise. For 140 of the entries, the standard deviations were less than 0.1 GU, and glycans with standard deviations greater than 0.1 were typically observed with very low fluorescent and MS signals or were sialylated and were observed to have longer retention times. It is therefore recommended to use a GU search window of ±0.2 GU to ensure that these glycans are reported when using this library.
To elucidate the structures of the N-linked glycans included in the library, exhaustive exoglycosidase array digestions were performed to release selected monosaccharides in a controlled and predictable manner and were performed as previously described.Citation20 To begin the array digestions, the sialic acids, both N-acetylneuraminic and N-glycolylneuraminic acids, were selectively removed, first with NAN1, a recombinant Streptococcus pneumoniae sialidase expressed in E. coli, to release α2,3-linked sialic acids. Next ABS, a recombinant Arthrobacter ureafaciens sialidase, also expressed in E. coli, was employed to remove α2,6-linked species. Subsequently, galactoses were removed with bovine testis β-galactosidase (BTG) and GlcNAcs were then removed with the hexosaminidase GUH. It should be noted that glycans labelled with RFMS showed some resistance to digestion with bovine kidney fucosidase (BKF); thus, it was not possible to completely digest α1,3 (core) fucosylated glycans to the chitobiose core, as show in Supplemental Fig. 2.
Following the treatment with each exoglycosidase, the digested sample was subsequently analyzed by HILIC-UPLC/FLR using the chromatographic conditions described in the Methods, and the diagnostic shifts in GU values revealed structural information. A relevant example for trastuzumab is shown in . This figure depicts the exoglycosidase digestion scheme used for structural characterization. Glycans are reconstructed from the core, explained later in this section; Panel A presents the FLR and QDa chromatogram and mass chromatogram following digestion with ABS, BTG, and GUH. This panel shows the afucosylated and fucosylated cores. The QDa information presented mass confirms these analytes. Panel B presents the FLR and QDa traces following digestion with ABS and BTG, and shows mass confirmed desialylated and degalactosylated components. Panel C shows the FLR chromatogram and the QDa trace for glycans that have been desiaylated and Panel D depicts the FLR chromatogram and QDa mass chromatogram for the undigested sample. Panel E is the FLR trace of a dextran ladder used to determine the GU values for the analytes in Traces A-D.
To determine each structure in the undigested profile, the core structures following digestion with the complete array, as shown in , were reconstructed. Comparing to , which shows the FLR chromatogram for samples digested with NAN1, ABS, and BTG, demonstrates that each GlcNAc adds 0.6 GU to the F(6)M3 core, thus the feature at a GU value of 5.85 in has a structure corresponding to F(6)A2. Similarly, a galactose attached to the α1,6 arm increases the GU value by ∼ 0.8 GU unit, while one associated with the α1,3 arm adds ∼0.85 GU, and a galactose added to each arm will add ∼1.6 GUs. Therefore, in the features at GU values of 6.58, 6.71, and 7.46 correspond to F(6)A2[6]G(4)1, F(6)A2[3]G(4)1, and F(6)A2G(4)2, respectively. Finally, α2,3 linked sialic acids increase the GU by ∼0.7. Using this procedure, the glycan at GU 8.14 in the undigested analysis shown in is F(6)A2G(4)2S(3)1 and that at a GU value of 8.90 is F(6)A2G(4)2S(3)2.
To further verify and validate the assignments based on GU value shifts for the undigested profiles and those treated with exoglycosidases, mass information was incorporated by using a QDa mass detector (MD). Adding mass information ensured a very high degree of certainty in assigning the correct glycan structure and exoglycosidase digestion product to each FLR peak; the mass data are shown as the insets of . This is demonstrated again with trastuzumab, which was subjected to a second exoglycosidase digestion array and reanalyzed by HILIC-UPLC/FLR/MD to verify the glycan assignments following the addition of each enzyme. Including orthogonal mass information confirms the preliminary assignments based on GU, thus adding confidence to the structural accuracy of the glycans in the library and in library searches.
Integrating library searching into a glycan analysis solution
The RFMS GU Scientific Library is one component in an overall solution aimed to streamline and simplify glycan analysis. The total solution includes an optimized Rapid PNGase F digestion, labelling with RFMS, HILIC-UPLC/FLR/QTOF MS measurements, and data processing and GU library searching. The need for a streamlined analytical solution is demonstrated in the biopharmaceutical setting, where the N-linked glycan profiles of hundreds of samples commonly must be analyzed with a short turnaround time, for example during cell line selection. Thus, the long sample preparation times associated with the traditionally used protocols, ranging from several hours to several days, may not be ideal for high throughput settings, nor is the time-consuming manual interpretation of the data.
