ABSTRACT
Respiratory syncytial virus (RSV) is a leading cause of lower respiratory tract infection in young children and older adults. Currently, no licensed vaccine is available, and therapeutic options are limited. The primary target of neutralizing antibodies to RSV is the surface fusion (F) glycoprotein. Understanding the recognition of antibodies with high neutralization potencies to RSV F antigen will provide critical insights in developing efficacious RSV antibodies and vaccines. In this study, we isolated and characterized a panel of monoclonal antibodies (mAbs) with high binding affinity to RSV prefusion F trimer and neutralization potency to RSV viruses. The mAbs were mapped to previously defined antigenic sites, and some that mapped to the same antigenic sites showed remarkable diversity in specificity, binding, and neutralization potencies. We found that the isolated site III mAbs shared highly conserved germline V-gene usage, but had different cross-reactivities to human metapneumovirus (hMPV), possibly due to the distinct modes/angles of interaction with RSV and hMPV F proteins. Furthermore, we identified a subset of potent RSV/hMPV cross-neutralizing mAbs that target antigenic site IV and the recently defined antigenic site V, while the majority of the mAbs targeting these two sites only neutralize RSV. Additionally, the isolated mAbs targeting site Ø were mono-specific for RSV and showed a wide range of neutralizing potencies on different RSV subtypes. Our data exemplify the diversity of anti-RSV mAbs and provide new insights into the immune recognition of respiratory viruses in the Pneumoviridae family.
Introduction
Human respiratory syncytial virus (RSV) and human metapneumovirus (hMPV) are genetically related single-stranded negative-sense RNA viruses that were recently reclassified into the newly created Pneumoviridae family from Paramyxoviridae family.Citation1 RSV is the most important pathogen of pediatric acute lower respiratory tract illness (ALRI) worldwide and the leading cause of pneumonia and bronchiolitis in infants.Citation2–Citation4 Furthermore, RSV can infect individuals of all ages and cause severe disease in the elderly and immunocompromised populations.Citation5–Citation8 No licensed RSV vaccine is currently available. Palivizumab (Synagis®), a monoclonal antibody (mAb) that targets RSV surface fusion protein F, is commercially available for RSV prophylaxis, but the application is restricted to only pediatric patients with high risk of RSV infection due to the limited effectiveness.Citation9,Citation10 hMPV is another major pathogen that causes respiratory-tract infection in children and adults worldwide, and no vaccines or therapeutics are available for hMPV infections.Citation11–Citation16 Hence, there is substantial unmet medical need for efficacious vaccines and potent neutralizing antibodies (nAbs) to provide protection to children and high-risk adults from RSV and hMPV infections.
The highly conserved RSV F protein has become the primary target for RSV prophylactic antibody discovery and vaccine development. RSV F is a type I transmembrane fusion protein that assembles as a trimer and mediates membrane fusion during virus entry, through a conformational change from a metastable prefusion (PreF) structure to a highly stable postfusion (PostF) structure.Citation17–Citation23 There are at least six major antigenic sites on RSV F reported in the literature (Ø, I, II, III, IV, V).Citation24 PreF-specific antibodies targeting the apex sites, including site Ø, are immunodominant and account for a large proportion of neutralizing activity in human sera.Citation25,Citation26 A recent study has reported that the PreF-specific site V is targeted by nearly half of the most potent nAbs isolated from healthy adults.Citation27 These studies suggested that as an RSV vaccine candidate, PreF might elicit higher nAb responses than PostF. Due to the metastability of the wild-type prefusion RSV F structure, F antigens stabilized in the PreF conformation have been engineered by structure-based rational design, resulting in some promising candidates of RSV subunit vaccines.Citation28,Citation29 Different from RSV, the immunogenicity potential of stabilized hMPV PreF and PostF appears to be similar, likely due to the large glycan shield that is only observed at the site Ø of hMPV F protein.Citation30
MAbs targeting various antigenic sites of RSV and hMPV have previously been isolated via different methods.Citation24,Citation27,Citation31–Citation45 Furthermore, the crystal structures of RSV and hMPV F proteins in both PreF and PostF conformations, as well as complex structures with mAbs targeting various antigenic sites, have been reported.Citation20,Citation21,Citation28–Citation35,Citation37,Citation45–Citation49 The structures of PreF and PostF antigens between RSV and hMPV are remarkably conserved, albeit only sharing about 35% sequence identity.Citation21,Citation29–Citation31,Citation46,Citation48 MAbs that cross-neutralize both RSV and hMPV have been reported. Most of these dual-nAbs recognize the highly conserved site III between the two viruses.Citation27,Citation43,Citation49 It was reported that site III mAbs were enriched in IGHV3-11/IGHV3-21: IGLV1-40 germline pairing in the immune repertoire of both infants and adults.Citation27,Citation35 In addition, several cross-reactive mAbs targeting a conserved epitope on site IV have been isolated,Citation36,Citation40,Citation46 likely due to different binding angles from the RSV-specific site IV mAbs.Citation36 MAbs recognizing other antigenic sites, including site Ø, I, II and V, only show mono-specific neutralizing activities and no cross-neutralizing mAb against these sites has been reported.Citation9,Citation27,Citation31–Citation34,Citation38,Citation39
In this study, we focused on characterization of a panel of nAbs isolated from adult human memory B cells with relatively high binding affinity to RSV F antigen and potent RSV neutralizing activities. We aimed to understand the antibody binding epitopes in relation to their binding specificities and neutralization potencies to different RSV and hMPV viruses. Our data revealed the diverse binding modes of anti-RSV mAbs and provided new insights into the mechanisms and breadth of immune recognition of RSV and related viruses.
Results
Potent RSV neutralizing mAbs were isolated from antigen-specific memory B cells and characterized
The memory B cells from healthy adult donors were isolated by flow cytometry using RSV or hMPV F antigen-specific single memory B cell sorting or memory B cell enrichment methods. Selected based on their high RSV neutralization potency, a panel of 23 mAbs was successfully cloned and expressed (). Most of the mAbs (18 of 23) were isolated from single memory B cell sorting using RSV F antigens, 3 mAbs were isolated from memory B cell sorting using hMPV F antigens, and 2 mAbs were derived from memory B cell enrichment approach (S2 Table). The average somatic hypermutation (SHM) of VH genes and lengths of complementarity-determining region H3 are 11–52 (median 25) nucleotides and 8–21 (median 13) amino acids (S1 Fig), respectively, which are comparable with a recent report on profiling of anti-RSV F antibody repertoires in adults.Citation27 The purified mAbs showed high binding affinity to RSV PreF (DS-Cav1) with EC50s ranging from 4.2 to 30.5 ng/mL (, S2 Fig). The neutralizing activities of the mAbs were determined with RSV A Long strain and RSV B Washington strain. The IC50s of all mAbs exhibited sub-nanomolar potency against RSV A Long strain, ranging from 4.8 to 106.6 ng/mL (). Most of the mAbs, except 139B8 and M1C7, also neutralized RSV B Washington strain in the nanomolar or sub-nanomolar range, with IC50s ranging from 1.2 to 231.9 ng/mL (). M1C7 neutralized RSV B Washington strain at a much weaker IC50 (1105 ng/mL). 139B8 did not neutralize RSV B Washington strain at the concentrations we tested. The data suggested that all 23 mAbs isolated from memory B cells are potent RSV nAbs, with some significantly more potent than palivizumab and comparable to D25 (). In addition, seven mAbs were found to bind to the hMPV F antigen with EC50s ranging from 6.0 to 290.6 ng/mL (). Of the seven mAbs, six were found to cross-neutralize hMPV with IC50s ranging from 8.3 to 828 ng/mL and 8.5 to 58.4 ng/mL against hMPV A and B viruses, respectively ().