To reduce the complexity and decrease the time investment associated with sample preparation, an optimized and simplified sample preparation protocol that utilizes Rapid PNGase F was developed.Citation12 The optimized protocol uses a 5-minute denaturing step at 95°C, enhanced with 1% RapiGest-SF surfactant, followed by a 5-minute Rapid PNGase F digestion at 55°C. Under these digestion conditions, N-linked glycans are quickly released with efficiencies that least match those of longer (overnight) PNGase F digestions. The short digestion time is critical for RMFS labelling, which selectively reacts with the glycosylamine. Therefore, this functionality must be preserved, and the short digestion time prevents its hydrolysis to a reducing end to any appreciable extent. Following the rapid digestion, glycosylamine labelling is achieved in 5 minutes, and the samples are then purified using a propylamine HILIC μ-elution solid phase extraction plate. Next, the labelled glycans are analyzed by HILIC-UPLC/FLR/MS.
The total analytical solution used in this study also addresses perhaps the greatest bottleneck in glycan analysis: a robust and automated data analysis and structural assignment. To address this area, an RFMS GU Scientific Library that can be searched within UNIFI Scientific Information System was developed. The library search consists of three main steps. First, the HILIC-UPLC retention times are converted to GU values. This is achieved by including an analysis of an RFMS-labeled dextran ladder with known GU values in the sample set, and then generating a calibration curve by applying a cubic spline fit of GU vs retention time. The resulting equation is then used to calculate GU values for the unknown samples from their retention times.Citation17 The second step is the compilation of a subset of candidate glycans contained in the library within a search tolerance (±0.2 GU value was used in this study). Since a GU tolerance window is needed, it is highly probably that multiple glycans will be included in the library subset for each FLR peak. The final step employs accurate mass data (± 5 ppm was used in this study) to cull the subset and confirm the structural assignment for a given FLR peak. Using GU values with accurate mass during characterization studies of therapeutic glycoproteins is a powerful approach when the glycan structures have not been rigorously determined. During the development of the library, a QDa with unit resolution was used because the glycan profiles for all of the proteins used for the development of the library had been previously characterized. However, for uncharacterized samples, the high mass accuracy becomes increasingly important. Consider M5A1G(4)1Sg(6)1 and F(6)M5A1G(4)1Ga(3)1, which would have triply-charged m/z values of 740.2847 and 740.6248. High resolution instruments will easily distinguish these structures.
The necessity of using mass information is exemplified by considering a GU value of 8.35. Using a GU searching tolerance of only 0.05 GU (8.30-8.40, or a difference of approximately only 10 seconds for this particular GU window), the library search will return a subset of 4 unique glycans: F(6)A2[3]G(4)1Sg(6)1, GU=8.31; A3G(4)2, GU=8.38; M5A1G(4)1Ga(3)1, GU=8.40; and A2G(4)2S(6)1, GU=8.40. Even with these tight parameters, it can be very difficult to determine the correct glycan based on GU values alone because the calculated standard deviations of all of these glycans are on the order of about 0.1 GU. Thus, the ability to use mass information to eliminate incorrect matches and to verify the correct glycan is a key component of the search algorithm to increase library search accuracy and robustness. Using mass information confidently confirmed the peak at GU=8.37 as A2G(4)1S(6)1 ().
Table 1. A list of the mass-confirmed N-linked glycans derived from the NIST RM mAb. Also included are the observed retention times, normalized abundance (% Amount), expected and observed GU values, expected and observed masses and m/z values, the predominant charge state, and the mass error expressed in parts-per-million (ppm). Glycans without %Amounts or mass data co-eluted and were mass-confirmed through manual inspection of the data. In the case of co-eluting glycans, the total peak area and % Amounts are reported (for example F(6)A1[3]G(4)1 and F(6)A3).
Library robustness: Identifying Gal-α1,3-Gal isomers
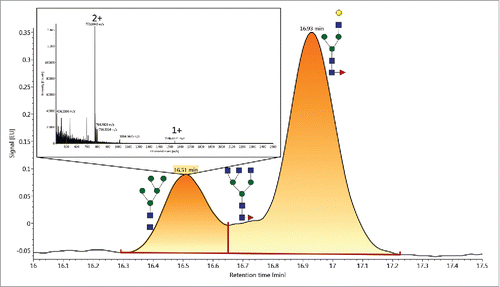
Due to the high performance of UPLC, numerous isomeric species are resolved and are included in the library. An example relevant to the biopharmaceutical industry is the set of galactosylated glycans: F(6)A2[6]G(4)1Ga(3)1, GU=7.39; F(6)A2G(4)2, GU=7.43; and F(6)A2[3]G(4)1Ga(3)1, GU=7.55. The separation and correct assignment of these glycans are shown in . Each of these glycans produced a 2+ ion observed at the monoisotopic m/z value of 1049.9186 (, , ). To verify the library assignments were correct, data-independent acquisition (DIA) was conducted on a XEVO G2-XS QTof MS controlled by UNIFI Scientific Information System software. The DIA or MSE data are shown in , , . Fragmentation produced a dominant peak at m/z=528.1917 (a B ion of Gal-Gal-GlcNAc) and confirmed the presence of the gal-α1,3-galCitation21 feature for the glycans with GU values of 7.35 and 7.53 ( and ). For a glycan with a GU value of 7.41, an intense ion at m/z=366.1353 (Gal-GlcNAc) () was detected and confirmed that one galactose was associated with each arm. MSE was only needed to confirm the library assignments, and was deemed necessary due to the similarity of their GU values. Since they were sufficiently resolved, MSE was an appropriate choice for this sample. However, a data-directed analysis (DDA) experiment should be performed for more complex samples in which more significant levels of co-elution occur and fragmentation data is needed for verification.