Table 1. Binding and neutralization properties of isolated mAbs.
To map the targeted antigenic sites of the discovered mAbs, we first categorized the mAbs into five competing groups by biolayer interferometry (BLI)-based epitope binning using reference antibodies with known epitopes (). Four of the groups represent sites V, II, III, Ø, as indicated by the reference mAbs as internal controls. The last group is likely to be site IV, as the mAbs in this group compete with 101F and AM14.
Figure 1. Epitope mapping of isolated mAbs. (a) Epitope binning with RSV PreF antigen. The heatmap shows epitope binning from the sandwich-based BLI binding assay, with darker green color and small number indicating more competition between the 1st and the 2nd antibodies. Gray sidebar on the left shows mAbs with PreF-preferred binding, determined by ELISA (without dashed box) or BLI (in dashed box); colored side-bar on the left shows mAbs of which epitopes were confirmed by site knock-out mutants. (b) Epitope binning with hMPV WT trimer antigen. Similarly, the heatmap shows epitope binning from the sandwich-based BLI binding assay, with darker green color and small number indicating more competition between the 1st and the 2nd antibodies.
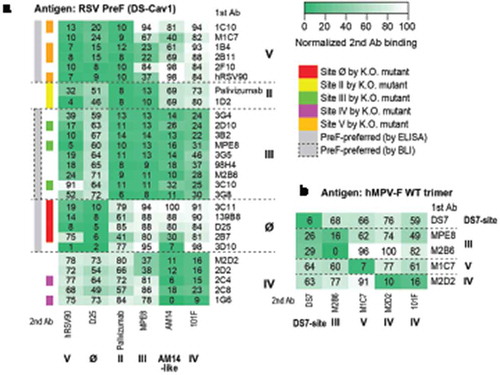
The mapped antigenic sites of some mAbs were further validated by the loss of enzyme-linked immunosorbent assay (ELISA) binding to the corresponding site knock-out mutants (, S3 Fig). Not every mAb was confirmed by site knock-out mutants, suggesting that the mAbs use multiple distinct binding modes to target the same antigenic sites. In addition, as expected, all mAbs mapped to sites V and Ø preferred to bind PreF as compared to PostF, whereas mAbs mapped as site II and IV showed similar binding to PreF and PostF in the ELISA assay (, S2 Fig). The mAbs mapped to site III showed binding to both PreF and PostF in the ELISA assay (, S2 Fig), but had preferred PreF binding by BLI (, S4 Fig, S5 Fig). This apparent discrepancy might result from different assay formats and immobilization methods. The binding preferences of these mAbs to PreF and PostF were consistent with previous reports,Citation22,Citation27,Citation34,Citation43,Citation50 further cross-validating the epitope mapping results. Taken together, combining multiple approaches, all 23 potent RSV nAbs were mapped into five major antigenic sites including sites Ø, II, III, IV, and V.
Site III mAbs exhibited multiple hMPV cross-binding and cross-neutralization modes
All eight antibodies mapped to site III utilized IGLV1-40 gene for light chain, and 7 of 8 used IGHV3-21 gene for heavy chain (, S3 Table), consistent with previous findings.Citation27,Citation35,Citation43 Interestingly, although they share very conserved germline gene usage, their binding and neutralization activities to hMPV were diversified. Three different groups of site III mAbs were classified, as determined by ELISA binding with hMPV F antigens and neutralization with hMPV A and B subtypes (, ): 3G5, 3G4 and 3B2 did not bind to hMPV F antigen and did not neutralize hMPV viruses (group 1); 98H4 bound to hMPV F antigen, but did not show detectable neutralization under tested concentrations (group 2); 3G8, 2D10, 3C10, and M2B6 bound to hMPV F antigens and neutralized both hMPV subtypes (group 3), similarly as previously discovered mAbs such as MPE8.Citation43 Consistent with the ELISA binding data, in a BLI-based binding assay, 3G8 and M2B6 (group 3) showed the tightest binding affinity to hMPV F (KD = 25.3 pM and 114 pM, respectively), 98H4 (group 2) showed lower binding affinity to hMPV F (KD = 4.3 nM), which was largely contributed by a higher dissociation rate, and 3G5 (group 1) did not show binding to hMPV F (S5 Fig).
Table 2. V gene germline usage and somatic hypermutation analysis of isolated site III mAbs.
Figure 2. Characterization of site III mAbs with different binding and neutralization modes to hMPV. (a, b) ELISA binding of site III mAbs to wild-type hMPV-B F antigen and the A314N mutant. Error bars represent the standard deviation of experiment duplicates. (c, d) Critical residues for binding of 3G4, 98H4, and 3G8 mAbs to RSV PreF antigen, determined by alanine scanning library. For each experiment, HEK293T cells expressing the RSV F mutation library were tested for immunoreactivity with mAb of interest, measured on an Intellicyt high-throughput flow cytometer. Clones with reactivity of <30% relative to that of wildtype RSV F (horizontal line) yet >70% reactivity for a control mAb (vertical line) were initially identified to be critical residues for mAb binding (red dots), and were verified using algorithms described elsewhere.Citation52 The mean binding reactivities (and ranges) are shown in the tables. The critical residues are visualized on RSV PreF structure.Citation28.
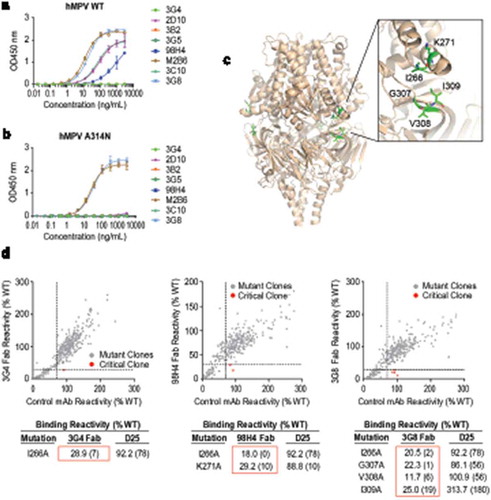
Sequence analysis showed that the differences of hMPV cross-reactivity within these groups of mAbs did not appear to be correlated with the nucleotide or amino acid substitutions in SHM (S6 Fig). To further understand the different binding and cross-neutralization modes among these site III antibodies, one representative mAb from each group was screened by an alanine scanning library of RSV F to determine the most critical residue(s) involved in binding to the antibody of interest. I266 was shown as a critical residue for antigen binding in all three groups (). K271 is only critical for a group 2 mAb, 98H4, which bound but did not potently neutralize hMPV (). Consistent with data previously reported for a site III cross-neutralizing mAb MPE8,Citation43 G307, V308, I309 are found to be critical for the binding of RSV/hMPV cross-neutralization mAbs in group 3, but not as critical for mAbs in groups 1 and 2 (). These results indicate that site III mAbs bind to the RSV PreF antigen in at least three different modes, which may affect their binding to hMPV F antigens and cross-neutralization potency to hMPV viruses. Furthermore, for mAbs in group 3, an A314N mutation on hMPV F antigen, which presumably altered antigenic site III on hMPV F, only disrupted the binding of 3C10 and 2D10, but did not affect the binding of 3G8 and M2B6 (), suggesting that even within the group of RSV/hMPV cross-neutralization mAbs, multiple binding modes exist and might be different for RSV vs. hMPV F binding.