Figure 2. A) FLR channel of a HILIC-UPLC/FLR/MS analysis of the NIST RM mAb showing chromatographic resolution of F(6)A2[6]G(4)2Ga(3)1, F(6)A2G(4)2, and F(6)A2[3]G(4)2Ga(3)1. The MS spectra for each FLR peak are shown in B, D, and F for F(6)A2[6]G(4)2Ga(3)1, F(6)A2G(4)2, and F(6)A2[3]G(4)2Ga(3)1, respectively. Each of these glycans produced a 2+ ion observed at m/z=1049.919. The MSE spectra for F(6)A2[6]G(4)2Ga(3)1, F(6)A2G(4)2, and F(6)A2[3]G(4)2Ga(3)1 are shown in C, E, and G respectively. The intense ion at m/z=528.192 in C and G confirms the presence of the gal-α1,3-gal moiety and the ion at 366.135 confirms a galactose is present on each arm. The loss of RFMS is recorded at m/z 313.162.
![Figure 2. A) FLR channel of a HILIC-UPLC/FLR/MS analysis of the NIST RM mAb showing chromatographic resolution of F(6)A2[6]G(4)2Ga(3)1, F(6)A2G(4)2, and F(6)A2[3]G(4)2Ga(3)1. The MS spectra for each FLR peak are shown in B, D, and F for F(6)A2[6]G(4)2Ga(3)1, F(6)A2G(4)2, and F(6)A2[3]G(4)2Ga(3)1, respectively. Each of these glycans produced a 2+ ion observed at m/z=1049.919. The MSE spectra for F(6)A2[6]G(4)2Ga(3)1, F(6)A2G(4)2, and F(6)A2[3]G(4)2Ga(3)1 are shown in C, E, and G respectively. The intense ion at m/z=528.192 in C and G confirms the presence of the gal-α1,3-gal moiety and the ion at 366.135 confirms a galactose is present on each arm. The loss of RFMS is recorded at m/z 313.162.](/cms/asset/e22447ba-06ab-4723-9c14-77428b0960b9/kmab_a_1377381_f0002_oc.gif)
Characterizing the NIST RM mAb with the glycan analysis solution
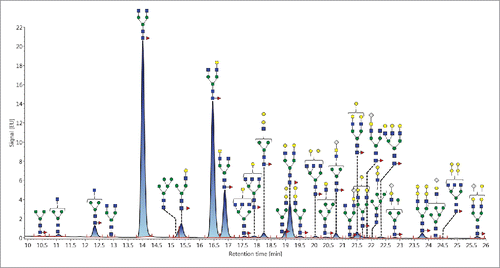
The total analytical solution described here was used to characterize the N-linked glycans associated with the NIST RM mAb 8761 using UNIFI software. In total, 35 mass-confirmed N-glycans were identified (). These glycans spanned a normalized abundance range from 0.1% to nearly 40%, as was calculated for F(6)A2. Complete co-elution was minimal and only five glycans that co-eluted with a more abundant species were identified and mass confirmed. These were F(6)A3, F(6)M4A1G(4)1 or F(6)M5A1 (isobaric species; collision-induced dissociation (CID) was unable to definitively determine the structure), M6, M7, and F(6)A2G(4)2Sg(6)1. Each of these glycans was mass confirmed through manual inspection of the MS data. In , the normalized abundance is expressed as the total area of the FLR peak and includes the contribution of co-eluting species, if they are present.
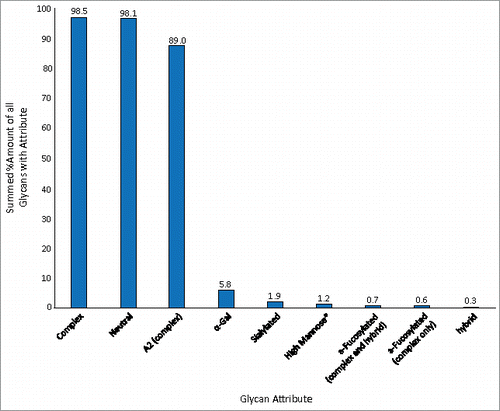
The abundance levels of various glycan attributes for the NIST RM mAb are shown in . The percentages calculated are the sum of the individual normalized amounts (found in ) for each glycan possessing a given attribute. The majority of the glycans associated with the NIST RM mAb were complex type; 98.5% of the total normalized FLR peak area was due to these glycans (neutral and sialylated). Most of this subset, 98.1% of the total normalized area, was composed of neutral glycans. Only 1.9% of the total abundance was due to sialylated glycans. Of the complex-type of glycans, biantennary structures were the predominant branched species; these types of glycans contributed 89% to the total FLR peak area. Similar to many other mAbs, afucosylated glycans were minor components. A total relative abundance of only 0.6% was associated with complex-type/afucosylated glycans; if hybrid and complex-type glycans were summed, 0.7% of the total abundance was due to afucosylated glycans.