MAbs targeting site IV and V can cross-neutralize hMPV
Most of the isolated mAbs targeting site IV and V only neutralized RSV. However, according to the results of epitope mapping with RSV PreF antigen, two mAbs that cross-neutralized hMPV appeared to target two distinct antigenic sites other than site III (, ). M2D2 was mapped to site IV and competed with 101F, partially competed with MPE8, and did not compete with palivizumab or hRSV90 (). In contrast, M1C7 showed more competition with D25, hRSV90, palivizumab, and less competition to MPE8 and least to 101F. This competition profile is highly similar to that of hRSV90, suggesting that M1C7 targets site V. However, we were not able to confirm the epitopes of these two mAbs with the site knock-out mutants we used (). To validate the epitope mapping results, we further conducted epitope binning experiment on an hMPV F antigen with M2D2, M1C7, two site III cross-neutralizing mAbs MPE8 and M2B6, a site IV cross-neutralizing mAb 101F, and an hMPV-specific mAb DS7 (). As expected, M2D2 shared a similar competition profile to 101F (), confirming that M2D2 targets site IV. M1C7 did not show significant competition with any of these tested mAbs (), suggesting that it indeed targets a distinct epitope from any other known cross-neutralizing mAb. These observations were also consistent with the ELISA and BLI binding results that M2D2 bound RSV PreF and PostF with high affinity, whereas M1C7 only bound to RSV PreF but not RSV PostF (, S5 Fig), as site IV exists in both PreF and PostF conformations, but site V is PreF-specific.
Figure 3. Characterization of two RSV/hMPV cross-neutralizing mAbs targeting site IV and V. (a) ELISA binding of M2D2 and M1C7 to wild-type hMPV-B2 F antigen. (b) Binding of M2D2 and M1C7 on RSV A PreF, RSV B PreF, and RSV B PostF antigens, determined by ELISA with Expi293 cell culture supernatants containing expressed antigens. (c) Neutralization assay of M2D2 and M1C7 to RSV A Long and B Washington strains. (d) Neutralization assay of M2D2 and M1C7 to hMPV A and B subtypes.
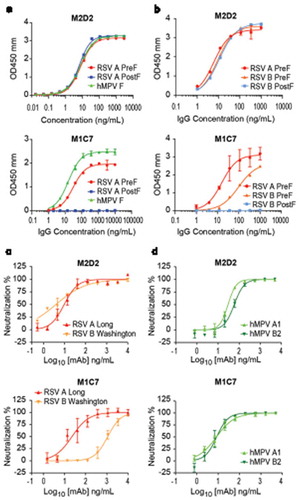
Unlike M2D2 which showed potent neutralization on all four subtypes of RSV and hMPV viruses, M1C7 showed significantly higher neutralization potency only on RSV A, hMPV A and hMPV B (, ). The neutralization potency of M1C7 to RSV B appeared to be significantly weaker (, ). Consistent with the neutralization results, the EC50 of M1C7 binding to an RSV PreF antigen derived from a B strain is 6-fold less than that to DS-Cav1 (98.7 ng/mL vs. 15.9 ng/mL), which is derived from an A strain (). To further investigate the discrepancy of binding and neutralizing activities of M1C7 on RSV A and B strains, we aligned the sequences of RSV A Long and B Washington strains, and found that residues 169 and 201 within the hRSV90 epitope are not conserved ().Citation49 Mutating S169 of RSV A to its equivalent residue N169 in RSV B did not significantly change the binding of M1C7, whereas mutating K201 of RSV A to its equivalent residue N201 in RSV B significantly reduced the binding of M1C7 by 6-fold (2.8 ng/ml vs 16.5 ng/ml, ). Therefore, residue 201 is likely within the epitope that directly interacts with M1C7.
Figure 4. Epitope mapping of M1C7. (a) Binding of M1C7 on RSV A PreF antigen and derived K201N, S169N mutants, determined by ELISA with Expi293 cell culture supernatants containing expressed antigens. Error bars represent the standard deviation of experiment duplicates. (b) Critical residues for M1C7 binding, determined by alanine scanning library. For each experiment, HEK293T cells expressing the RSV F mutation library were tested for immunoreactivity with mAb of interest, measured on an Intellicyt high-throughput flow cytometer. Clones with reactivity of <10% relative to that of wildtype RSV F (horizontal line) yet >70% reactivity for a control mAb (vertical line) for M1C7 were initially identified to be critical residues for mAb binding (red dots), and were verified using algorithms described elsewhere.Citation52 The mean binding reactivities (and ranges) are shown in the tables. (c) The critical residues are visualized on RSV PreF structure.Citation28 (d) Sequence alignment of F antigens in hMPV A2, B2, RSV-A Long, RSV-B Washington strains near the epitope of M1C7. Numbers above sequences indicate the original residue number in the full-length sequence. Arrows indicate the five residues critical for M1C7 binding. The residues were colored in Crystal-X style by MOE.
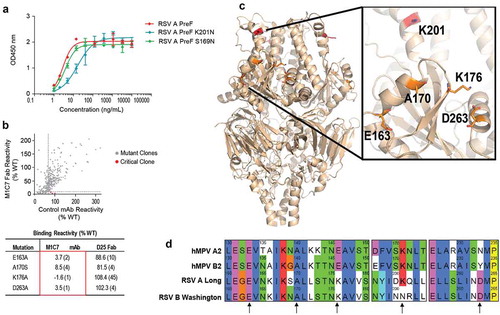
An alanine scanning library of RSV PreF was used to further map the epitopes of M2D2 and M1C7. We showed that G430, I431, and I432 were the most critical residues for M2D2 binding (S7 Fig), suggesting that M2D2 shares the conserved epitope with previously reported cross-neutralizing site IV mAbs 101F, 54G10, and 17E10,Citation36,Citation40,Citation46 as all three residues are conserved between RSV and hMPV.Citation36 E163, A170, K176, and D263 are critical residues for M1C7 to bind RSV F (), further confirming that M1C7 targets antigenic site V. Interestingly, only E163 and A170 are conserved between RSV and hMPV F proteins, whereas K176 and D263 in RSV F are E146 and N/Y233 in HMPV F (). In the crystal structure of RSV PreF,Citation31 K176 interacts with D263. Similarly, in the crystal structure of hMPV PreF,Citation30,Citation48 E146 may pack with N/Y233. Therefore, K176 and D263 might affect the binding of M1C7 through allosteric effects instead of direct binding to the antibody. In addition, alanine mutation on residue K201 did not significantly affect the binding of M1C7 as compared to the K201N mutation described above. High-resolution structural information would be needed to clarify the exact binding interactions between the antigen and antibody. Taken together, our results show the antibody M1C7 targets a novel RSV/hMPV cross-neutralizing epitope belonging to site V.
Site ø mAbs showed a wide range of neutralization potencies on different RSV subtypes
Contrary to site III, IV, and V, the isolated mAbs targeting site II and Ø only neutralized RSV, but not hMPV. Furthermore, site Ø mAbs showed a wide range of potencies on RSV A and RSV B subtypes. For most of the mAbs including sites II, III, IV, and V, except M1C7, the IC50 values for RSV A and RSV B were both below 50 ng/mL () and the ratios of the neutralization IC50 between RSV A Long and RSV B Washington strains were in the range of 0.2 to 1.6 (), indicating that they are equally potent against both RSV subtypes. In contrast, the IC50 ratios of the isolated four site Ø mAbs against RSV A and B ranged from 0.1 to 19.5, suggesting that the neutralization potencies of these site Ø mAbs are more variable on different RSV subtypes. Three of four site Ø mAbs showed significantly weaker neutralization against RSV B Washington strain than RSV A Long strain (). Furthermore, 139B8 is an RSV A strain-specific neutralizing mAb that did not show any detectable neutralization on RSV B Washington strain in the tested concentration range (). This subtype-specificity is determined by a single residue difference at position 201, as we demonstrated by mutating the residue K201 in RSV A PreF to its equivalent residue N201 in RSV B or an alanine, which completely abolished the binding of 139B8 (S8 Fig). The previously reported RSV A-specific mAb 5C4, which was derived from mice, also depends on K201 for its RSV A specificity.Citation47 Our results showed that a similar type of recognition could occur in human B cell repertoire as well.