Figure 3. Abundance levels of various glycan attributes. The values were calculated by summing the normalized abundance levels from for each glycan that possess a given attribute.
The N-linked glycan profile of the NIST RM mAb () largely resembles that of other mAbs. In total, 35 glycans were mass confirmed. Of these, 25 were complex, 4 were high mannose, and 6 were hybrid. The majority of the complex type glycans were biantennary, 14 such glycans were observed and 13 of these were core α1,6-fucosylated. The top four most abundant glycans belonged to this biantennary/fucosylated group. F(6)A2 was the most abundant and accounted for ∼ 40% of the total normalized area. The next most abundant glycans were the isomeric structures F(6)A2[6]G(4)1 (∼ 28%) and F(6)A2[3]G(4)1 (nearly 10%) and F(6)A2G(4)2 (∼ 7%). A total of 7 sialylated glycans were confirmed; all were α2,6-linked and all contained N-acetylneuraminic acid. Only two complex type glycans were afucosylated.
Figure 4. Annotated FLR trace depicting the N-linked glycans derived from the NIST RM mAb. In total, 35 glycans were mass confirmed.
Of particular interest to the biopharmaceutical industry are glycan structural attributes pertaining to the level of core fucosylation, the amounts of the nonhuman moieties, (N-glycolylneuraminic acid and gal-α1,3-gal that are associated with product safety) and the abundance of M5. Based on the results collected in this study, the total amount of afucosylated glycans, including the M5, is ∼ 1.5%. If only complex glycans are included, the value is about 0.7%. The amount of sialylation was associated only with N-glycolylneuraminic acid; no glycans terminated with N-acetylneuraminic acid were observed. These glycans accounted for ∼ 2% of the total normalized abundance, with F(6)A1G(4)1Sg(6)1 being the most abundant at 1%. Up to 10 glycans with the gal-α1,3-gal moiety were identified, including one that co-eluted and could not be mass confirmed nor verified through MSE experiments. However, the remaining 9 were mass confirmed and further verified through CID fragmentation experiments. The glycans with gal-α1,3-gal group contributed nearly 6% to the total normalized abundance. Three of these were determined to be greater than 1%, (F(6)A1G(4)1Ga(3)1, F(6)A2G(4)2Ga(3)1, and F(6)A2G(4)2Ga(3)2), and a fourth, F(6)A2[6]G(4)1Ga(3)1, was at 0.9%. The remaining glycans were observed to be between 0.1% and 0.5%.
To determine accurately the level of M5, a slightly different gradient was employed due to co-elution with F(6)A1[3]G(4)1. To resolve these two glycans, the column temperature was decreased to 45°C and a segmented gradient from 80% to 73% B over 3 min at 0.5 mL/min, then to 63% B over 32 min, was employed. As depicted in , the resolution between M5 and F(6)A1[3]G(4)1 was greatly improved. When integrated using a baseline-to-valley algorithm, the relative abundance of M5 was determined to be between 0.95 ± 0.04%. For triplicate sample preparations, the %RSD for M5 was 4%.
Figure 5. The FLR trace showing the enhanced resolution of M5 from other glycans with similar retention times. This was achieved using a slightly different set of separation conditions (see the experimental section) to achieve a more accurate abundance level of the M5 glycan. Using these altered conditions, M5 was determined to have a normalized abundance level of 0.95 ± 0.04%.
Overall, the structures identified and their relative amounts correlate quite closely to a previous multi-laboratory, multi-approach characterization study of the NIST mAb.Citation22
Discussion
During the development of our overall analytical solution, one goal was to decrease the overall time investment for N-linked glycan sample preparation. Typical glycan sample preparation workflows used in many laboratories tend to be time consuming and laborious, require multiple steps, including denaturation, digestion, sample purification, labelling, and repurification prior to analytical analysis. Regardless of the method used, whether a more traditional approach to sample preparation, or the method presented here, the highest digestion efficiency is achieved when proteins are first denatured. In many procedures, sodium dodecyl sulfate (SDS) is commonly employed as a denaturant at elevated temperatures; however, to maintain enzymatic activity, a nonionic surfactant like NP-40 must be added at a concentration above its critical micellular concentration prior to the addition of PNGase F. Above this concentration, SDS partitions into the micelles and does not reduce digestion efficiency. In the method described here, RapiGest-SF is used to induce protein denaturation at temperatures between 90–100°C for 5 min. The presence of 1% RapiGest-SF does not negatively influence the Rapid PNGase F digestion efficiency, thus no additional reagents need to be added to preserve enzymatic activity. Historically, deglycosylation generally requires several hours to overnight. However, by utilizing Rapid PNGase F, a recombinant form of the enzyme that can tolerate higher temperatures, the digestion time has been reduced to 5 min at 55°C. Even under these digestion conditions, Rapid PNGase F can facilitate removal of N-linked glycans that is just as efficient as overnight digestions.Citation12 Following N-glycan release, traditional procedures often require sample purification, solid-phase extraction (SPE) with either HILIC or graphite media is a popular method, followed by vacuum centrifugation prior to labelling. In the protocol described in this work, the components in the sample do not interfere with labelling, thus this purification step was eliminated. Subsequent labelling is often achieved through a reductive amination procedure and requires 2–4 hoursCitation10 or is performed overnight.Citation23 Permethylation is another popular approach and can be completed in 1–2 hours.Citation24 Conversely, the rapid labelling kinetics of NHS carbamates, like RFMS, towards amino functionalities enables glycan labelling to be accomplished in only 5 mins. Following derivatization, it is recommended to purify the samples prior to analysis and many procedures require vacuum centrifugation. In this study, HILIC SPE was used for sample purification. However, the samples were not dried, rather the aqueous eluent is diluted with dimethylformamide (DMF) and acetonitrile (ACN) to adjust the organic solvent composition to an appropriate level for injection onto a HILIC column. Overall, the methods historically used may require 1–2 days for sample preparation, while the method utilized in this work may be accomplished in 30 minutes by employing Rapid PNGase F, labelling with RapiFluor-MS, and minimizing the number of sample purification/dry down steps.
Once prepared, glycans are a particularly challenging class of molecules to fully characterize. An initial challenge is their relatively insensitive nature towards optical detection methods (FLR and UV) and MS in their native state, which can be now overcome through high-performance rapid labelling methods like RFMS. Additionally, glycans exhibit extraordinary structural diversity through different monosaccharide linkages and positions, and frequently isomeric (defined as analytes with the same monosaccharide composition but different structures) and isobaric (those with the same mass but different compositions) glycans are present on therapeutic proteins. Using MS-only methods, isomeric and isobaric glycans present in a mixture are difficult to confidently identify and quantitate. Fortunately, even though isomeric glycans have the same monosaccharide composition, their overall structures are often sufficiently different enough that they exhibit different chromatographic retention times. Similarly, isobaric species can be resolved chromatographically. Therefore, a rigorous characterization requires UPLC coupled to MS for confident identification and precise quantitation.
The high resolving power of UPLC is evident for isomeric/isobaric glycans, and these types of analytes can be very important to correctly identify and quantitate. This is exemplified by the isobaric pair A2G(4)2S1 and F(6)A2[6]G(4)1Sg(6)1, where the latter is a potentially immunogenic glycan. Both glycans have a monoisotopic mass of 2242.8622 amu following derivatization with RFMS. However, they have GU values of 8.40 and 8.16, respectively, and thus can be resolved chromatographically and mass confirmed as discrete analytes. In this study, an analyte was observed at GU=8.15 and was mass confirmed as F(6)A2[6]G(4)1Sg(6)1. Even though an analyte was present at GU=8.41, mass confirmation proved that its identity was M5A1G(4)1Ga(3)1, and not A2G(4)2S1. The isomeric set of glycans F(6)A2[6]G(4)1Ga(3)1 (GU=7.40), F(6)A2[3]G(4)1Ga(3)1 (GU=7.55), and F(6)A2G(4)2 (GU=7.43) comprise a second example highlighting the need to combine both GU and MS data. After derivatization with RFMS, these glycans all have a monoisotopic mass of 2097.825, but they have very different overall structures; the first and second glycans contain the potentially immunogenic gal-α1,3-gal group located on the α1,6-branched core mannose or the α1,3-branched core mannose, respectively. For the third glycan, one galactose is attached to each arm. Thus, due to their structural differences, GU values of 7.35, 7.55, and 7.41, respectively, were observed. Mass confirmation proved that these glycans were F(6)A2[6]G(4)1Ga(3)1 (GU=7.40), F(6)A2[3]G(4)1Ga(3)1 (GU=7.55), and F(6)A2G(4)2 (GU=7.43), and CID experiments verified the identifications. These examples of isobaric and isomeric glycans demonstrate that, for robust glycan identification and relative quantification, both UPLC normalized retention times and MS data are needed. All of these data are incorporated into the RFMS GU Scientific Library to ensure search robustness and accuracy.