Discussion
Highly potent nAbs and vaccines that elicit such nAbs are proposed as two potential means for RSV prophylaxis. Therefore, understanding the recognition of such highly potent nAbs to RSV F antigen will provide critical insights in developing efficacious RSV nAbs and vaccines. In this study, we focused on the characterization of a panel of highly potent RSV nAbs isolated from healthy adults. Different from the previous profiling studies, where conjugated quadrivalent RSV F trimers were used,Citation27,Citation35 unconjugated monovalent RSV or hMPV F antigens were used in memory B cell sorting, thus only mAbs with high binding affinity to the antigens were selected. Although these mAbs were mapped to the pre-defined antigenic sites, our characterizations suggested that some of these mAbs target diverse epitopes that were not previously reported.
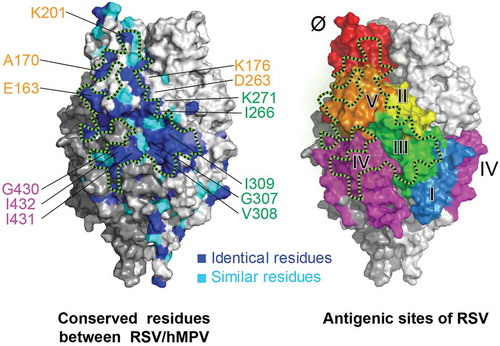
A recent profiling study revealed that the antibody responses in infants younger than 3 months were dominant against site III with highly convergent germline usage in the absence of somatic hypermutations.Citation35 Therefore, vaccination of infants may elicit site III-focused antibody responses at the very young age. Site III is also the most conserved site between RSV and hMPV, targeted by mAbs that cross-neutralize hMPV.Citation27,Citation43,Citation49 It remains unclear whether this site III-focused immune response could also provide cross-protection against hMPV infection. Here, we reported a panel of site III mAbs shared the same germline usage with the previously reported site III mAbs. Interestingly, we found that even with highly convergent germline usage and similarly high-binding affinity to RSV F antigen, these site III mAbs exhibited remarkably different binding and neutralizing specificities to hMPV. Based on screening with an alanine scanning mutant library of RSV F, residue I266 has been identified as a residue likely critical for all site III mAbs we report in this study. This residue was previously identified as part of antigenic site II,Citation53 and our data demonstrated that it also contributes to the binding of various site III mAbs, probably due to the closeness of these two sites in the 3-dimentional structures of F protein. Furthermore, we found that residue K271, which was also previously identified as part of antigenic site II similarly as I266, is a critical residue for a mAb that only binds but does not neutralize hMPV. In addition to I266, residues G307, V308, and I309 appear to be critical for the cross-neutralizing site III mAbs and are located on the opposite side of I266 to K271 (). Therefore, the observed diverse binding and neutralization modes are likely determined by the differences of binding epitopes and angles between site III mAbs and the F antigens. These results suggested that modification of the residues around sites II and III on the F antigen might provide potential improvement for cross-protection against hMPV.
In addition to site III mAbs, we isolated two RSV/hMPV cross-neutralizing mAbs targeting site IV and site V, respectively. Different from previously discovered human mAbs 17E10 and 54G10, which showed potent neutralization on hMPV with weaker neutralization on RSV,Citation36,Citation40 the newly-isolated site IV mAb M2D2 has similarly highly-potent neutralization on all four tested RSV and hMPV subtypes. Although the epitope of M2D2 seems to be similar to 17E10 and the mouse-derived 101F based on mutational studies, the exact interactions between antigen and antibody could be slightly different among these cross-reactive mAbs.Citation36 High resolution structure information might provide additional insights on the different neutralizing potencies of these mAbs to RSV. To our knowledge, the newly isolated PreF-specific mAb M1C7 is the first reported cross-neutralizing mAb that targets antigenic site V. K201, E163, and A170 have been shown as critical residues for M1C7 binding to RSV PreF, and all three residues are conserved in hMPV (). Interestingly, the same K201 residue was identified as the key contact residue for both the site Ø mAb 139B8 and the site V mAb M1C7, highlighting the importance of this residue in RSV A vs. B neutralization specificity. The atomic details of M1C7 epitope through structural determination is warranted to further understand the interactions of the antibodies against different RSV or hMPV antigens.
A limitation of our study is the small number of mAbs generated, as we only focused on highly potent antibodies against RSV. It remains unclear about the frequency of M2D2-like and M1C7-like mAbs in the human repertoire, although a previous study has shown that site IV and site V mAbs are two of the most prevalent antibody classes in the human repertoire against RSV F.Citation27 Interestingly, although many site V RSV nAbs have been isolated,Citation27,Citation34 none of them has been reported to neutralize hMPV. In our study, the site V dual nAb M1C7 was discovered through the isolation of memory B cells using an hMPV antigen (S2 Table). However, the binding affinity of M1C7 to RSV F vs. hMPV F are similar (, S5 Fig.); therefore, one should be able to obtain such an antibody using an RSV F antigen in the B cell sorting, in theory. One possible explanation is the diversity of hMPV neutralizing antibody repertoire among donors in different studies. We did not have records of the history of RSV and hMPV infections for the donors. However, the donor from which M1C7 was derived had very high serum hMPV neutralization titers (S2 Table), suggesting that this individual might have had a recent hMPV infection. This might have resulted in an antibody repertoire enriched with a diverse panel of potent nAbs against hMPV at the time of peripheral blood mononuclear cell (PBMC) collection, increasing the chance of obtaining RSV/hMPV cross-neutralizing antibodies against different epitopes. Further study of profiling hMPV mAbs from multiple donors with high hMPV neutralizing titers will help answer this question.
Taken together, our results are well correlated with the large surface patch formed by conserved residues across sites III, IV, and V for cross-reactive mAbs recognition (). Epitope-focusing on such a surface patch may lead to the design of vaccines that provide broad protections against viruses in Pneumoviridae family. Outside this conserved patch, previous analysis of a large number of sequences revealed that site Ø of RSV F is the most variable site with divergent sequences.Citation54 A recent report of two new site Ø mAbs with different RSV A and B specificities also highlighted the structural variations of this antigenic site.Citation45 Among the isolated highly potent mAbs in this study, we indeed found that mAbs targeting site Ø showed the most variations in neutralization potencies between the two different RSV subtypes, including a strain A-specific mAb 139B8. Similar mAbs were also reported previously from immunized mice and from human adults.Citation27,Citation47 Most of the site Ø mAbs we isolated had a significantly skewed neutralizing potency toward RSV A Long strain rather than RSV B Washington strain. One possible explanation was that most of these mAbs were sorted with the prefusion form of F protein derived from RSV A2 strain, DS-Cav1.Citation28 Our results suggested that additional considerations may need to be given for the vaccinations of different populations with a monovalent subunit vaccine. Vaccination of adults, who have likely been previously exposed to RSV and established immunologic memories, by a monovalent F antigen derived from one subtype is likely to boost existing memory responses. Therefore, the different immune history of each individual might influence the responses elicited by the vaccine. In the infant population who start to exhibit PreF-focused antibody response from naïve immune repertoire after six months,Citation35 vaccination with a monovalent subunit vaccine with a single virus subtype may elicit differences in subtype potencies.