Methods and materials
Materials
In this study, the NIST mAb Reference Material 8761 was obtained from the National Institute of Standards and Technology. Trastuzumab (item number 48682, a product of Genentech), infliximab (item number 48115, a product of Pfizer), and etanercept (item number 25479, a product of Amgen) were acquired from Besse Medical under an approved license. RFMS-labelled human IgG (RFMS Glycan Performance Test Standard, product number 186007983) and mouse IgG (Intact mAb Mass Check Standard, product number 186006552) were purchased from Waters Corp. Erythropoietin, EU pharmacopeia standard (product number E1515000) was obtained from the European Directorate for the Quality of Medicines, and bovine fetuin (product number F2379), and yeast invertase (product number I0408-500UG) were products of Sigma-Aldrich Corp. Ammonium formate solution, Glycoworks® RapiFluor-MS Performance Test Standard, Glycoworks RapiFluor-MS dextran calibration ladder, and a Glycoworks RapiFluor-MS 24-sample N-Glycan Kit were obtained from Waters Corp. The complete kit consists of three modules: deglycosylation, labeling, and clean-up. When combined, the kits contain all of the supplies needed to complete the sample preparation, and all three were utilized in this project. The Glycoworks Deglycosylation Module contained Intact mAb Mass Check Standard, Glycoworks Rapid PNGase F and Buffer, and RapiGest™ SF. Glycoworks® RapiFluor-MS™ Labeling Module contained the Glycoworks® RapiFluor-MS™ Reagent and Glycoworks® Reagent Solvent Anhydrous DMF. The Glycoworks® RapiFluor-MS™ Clean-up Module contained a Glycoworks® HILIC μ-Elution Plate, Glycoworks® SPE elution buffer (200 mM ammonium acetate, 5% ACN), and Glycoworks® Sample Diluent (DMF). The Glycoworks® RapiFluor-MS™ Sample Collection Module contained sample collection tubes, a waste tray, and a collection tray. LC-MS grade water and ACN and formic acid were purchased from Sigma-Aldrich Co. The exoglycosidases used in this study were acquired from Prozyme, Inc.
Methods
N-Glycan release
N-linked glycans derived from the NIST mAb Reference Material 8671 were prepared in triplicate as previously described.Citation12 The N-linked glycans from proteins used to create the RFMS GU Scientific Library were similarly prepared. Briefly, the samples were diluted to a concentration of 2 μg/μL in a phosphate-buffered saline solution (100 mM sodium phosphate, 150 mM sodium chloride, pH=7.2) and a 7.5-μL aliquot (15 μg) was added to 15.3 μL of water and 6 μL of a 5% RapiGest solution. The proteins were denatured at 95°C for 5 min and then cooled to the ambient temperature. Next, a 1.2-μL aliquot of Rapid PNGase F was added and the samples were incubated at 55°C for 5 minutes to release the N-linked glycans as their glycosylamines. This short digestion time maintained the released glycans in their glycosylaminyl state and did not allow the amino group to be hydrolyzed to a hydroxyl group, which is a critical feature for labelling with RFMS.
Glycan labeling
Following the digestion, the amino group of the released glycosylamines was labeled with RFMS. A 24-reaction kit was used and the RFMS reagent (9 mg) was dissolved in 131 μL of anhydrous DMF. A 12-μL aliquot of this solution was added to each glycan sample and allowed to react at the ambient temperature for 5 minutes. The reaction was then quenched with the addition of a 360-μL aliquot of ACN, which also adjusted the solution to an appropriate organic solvent concentration for HILIC-based purification.
Glycan purification
The derivatized N-linked glycans were purified using a HILIC μ-elution plate. The medium was first washed with three 300-μL aliquots of water, followed by three 300-μL aliquots of an 85%/15% ACN/water solution. The samples were then loaded onto the medium and washed with three 600-μL aliquots of a 90%/9%/1% solution of ACN/water/FA. The glycans were eluted by three 30-μL aliquots of an SPE Elution Buffer (200 mM ammonium acetate in 5% ACN).
Exoglycosidase digestions for structural characterization
The glycans included in the library were subjected to exoglycosidase array digestions for rigorous structural verification. All enzymes were purchased from Prozyme. RFMS-labeled glycans were digested in 10 μL of a 50 mM sodium acetate buffer, pH 5.5 for 18 h at 37°C, using arrays of the following enzymes: NAN1 (recombinant sialidase, EC 3.2.1.18, to release α2-3 linked non-reducing terminal sialic acids); ABS (Arthrobacter ureafaciens sialidase, EC 3.2.1.18, to release α2-3,6,8-linked sialic acids); BKF (bovine kidney α-fucosidase, EC 3.2.1.51, to release α1-2,6-linked fucose residues); BTG (bovine testes β-galactosidase, EC 3.2.1.23, to hydrolyze β1-3- and β1-4- linked galactoses); and GUH (β-N-acetylhexosaminidase cloned from Streptococcus pneumoniae expressed in E. coli, EC 3.2.1.30, to release GlcNAc residues but not a bisecting GlcNAc linked to a core mannose). After incubation, the enzymes were removed by filtration using 10 kDa spin filters (Pall Corp.). Following the addition of each exoglycosidase, the digested N-linked glycans were analyzed by HILIC-UPLC.
UPLC-MS/MSE
RFMS-labelled glycans and those subjected to exoglycosidase digestions were diluted with 100 μL of DMF and 210 μL of ACN to adjust the solution to an appropriate level of organic solvent for HILIC separations. In this study, a Waters ACQUITY H-class Bio UPLC system, consisting of a bio-quaternary solvent manager, a sample manager, set to 10°C, a column manager, operating at 60°C, and a fluorescence (FLR) detector (excitation wavelength = 265 nm, emission wavelength = 425 nm, data collection rate of 2 Hz) was used for the separations. The glycans were separated with a Waters ACQUITY UPLC Glycan BEH Amide column (2.1 × 150 mm, 1.7 μm particle size, 130 Å pore size). Mobile Phase A was a 50 mM ammonium formate solution (pH=4.4) and Mobile Phase B was neat ACN. Analyte separation was accomplished by gradient elution using a gradient running from 75%-54% Mobile Phase B over 35 minutes at a flow rate of 0.4 mL/min. For the NIST mAb analysis, the UPLC system was coupled to a Waters XEVO G2-XS QTof MS and was operated in its positive sensitivity mode to monitor the m/z range from 300–2500 at a scan rate of 2 Hz. The capillary voltage was set to 3 kV and a cone voltage of 80 V was used. The source temperature was 125°C and the desolvation temperature was set at 425°C. MSE (data independent acquisition) experiments were performed to verify library assignments. For these experiments, the low energy channel was set to 6 V and the high energy was ramped from 15–40 V to induce fragmentation. All other parameters were the same as for the MS experiments. For all MS experiments, a 100 pmol/μL solution of [GluCitation1]-fibrinopeptide B in 50%/50%/0.1% water/ACN/formic acid was used for Lockspray calibration and the 2+ ion at m/z=785.8421 was used for calibration. For mass verification of exoglycosidase digests, a QDa mass detector (Waters Corp.) was used. For these experiments, the m/z range from 500–1250 was monitored. The cone voltage was set to 15 V and a capillary voltage 1.5 kV was utilized.
Generating the RFMS GU library
The N-linked glycans from nine glycoproteins (trastuzumab, infliximab, etanercept, NIST mAb, human IgG, mouse IgG, erythropoietin, bovine fetuin, and yeast invertase) were prepared as previously described and analyzed by HILIC-UPLC/FLR(/QDA MD). The glycans derived from each protein were analyzed a minimum of 3 times to precisely determine the GU value for each glycan. The average GU for all measurements of a given glycan were used in the library. The structures were verified through exoglycosidase digestions as described and further confirmed through data acquired using a QDa mass detector.
Data analysis
HILIC-UPLC/FLR/MS data were processed and analyzed using the Glycan Assay (FLR with MS confirmation) workflow within UNIFI. This workflow first converted the retention times of the labelled glycans samples to GU based on a cubic spline calibration curve against a dextran ladder labeled with RFMS. These GU values were then searched against the RFMS Glycan GU Scientific Library housed within UNIFI for glycan structural identification. The library searches used a GU tolerance of 0.2 GU and a mass error of 5 ppm. For relative quantitation, the FLR peak area for each glycan was expressed as a percentage of the total summed peak area for all of the glycans identified.
Suppl_Mat.zip
Download Zip (365.5 KB)Acknowledgments
This work was supported by an Enterprise Ireland grant under the Innovation Partnership Program in collaboration with Waters (Grant No: IP/2014/0326).
References
- Yu M, Brown D, Reed C, Chung S, Lutman J, Stefanich E, Wong A, Stephan JP, Bayer R. Production, characterization and pharmacokinetic properties of antibodies with N-linked Mannose-5 glycans. MAbs. 2012;4(4):475-87. https://doi.org/10.4161/mabs.20737 PMID:22699308
- Ghaderi D, Taylor RE, Padler-Karavani V, Diaz S, Varki A. Implications of the presence of N-glycolylneuraminic acid in recombinant therapeutic glycoproteins. Nat Biotech. 2010;28(8):863-67. https://doi.org/10.1038/nbt.1651
- Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-α-1,3-galactose. N Engl J Med. 2008;358(11):1109-17. https://doi.org/10.1056/NEJMoa074943 PMID:18337601
- Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. Lack of fucose on human IgG1 N-Linked oligosaccharide improves binding to human Fc gamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277(30):26733-40. https://doi.org/10.1074/jbc.M202069200 PMID:11986321
- Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J Bio Chem. 2003;278(5):3466-73. https://doi.org/10.1074/jbc.M210665200
- Beck A. Biosimilar, biobetter and next generation therapeutic antibodies. MAbs. 2011;3(2):107-10. https://doi.org/10.4161/mabs.3.2.14785 PMID:21285536