Figure 5. A conserved surface patch on F protein for RSV/hMPV cross-neutralization. Crystal structure of RSV PreF,Citation28 colored by conserved residues (blue and cyan) between RSV and hMPV (left) and antigenic sites (right). Black dashed lines circle a conserved surface patch between RSV/hMPV F antigens. Critical residues identified for antibody binding were labeled and color-coded according to the corresponding antigenic sites.
In summary, we reported here a panel of highly potent human RSV antibodies isolated from healthy adults that could be used in prophylactic or therapeutic applications. Characterization of these antibodies adds to the previously reported diversity among site Ø and IV binding mAbsCitation36,Citation45,Citation47 and extends to additional antigenic sites (III and V). Our data exemplified the diversity of anti-RSV mAbs and provided novel understanding into immune recognition mechanisms of RSV. Our data offers new insights into the development of novel antibodies or vaccines for prophylaxis against respiratory viruses RSV and hMPV.
Materials and methods
Ethics statement
Blood samples from healthy adults were collected with informed written consent obtained in accordance with the Helsinki Declaration of 1975 (approved by the Institutional Review Board of Merck & Co., Inc., Kenilworth, NJ, USA).
Production and purification of recombinant proteins
The plasmid construction and production of RSV PreF (DS-Cav1), including expression and purification, was performed as previously described.Citation28,Citation55 The RSV PreF from a B strain sequence with the same set of DS-Cav1 mutations was also produced similarly (patent application: WO 2014/160463, PCT/US2014/026714). The sequence of RSV PostF antigen was adopted from a previous report.Citation21 The hMPV F trimeric antigen was derived from a previously published sequence of B2 strain with a C-terminal GCN4 trimerization domain, which adopted a PreF-like structure.Citation48 The monomeric hMPV F antigen was derived from the same B2 strain, but without the GCN4 domain. The antigenic site knock-out mutants were based on constructs DS-Cav1 or the hMPV trimeric antigen with additional mutations (S1 Table). All constructs had a C-terminal 6xHis tag and were codon-optimized for mammalian expression (Life Technologies and Genewiz), cloned into an expression vector, and transiently transfected into Expi293 suspension cells (Life Technologies). At day 3 to 7 post transfection, supernatants were harvested for western blot to confirm expression, for direct ELISA binding assay, and for large-scale purification. The purification of all antigens was similar as previously described.Citation55 Briefly, harvested supernatants with His-tagged proteins were captured by Ni-Sepharose chromatography (GE Healthcare) and eluted by high imidazole concentration. After an overnight dialysis in the presence of thrombin, the His-tag was cleaved, and concentration of imidazole was reduced. Cleaved His-tag and uncleaved His-tag products, as well as initial Ni-Sepharose binding impurities, were removed by negative Ni-Sepharose chromatography (product in flow-through). The antigen proteins were further purified by size-exclusion chromatography (Superdex 200, GE Healthcare) and stored in a buffer of 50 mM HEPES pH 7.5 with 300 mM NaCl.
Human subjects and PBMC preparation
Blood samples from healthy adults were collected with informed written consent obtained in accordance with the Helsinki Declaration of 1975 (approved by the Institutional Review Board of Merck & Co., Inc., Kenilworth, NJ, USA). Serum samples from these donors were screened by RSV F ELISA binding and an RSV microneutralization assay. Most of the mAbs (except M2D2, 98H4, and 139B8) discussed here were isolated from donors that showed high serum RSV neutralizing titers (S2 Table). M2D2, 98H4, and 139B8 were isolated from donors whose RSV neutralization titers were not screened. PBMCs were purified from blood collected in EDTA tubes by density gradient centrifugation in histopaque over AccuspinTM tubes (Sigma Aldrich, Cat. No: A2055) according to the manufacturer’s instructions. PBMCs were then frozen in 90% heat-inactivated fetal bovine serum (FBS) supplemented with 10% dimethyl sulfoxide and stored in liquid nitrogen until thawed for use in assays.
Memory B cell enrichment, single B cell sorting, and culture
Human memory B cells from selected donors were either enriched from PBMCs by depleting non-B cells plus IgD+/IgM+ B cells using human memory B cell enrichment kit from STEMCELL Technologies (Cambridge, Cat. No: 19054) following the manufacturer’s instructions or single-cell sorted with specific antigens as previously described.Citation56 For single-cell sorting, the purified recombinant RSV PreF (DS-Cav1), hMPV F trimer and monomer antigen proteins were biotinylated (Thermo, Cat. No: PI21435). Cryopreserved PBMCs were thawed on the day of sorting and first stained with biotinylated F antigens (for the single cell sorting method), and then followed by staining with a panel of commercially available mAbs including anti-CD3 mAb-PE-CyTm 7 (BD Biosciences, Cat. No: 563423), anti-CD19-FITC (BD Biosciences, Cat. No: 561295), anti-human IgG-APC (BD Biosciences, Cat. No: 550931), and PE-streptavidin (BD Biosciences, Cat. No: 349023). CD3−/CD19+/IgG+/F+ cells were sorted with a BD FACS Jazz in single cell mode into a 96-well plate (S2 Table). The enriched or sorted memory B cells were then cultured for 14 days at 37°C, 5% CO2 for conversion into IgG-secreting cells (plasmablasts) as previously described.Citation56 At the end of cultures, plates were centrifuged at 2,000 rpm for 10 min. The culture supernatants were then transferred to new 96-well plates and screened for antigen specificity in ELISA and assessed for RSV and/or hMPV neutralization. The cell pellets were lysed in 50 μl of RLT buffer (Qiagen, Cat. No: 79216) supplemented with 1% 2-mercaptoethanol (Sigma) and stored at −80°C for immunoglobulin gene sequencing.
Recovery of antibody sequences and antibody production
The antibody sequences from the wells with both positive binding and neutralization of RSV or hMPV were further recovered by the method as previously described.Citation56 The naturally paired heavy and light chain variable region sequences obtained from single sorted human memory B cell cultures were sub-cloned into pTT5 vector as full-length human IgG1 for expression in CHO-3E7 cells (performed at GenScript). The recombinant plasmids encoding heavy and light chains of each antibody were transiently co‐transfected into 100 ml suspension CHO‐3E7 cell cultures. The culture supernatants collected on day 6 were used for purification with a Protein A CIP column (GenScript). Monovalent antigen-binding fragments (Fabs) were either expressed and purified directly from CHO-3E7 cells (performed at GenScript) or generated from full-length IgG by papain cleavage (Thermo #44985). The purified antibodies were quality-control tested by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and western blot analysis.
ELISA assay
ELISA assays with purified antigens were conducted with the recombinant purified F proteins coated at 1 μg/mL in phosphate-buffered saline (PBS), on 96-well Nunc-Immuno MaxiSorp plates at 4°C overnight. Plates were then blocked with 3% nonfat milk in PBS with 0.05% Tween-20 (PBST). For ELISA assays with unpurified antigens in Expi293 supernatants, 96-well Ni-NTA coated plates (Thermo Scientific, Cat. No: 15442) were coated with cell culture supernatants for 2 h at room temperature. Plates were then blocked by addition of 2% (v/v) bovine serum albumin (BSA) in PBS. After the blocking step, plates were incubated with serial dilutions of antibodies at room temperature for 90 min. Plates were washed by PBST and then incubated with horseradish peroxidase-conjugated goat anti-human IgG (1: 2,000, Southern Biotech, Cat. no: 2040–05) for 45 min. Plates were washed again and developed with TMB solution (Virolabs, Cat. no: TMB-T-100). Absorbance at 450 nm was read on a plate reader (Victor III; Perkin-Elmer). EC50 values were calculated with Hill slope curve fitting using GraphPad Prism software.
RSV microneutralization assay
The RSV microneutralization assay was performed as previously described.Citation41 Antibodies in serial dilutions with EMEM containing 2% FBS (heated inactivated) were mixed with equal volume (100 pfu) of RSV A Long strain or RSV B Washington strain and incubated for 1 h at 37°C. At the end of incubation, HEp-2 cells (ATCC) were added to the plates at 1.5 × 104 cells/well and the plates were incubated at 37°C with 5% CO2 for 3 days. Afterward, the cells were washed and fixed with ice-cold 80% acetone (Sigma Aldrich, Cat. No: 270725) for 15 min. Mouse anti-RSV-F and anti-RSV-N mAbs (in-house generated, clone 143-F3-1B8 and 34C9, respectively) at 1.25 μg/mL each were added to the plates and incubated for 1 h at room temperature. Plates were then washed and biotinylated horse anti-mouse IgG (Vector Laboratories, Cat. No: Cat# BA-2000) at 1:200 dilution was added. One hour later, the plates were washed and developed by a dual channel near infrared detection system (Licor Odyssey Sa). Infrared Streptavidin-dye to detect RSV specific signal and two cell stains for assay normalization were added to the 96-well plates. Plates were incubated for 1 hr in the dark, washed and dried in the dark for 20 min. Plates were then read on the Licor Aerius® Automated Imaging System utilizing a 700-channel laser for cell normalization and an 800-channel laser for detection of RSV specific signal. 800/700 ratios were calculated, and serum neutralizing titers were determined by 4-parameter curve fit with GraphPad Prism software.
hMPV plaque reduction neutralization assay
A plaque reduction neutralization assay was developed for hMPV in a manner similar to that described previously for RSV.Citation57 Briefly, antibodies in serial dilutions with OptiMEM medium (Gibco, Cat. No: 31985–070) were first added to Poly-D coated 96-well flat bottom plates (Corning Costar, Cat. No: 354640) at 50 μl per well. The hMPV A1, A2, and B2 viruses (ZeptoMetrix Corp) at 2000 pfu/ml were mixed with antibody at 50 μl per well and incubated for 1 h at 37°C with 5% CO2. The LLC-MK2 cells (ATCC) at 0.8 × 106 to 1.2 × 106 cells/ml in OptiMEM medium were then added to the antibody/virus mixtures at 25 μl per well. After 1 h incubation at 37 °C with 5% CO2, the plates were centrifuged at 1,200 rpm for 10 min. 125 μl of OptiMEM medium supplemented with 1% methylcellulose was overlaid in each well. Plates were incubated at 37 °C with 5% CO2 for 4 days. Cells were then fixed with 10% formalin (Fisher Scientific, Cat. No: SF1004) at 100 μl per well for 30 min at room temperature. The plates were dried for 20 min before washing with PBST. Fixed cells were blocked with blocking buffer (Odyssey) for 30 min, followed by 2 h incubation with a mouse anti-hMPV mAb (1:1000, EMD Millipore, Cat. No: MAB80124) diluted in blocking buffer at 50 μl per well. Plates were washed again by PBST and then incubated with an anti-mouse IgG Alexa 488-conjugated secondary antibody (1:500, Invitrogen, Cat. No: A11017) in assay diluent at 50 μl per well for 1 h. After washing off the excessive secondary antibodies by PBST, the plates were read and counted by EnSight (PerkinElmer). The IC50 were calculated from a 4-parameter nonlinear fitting algorithm using Graph Pad Prism software.
Biolayer interferometry-based competition assay and epitope binning
BLI-based competition assays were performed using an Octet Red 96 instrument (ForteBio). Recombinant D25,Citation31 palivizumab,Citation9 MPE8,Citation43 101F,Citation37 hRSV90,Citation34 AM14,Citation33 and DS7Citation48 were used as reference mAbs for antigenic sites Ø, site II, site III, site IV, site V, the quaternary epitope of RSV, and DS7-site of hMPV, respectively. Recombinant DS-Cav1 or hMPV F trimer were used as antigens. All the protein samples were diluted in the kinetics buffer (ForteBio, Cat. No: 18–1105). The antibodies to be characterized or the reference mAbs to be used as internal controls (5 μg/mL) were first individually immobilized on anti-human Fc capture (AHC) biosensors (ForteBio, Cat. No: 18–5064) for 600 sec, followed by blocking with an irrelevant mAb (5 μg/mL) for 600 sec. The biosensors were then immersed into wells containing the F antigen (2 μg/mL). Finally, the antigen-loaded biosensors were immersed into wells containing the second reference mAbs (5 μg/mL) and the binding responses were measured. All experiments were performed at 30°C with shaking at 1,000 rpm. All the binding responses were combined and normalized into 1–100 scales, with the highest response as 100 and lowest response as 0. The normalized numbers were then plotted as clustered heatmaps by heatmap.2 package in R Studio 1.0.153. MAbs with similar competition profiles as the reference mAbs were clustered into the same categories. The binding of RSV PreF/PostF and site knock-out mutants were plotted as sidebars in the heatmaps.
BLI-based assays to measure mAb binding to RSV and hMPV proteins
BLI assays were performed on the Octet Red 96 instrument (ForteBio). IgGs (25 µg/mL) were immobilized onto AHC biosensors (ForteBio, Cat. No: 18–5064). The biosensors were then immersed into wells containing either RSV PreF protein (2 µg/mL) or RSV PostF protein (2 µg/mL or 20 µg/mL). After each binding step, the biosensors were immersed in kinetics buffer (ForteBio, Cat. No: 18–1105) and the baseline interference was read for 60 s in the kinetics buffer. All experiments were performed at 30°C with shaking at 1000 rpm. To measure antibody binding kinetics, BLI assays were performed on the Octet Red 96e instrument (ForteBio). Fabs (M2D2: 0.5 µg/mL, M2B6: 5 µg/mL, 98H4: 5 µg/mL, 3G8: 5 µg/mL, 3G5: 10 µg/mL, M1C7: 10 µg/mL) in 10 mM sodium acetate at pH 5 were immobilized onto AR2G biosensors (ForteBio, Cat. No: 18–5092) by the amine coupling method based on the manufacturer’s instructions, and then equilibrated into a binding buffer (kinetics buffer ForteBio, Cat. No: 18–1105 diluted in PBS + 0.1% BSA). In the association step, the biosensors were immersed into wells containing RSV PreF protein, RSV PostF protein, and hMPV F protein for 300 s, then the biosensors were immersed in the binding buffer for dissociation (600 s or longer). All experiments were performed at 30°C with shaking at 1000 rpm. The binding curves were fit into a 1:1 binding model.
Epitope mapping by alanine scanning library (shotgun mutagenesis)
Epitope mapping for antibodies 3G4, 98H4, 3G8, M2D2, and M1C7 was performed with an RSV F alanine scanning library as previously described by Integral Molecular, Inc. (Philadelphia, USA).Citation41 For screening, the library was expressed in HEK-293T cells (ATCC) and assayed for mAb binding to each clone by immunofluorescence. D25 was used as the control antibody. MAbs were in some cases converted to Fabs by papain digestion for better differentiation of binding. Antibody reactivity against each mutant protein clone was calculated relative to reactivity against the wild-type protein by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type protein-transfected controls. Mutations within clones were identified as critical to the mAb epitope if they did not support reactivity of the test mAb but supported reactivity of other antibodies. This counter-screen strategy facilitated the exclusion of mutants that were misfolded or had an expression defect. The detailed algorithms used to interpret shotgun mutagenesis data are described elsewhere.Citation52
Computational sequence and structural analysis
The V gene germline usage and SHM were analyzed by IgBlastCitation58 based on the Kabat delineation system.Citation51 Sequence alignments of RSV and hMPV F proteins were conducted by MOE 2018.01.01 (Chemical Computing Group). Structural visualization was generated by PyMOL 1.7.05 (Schrödinger).