- Beck A, Sanglier-Cianferani S, Van Dorsselaer A. Biosimilar, biobetter, and next generation antibody characterization by mass spectrometry. Anal Chem. 2012;84(11):4637-46. https://doi.org/10.1021/ac3002885 PMID:22510259
- Fang J, Doneanu C E, Alley W R, Yu Y Q, Beck A, Chen W. Advanced assessment of the physicochemical characteristics of Remicade® and Inflectra® by sensitive LC/MS techniques. MAbs. 2016;8(6):1021-34. https://doi.org/10.1080/19420862.2016.1193661 PMID:27260215
- Xie H, Chakraborty A, Ahn J, Yu Y Q, Dakshinamoorthy DP, Gilar M, Chen W, Skilton SJ, Mazzeo JR. Rapid comparison of a candidate biosimilar to an innovator monoclonal antibody with advanced liquid chromatography and mass spectrometry technologies. MAbs. 2010;2(4):379-94. https://doi.org/10.4161/mabs.11986 PMID:20458189
- Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230(2):229-38. https://doi.org/10.1006/abio.1995.1468 PMID:7503412
- Yoshino K, Takao T, Murata H, Shimonishi Y. Use of the derivatizing agent 4-aminobenzoic acid 2-(diethylamino)ethyl ester for high-sensitivity detection of oligosaccharides by electrospray ionization mass spectrometry. Anal Chem. 1995;67(21):4028-31. https://doi.org/10.1021/ac00117a034 PMID:8633763
- Lauber MA, Yu YQ, Brousmiche DW, Hua Z, Koza SM, Magnelli P, Guthrie E, Taron CH, Fountain KJ. Rapid preparation of released N-glycans for HILIC analysis using a labeling reagent that facilitates sensitive fluorescence and ESI-MS detection. Anal Chem. 2015;87(10):5401-9. https://doi.org/10.1021/acs.analchem.5b00758 PMID:25927596
- Ahn J, Bones J, Yu YQ, Rudd PM, Gilar M. Separation of 2-aminobenzamide labeled glycans using hydrophilic interaction chromatography columns packed with 1.7 microm sorbent. J Chromatogr B. 2010;878(3–4):403-8. https://doi.org/10.1016/j.jchromb.2009.12.013
- Aldredge D, An H J, Tang N, Waddell K, Lebrilla C B. Annotation of a serum N-glycan library for rapid identification of structures. J Proteome Res. 2012;11(3):1958-68. https://doi.org/10.1021/pr2011439 PMID:22320385
- Kronewitter SR, De Leoz ML, Strum JS, An HJ, Dimapasoc LM, Guerrero A, Miyamoto S, Lebrilla CB, Leiserowitz GS. The Glycolyzer: Automated glycan annotation software for high performance mass spectrometry and its application to ovarian cancer glycan biomarker discovery. Proteomics. 2012;12(15–16):2523-38. https://doi.org/10.1002/pmic.201100273 PMID:22903841
- Schiel JE, Rogstad SM, Boyne MT 2nd. Comparison of traditional 2-AB fluorescence LC-MS/MS and automated LC-MS for the comparative glycan analysis of monoclonal antibodies. J Pharm Sci. 2015;104(8):2464-72. https://doi.org/10.1002/jps.24522 PMID:26053232
- Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: Tools for HPLC-based glycan analysis. Bioinformatics. 2008;24(9):1214-6. https://doi.org/10.1093/bioinformatics/btn090 PMID:18344517
- Guile GR, Rudd PM, Wing DR, Prime SB, Dwek RA. A rapid high-resolution high-performance liquid chromatographic method for separating glycan mixtures and analyzing oligosaccharide profiles. Anal Biochem. 1996;240(2):210-26. https://doi.org/10.1006/abio.1996.0351 PMID:8811911
- Pabst M, Bondili JS, Stadlmann J, Mach L, Altmann F. Mass + retention time = structure: A strategy for the analysis of N-glycans by carbon LC-ESI-MS and its application to fibrin N-glycans. Anal Chem. 2007;79(13):5051-57. https://doi.org/10.1021/ac070363i PMID:17539604
- Houel S, Hilliard M, Yu YQ, McLoughlin N, Martin SM, Rudd PM, Williams JP, Chen W. N- and O-glycosylation analysis of Etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal Chem. 2014;86(1):576-84. https://doi.org/10.1021/ac402726h PMID:24308717
- Bosques CJ, Collins BE, Meador JW, Sarvaiya H, Murphy JL, DelloRusso G, Bulik DA, Hsu IH, Washburn N, Sipsey SF, et al. Chinese hamster ovary cells can produce galactose-[alpha]-1,3-galactose antigens on proteins. Nat Biotech. 2010;28(11):1153-56. https://doi.org/10.1038/nbt1110-1153
- Prien JM, Stöckmann H, Albrecht S, Martin SM, Varatta M, Furtado M, Hosselet S, Wang M, Formolo T, Rudd PM, Schiel JE. Orthogonal technologies for NISTmAb N-glycan structure elucidation and quantitation. In Schiel JE, Davis DL, Borisov OV, editors. State-of-the-art and emerging technologies for therapeutic monoclonal antibody characterization Volume 2. Biopharmaceutical characterization: The NISTmAb case study. Washington, D.C.: American Chemical Society; 2015. pp 185-235. https://doi.org/10.1021/bk-2015-1201.ch004.
- Ruhaak LR, Zauner G, Huhn C, Bruggink C, Deelder AM, Wuhrer M. Glycan labeling strategies and their use in identification and quantification. Anal Bioanal Chem. 2010;397(8):3457-81. https://doi.org/10.1007/s00216-010-3532-z PMID:20225063
- Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22(5):721-34. https://doi.org/10.1002/rcm.3395 PMID:18265433