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the authors.
Supplemental Material
Download MS Word (1.6 MB)Acknowledgments
The authors would like to acknowledge Drs. Barney Graham and Peter Kwong (Vaccine Research Center, National Institute of Allergy and Infectious Diseases) for providing the plasmid encoding RSV-A PreF (DS-Cav1) and RSV-B PreF constructs. We also thank Daniel J. DiStefano, Drs. James C. Cook, Bonnie J. Howell, and MRL Postdoctoral Research Program for their support of this work, and Drs. Amy S. Espeseth and Daria J. Hazuda for helpful discussions and review of this manuscript.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- Afonso CL, Amarasinghe GK, Banyai K, Bao Y, Basler CF, Bavari S, Bejerman N, Blasdell KR, Briand F-X, Briese T, et al. Taxonomy of the order mononegavirales: update 2016. Arch Virol. 2016;161:2351–60. doi:10.1007/s00705-016-2880-1.
- Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, Auinger P, Griffin MR, Poehling KA, Erdman D, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009;360:588–98. doi:10.1056/NEJMoa0804877.
- Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O’Brien KL, Roca A, Wright PF, Bruce N, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–55. doi:10.1016/S0140-6736(10)60206-1.
- Shefali-Patel D, Paris MA, Watson F, Peacock JL, Campbell M, Greenough A. RSV hospitalisation and healthcare utilisation in moderately prematurely born infants. Eur J Pediatr. 2012;171:1055–61. doi:10.1007/s00431-012-1673-0.
- Dowell SF, Anderson LJ, Gary HE Jr., Erdman DD, Plouffe JF, File TM Jr., Marston BJ, Breiman RF. Respiratory syncytial virus is an important cause of community-acquired lower respiratory infection among hospitalized adults. J Infect Dis. 1996;174:456–62. doi:10.1093/infdis/174.3.456.
- Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–59. doi:10.1056/NEJMoa043951.
- Hall CB, Powell KR, MacDonald NE, Gala CL, Menegus ME, Suffin SC, Cohen HJ. Respiratory syncytial viral infection in children with compromised immune function. N Engl J Med. 1986;315:77–81. doi:10.1056/NEJM198607103150201.
- Whimbey E, Ghosh S. Respiratory syncytial virus infections in immunocompromised adults. Curr Clin Top Infect Dis. 2000;20:232–55.
- Prevention of respiratory syncytial virus infections: indications for the use of palivizumab and update on the use of RSV-IGIV. Committee on Infectious Diseases and Committee on Fetus and Newborn, American academy of pediatrics. Pediatrics. 1998; 102: 1211–16. doi:10.1542/peds.102.5.1211.
- Homaira N, Rawlinson W, Snelling TL, Jaffe A. Effectiveness of palivizumab in preventing RSV hospitalization in high risk children: a real-world perspective. Int J Pediatr. 2014;2014:571609. doi:10.1155/2014/928529.
- Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, Wright PF, Crowe JE. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350:443–50. doi:10.1056/NEJMoa025472.
- Williams JV, Tollefson SJ, Heymann PW, Carper HT, Patrie J, Crowe JE. Human metapneumovirus infection in children hospitalized for wheezing. J Allergy Clin Immunol. 2005;115:1311–12. doi:10.1016/j.jaci.2005.02.001.
- van Den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. doi:10.1038/89098.
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785–90. doi:10.1086/367901.
- Esper F, Martinello RA, Boucher D, Weibel C, Ferguson D, Landry ML, Kahn JS. A 1-year experience with human metapneumovirus in children aged <5. J Infect Dis. 2004;189:1388–96. doi:10.1086/382482.
- Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, Peret T, Erdman D, Anderson L. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis. 2002;186:1330–34. doi:10.1086/jid.2002.186.issue-9.
- Graham BS. Biological challenges and technological opportunities for respiratory syncytial virus vaccine development. Immunol Rev. 2011;239:149–66. doi:10.1111/j.1600-065X.2010.00972.x.
- Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–19. doi:10.1038/nrm1076.
- Earp LJ, Delos SE, Park HE, White JM. The many mechanisms of viral membrane fusion proteins. Curr Top Microbiol Immunol. 2005;285:25–66.
- Swanson KA, Settembre EC, Shaw CA, Dey AK, Rappuoli R, Mandl CW, Dormitzer PR, Carfi A. Structural basis for immunization with postfusion respiratory syncytial virus fusion F glycoprotein (RSV F) to elicit high neutralizing antibody titers. Proc Natl Acad Sci U S A. 2011;108:9619–24. doi:10.1073/pnas.1106536108.
- McLellan JS, Yang Y, Graham BS, Kwong PD. Structure of respiratory syncytial virus fusion glycoprotein in the postfusion conformation reveals preservation of neutralizing epitopes. J Virol. 2011;85:7788–96. doi:10.1128/JVI.00555-11.
- McLellan JS, Ray WC, Peeples ME. Structure and function of respiratory syncytial virus surface glycoproteins. Curr Top Microbiol Immunol. 2013;372:83–104. doi:10.1007/978-3-642-38919-1_4.
- Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc Natl Acad Sci U S A. 2013;110:11133–38. doi:10.1073/pnas.1309070110.
- Rossey I, McLellan JS, Saelens X, Schepens B. Clinical Potential of Prefusion RSV F-specific Antibodies. Trends Microbiol. 2018;26:209–19. doi:10.1016/j.tim.2017.09.009.
- Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang G-Y, et al. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med. 2015;7:309ra162. doi:10.1126/scitranslmed.aad3106.
- Magro M, Mas V, Chappell K, Vazquez M, Cano O, Luque D, Terrón MC, Melero JA, Palomo C. Neutralizing antibodies against the preactive form of respiratory syncytial virus fusion protein offer unique possibilities for clinical intervention. Proc Natl Acad Sci U S A. 2012;109:3089–94. doi:10.1073/pnas.1115941109.
- Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, Mas V, Melero JA, Wright PF, Graham BS, et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol. 2016;1:6. doi:10.1126/sciimmunol.aaj1879.
- McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–98. doi:10.1126/science.1243283.
- Krarup A, Truan D, Furmanova-Hollenstein P, Bogaert L, Bouchier P, Bisschop IJ, Widjojoatmodjo MN, Zahn R, Schuitemaker H, McLellan JS, et al. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun. 2015;6:8143. doi:10.1038/ncomms9143.
- Battles MB, Mas V, Olmedillas E, Cano O, Vazquez M, Rodriguez L, Melero JA, McLellan JS. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun. 2017;8:1528. doi:10.1038/s41467-017-01708-9.
- McLellan JS, Chen M, Leung S, Graepel KW, Du X, Yang Y, Zhou T, Baxa U, Yasuda E, Beaumont T, et al. Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science. 2013;340:1113–17. doi:10.1126/science.1234914.
- Zhu Q, McLellan JS, Kallewaard NL, Ulbrandt ND, Palaszynski S, Zhang J, Moldt B, Khan A, Svabek C, McAuliffe JM, et al. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci Transl Med. 2017;9:388. doi:10.1126/scitranslmed.aaj1928.
- Gilman MS, Moin SM, Mas V, Chen M, Patel NK, Kramer K, Zhu Q, Kabeche SC, Kumar A, Palomo C, et al. Characterization of a prefusion-specific antibody that recognizes a quaternary, cleavage-dependent epitope on the RSV fusion glycoprotein. PLoS Pathog. 2015;11:e1005035. doi:10.1371/journal.ppat.1005035.
- Mousa JJ, Kose N, Matta P, Gilchuk P, Crowe JE Jr. A novel pre-fusion conformation-specific neutralizing epitope on the respiratory syncytial virus fusion protein. Nat Microbiol. 2017;2:16271. doi:10.1038/nmicrobiol.2016.251.
- Goodwin E, Gilman MSA, Wrapp D, Chen M, Ngwuta JO, Moin SM, Bai P, Sivasubramanian A, Connor RI, Wright PF, et al. Infants infected with respiratory syncytial virus generate potent neutralizing antibodies that lack somatic hypermutation. Immunity. 2018;48:339–49 e5. doi:10.1016/j.immuni.2018.01.005.
- Mousa JJ, Binshtein E, Human S, Fong RH, Alvarado G, Doranz BJ, Moore ML, Ohi MD, Crowe JE, Subbarao K. Human antibody recognition of antigenic site IV on Pneumovirus fusion proteins. PLoS Pathog. 2018;14:e1006837. doi:10.1371/journal.ppat.1006837.
- McLellan JS, Chen M, Chang JS, Yang Y, Kim A, Graham BS, Kwong PD. Structure of a major antigenic site on the respiratory syncytial virus fusion glycoprotein in complex with neutralizing antibody 101F. J Virol. 2010;84:12236–44. doi:10.1128/JVI.01579-10.
- Ulbrandt ND, Ji H, Patel NK, Riggs JM, Brewah YA, Ready S, Donacki NE, Folliot K, Barnes AS, Senthil K, et al. Isolation and characterization of monoclonal antibodies which neutralize human metapneumovirus in vitro and in vivo. J Virol. 2006;80:7799–806. doi:10.1128/JVI.00318-06.
- Williams JV, Chen Z, Cseke G, Wright DW, Keefer CJ, Tollefson SJ, Hessell A, Podsiad A, Shepherd BE, Sanna PP, et al. A recombinant human monoclonal antibody to human metapneumovirus fusion protein that neutralizes virus in vitro and is effective therapeutically in vivo. J Virol. 2007;81:8315–24. doi:10.1128/JVI.00106-07.
- Schuster JE, Cox RG, Hastings AK, Boyd KL, Wadia J, Chen Z, Burton DR, Williamson RA, Williams JV. A broadly neutralizing human monoclonal antibody exhibits in vivo efficacy against both human metapneumovirus and respiratory syncytial virus. J Infect Dis. 2015;211:216–25. doi:10.1093/infdis/jiu307.
- Chen Z, Zhang L, Tang A, Callahan C, Pristatsky P, Swoyer R, Cejas P, Nahas D, Galli J, Cosmi S, et al. Discovery and characterization of phage display-derived human monoclonal antibodies against RSV F glycoprotein. PLoS One. 2016;11:e0156798. doi:10.1371/journal.pone.0156798.
- Johnson S, Oliver C, Prince GA, Hemming VG, Pfarr DS, Wang SC, Dormitzer M, O’Grady J, Koenig S, Tamura JK, et al. Development of a humanized monoclonal antibody (MEDI-493) with potent in vitro and in vivo activity against respiratory syncytial virus. J Infect Dis. 1997;176:1215–24. doi:10.1086/514115.
- Corti D, Bianchi S, Vanzetta F, Minola A, Perez L, Agatic G, Guarino B, Silacci C, Marcandalli J, Marsland BJ, et al. Cross-neutralization of four paramyxoviruses by a human monoclonal antibody. Nature. 2013;501:439–43. doi:10.1038/nature12442.
- Bar-Peled Y, Diaz D, Pena-Briseno A, Murray J, Huang J, Tripp RA, Mousa JJ. A potent neutralizing site III-specific human antibody neutralizes human metapneumovirus in vivo. J Virol. 2019. doi:10.1128/JVI.00342-19.
- Jones HG, Battles MB, Lin CC, Bianchi S, Corti D, McLellan JS, Crowe JE. Alternative conformations of a major antigenic site on RSV F. PLoS Pathog. 2019;15:e1007944. doi:10.1371/journal.ppat.1007944.
- Mas V, Rodriguez L, Olmedillas E, Cano O, Palomo C, Terron MC, Luque D, Melero JA, McLellan JS, Plemper RK. Engineering, structure and immunogenicity of the human metapneumovirus F protein in the postfusion conformation. PLoS Pathog. 2016;12:e1005859. doi:10.1371/journal.ppat.1005859.
- Tian D, Battles MB, Moin SM, Chen M, Modjarrad K, Kumar A, Kanekiyo M, Graepel KW, Taher NM, Hotard AL, et al. Structural basis of respiratory syncytial virus subtype-dependent neutralization by an antibody targeting the fusion glycoprotein. Nat Commun. 2017;8:1877. doi:10.1038/s41467-017-01858-w.
- Wen X, Krause JC, Leser GP, Cox RG, Lamb RA, Williams JV, Crowe JE, Jardetzky TS. Structure of the human metapneumovirus fusion protein with neutralizing antibody identifies a pneumovirus antigenic site. Nat Struct Mol Biol. 2012;19:461–63. doi:10.1038/nsmb.2250.
- Wen X, Mousa JJ, Bates JT, Lamb RA, Crowe JE Jr., Jardetzky TS, Reyes JCC, Russell DA, Adair T, Alvey R, et al. Structural basis for antibody cross-neutralization of respiratory syncytial virus and human metapneumovirus. Nat Microbiol. 2017;2:16272. doi:10.1038/nmicrobiol.2016.251.
- Palomo C, Mas V, Thom M, Vazquez M, Cano O, Terron MC, Luque D, Taylor G, Melero JA. Influence of respiratory syncytial virus F glycoprotein conformation on induction of protective immune responses. J Virol. 2016;90:5485–98. doi:10.1128/JVI.00338-16.
- Kabat EA, Wu TT. Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J Immunol. 1991;147:1709–19.
- Davidson E, Doranz BJ. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi:10.1111/imm.12323.
- McLellan JS, Chen M, Kim A, Yang Y, Graham BS, Kwong PD. Structural basis of respiratory syncytial virus neutralization by motavizumab. Nat Struct Mol Biol. 2010;17:248–50. doi:10.1038/nsmb.1723.
- Mas V, Nair H, Campbell H, Melero JA, Williams TC. Antigenic and sequence variability of the human respiratory syncytial virus F glycoprotein compared to related viruses in a comprehensive dataset. Vaccine. 2018;36:6660–73. doi:10.1016/j.vaccine.2018.09.056.
- Zhang L, Durr E, Galli JD, Cosmi S, Cejas PJ, Luo B, Touch S, Parmet P, Fridman A, Espeseth AS, et al. Design and characterization of a fusion glycoprotein vaccine for respiratory syncytial virus with improved stability. Vaccine. 2018;36:8119–30. doi:10.1016/j.vaccine.2018.10.032.
- Cox KS, Tang A, Chen Z, Horton MS, Yan H, Wang XM, Dubey SA, DiStefano DJ, Ettenger A, Fong RH, et al. Rapid isolation of dengue-neutralizing antibodies from single cell-sorted human antigen-specific memory B-cell cultures. MAbs. 2016;8:129–40. doi:10.1080/19420862.2015.1109757.
- Wen Z, Citron M, Bett AJ, Espeseth AS, Vora KA, Zhang L, DiStefano DJ. Development and application of a higher throughput RSV plaque assay by immunofluorescent imaging. J Virol Methods. 2019;263:88–95. doi:10.1016/j.jviromet.2018.10.022.
- Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–40. doi:10.1093/nar/gkt382.
