ABSTRACT
The respiratory virus responsible for coronavirus disease 2019 (COVID-19), severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has affected nearly every aspect of life worldwide, claiming the lives of over 3.9 million people globally, at the time of this publication. Neutralizing humanized nanobody (VHH)-based antibodies (VHH-huFc) represent a promising therapeutic intervention strategy to address the current SARS-CoV-2 pandemic and provide a powerful toolkit to address future virus outbreaks. Using a synthetic, high-diversity VHH bacteriophage library, several potent neutralizing VHH-huFc antibodies were identified and evaluated for their capacity to tightly bind to the SARS-CoV-2 receptor-binding domain, to prevent binding of SARS-CoV-2 spike (S) to the cellular receptor angiotensin-converting enzyme 2, and to neutralize viral infection. Preliminary preclinical evaluation of multiple VHH-huFc antibody candidates demonstrate that they are prophylactically and therapeutically effective in vivo against wildtype SARS-CoV-2. The identified and characterized VHH-huFc antibodies described herein represent viable candidates for further preclinical evaluation and another tool to add to our therapeutic arsenal to address the COVID-19 pandemic.
Introduction
The novel respiratory virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has infected over 146 million and killed over 3 million people worldwide,Citation1 causing the worst global health crisis since the 1918–1919 influenza pandemic. In addition to the lives lost to COVID-19, this virus has wreaked havoc on the global economy and highlighted the threat that emerging diseases pose to global security.Citation2 It is important not only to work to develop effective vaccines for pre-exposure prophylaxis, but to create new treatments to prevent and mitigate severe disease.
Viral neutralizing antibodies are an effective therapeutic intervention for COVID-19, as the current pandemic response has shown. High titer convalescent plasma has emergency use authorization by the U.S. Food and Drug Administration (FDA) for the treatment of hospitalized patients early in the disease course and/or with impaired humoral immunity; however, batch-to-batch variability results in various levels of success, limiting its reliability as a treatment.Citation3 Monoclonal antibody therapies, like convalescent plasma, block cell entry, the first step of virus infection, but consist only of highly neutralizing antibodies with high target specificity, and more favorable pharmacokinetics.Citation4–7 Recently, promising clinical trial data demonstrated that a single intravenous infusion of monoclonal antibody (mAb) cocktail significantly reduced COVID-19 related hospitalization and death in comparison to placebo.Citation8,Citation9 These results and other clinical trial findings supported FDA’s emergency use authorization of three mAb cocktails for people 12 y and older who test positive for SARS-CoV-2 and are at high risk for progressing to severe COVID-19.Citation8,Citation9 Methods for improved development and characterization of novel neutralizing antibodies can be part of a critical toolset to combat the COVID-19 pandemic and future epidemics, by providing lower cost, easier manufacturability, and diverse functionality, including response to emerging variants.
Nanobodies (VHH) are the variable region of single-domain heavy chain only antibodies (sdAbs), which lack the light chain and the CH1 domain, are derived from camelids, and are smaller (~75 kDa) than human or murine immunoglobulin G (IgG) antibodies (150kDa).Citation10 VHH antibodies are highly soluble, stable, extremely versatile, and have unique structural attributes in their complementarity-determining region 3 (CDR3) loop that can facilitate binding to antigen sites inaccessible to traditional IgG antibodies.Citation11,Citation12 Neutralization of SARS-CoV-2 is achieved by targeting antibodies to the spike (S) receptor-binding domain (RBD), which engages the angiotensin-converting enzyme (ACE2) receptor to facilitate cell entry. The S protein is heavily glycosylated, limiting viable epitope availability with potential for therapeutic efficacy.Citation13 Compact VHH-based antibodies may have access to alternate epitopes on the SARS-CoV-2 S protein that are sterically inaccessible to traditional antibodies.
Herein, we describe a rapid discovery process for SARS-CoV-2 VHH-based antibody therapeutics, from molecular discovery of high-affinity variable regions to demonstration of therapeutic efficacy of fully humanized VHH-based antibodies in mice infected with SARS-CoV-2. We describe the construction of a high-diversity synthetic humanized VHH phage library (~3.2 x 1010) that was used to identify several VHH antibodies that are high-affinity binders to SARS-CoV-2 S protein and RBD. The top VHH candidates were produced as human sdAbs with the crystallizable fragment (Fc) domain of a human IgG1 (VHH-huFc) and were evaluated for their ability to block the interaction between purified SARS-CoV-2 S and the ACE2 receptor. Effective VHH-huFc antibodies were then screened for their ability to block infection of Vero cells with SARS-CoV-2 pseudotyped vesicular stomatitis virus (VSV-SARS-CoV-2-GFP), followed by verification of neutralization of wildtype (wt) SARS-CoV-2 (USA-WA 1/2020). The epitope targets were mapped by sequencing neutralization escape mutants generated in the presence of each VHH-huFc. The binding affinity of the VHH-huFc antibodies to RBD with mutations generated in this study, including the L452R mutation found in the Epsilon (B.1.427 and B.1.429), Kappa (B.1.617.1) and Delta (B.1.617.2) variants, or from the Alpha (B.1.1.7) and Beta (B.1.351) variants, was assessed with kinetic studies using biolayer interferometry (BLI). We also demonstrate that two of these VHH-huFc antibodies show efficacy in vivo both prophylactically and therapeutically against challenge with fully virulent wt SARS-CoV-2.
Results
Library construction
The VHH library incorporated both the diversity and prevalence of amino acids at key positions in each of the CDR1 and CDR2 derived from a single domain antibody (sdAb) database ().Citation14 This database contains sequences of validated sdAbs or nanobodies from both synthetic and natural sources from both naïve and immune repertoires. This database has been curated using sequences derived from existing databases (Protein Data Bank and NCBI) and publications. We examined the amino acid diversity and prevalence at each position in CDR 1 and 2 from 670 of these nanobodies and incorporated this into our custom library synthesis. FCDR3, full amino acid diversity was included, and three different lengths were used: 9-, 12-, and 15-amino acids. The length for each CDR 1 and 2 was based on the predominant lengths of these loops observed in the functional sdAb database. (Figure S1). Cysteines and methionines were omitted from all CDRs. The sequence used for the framework to house the custom made CDRs, hs2dAb, was derived from Moutel et al. In this framework, multiple residues are changed such that the framework more closely mirrors germline human VH3 immunoglobin.Citation15 The library was synthetically produced and the observed prevalence of amino acids at each of the CDR loops paralleled the desired prevalence as determined by next-generation sequencing (NGS; data not shown).
Figure 1. Development of high-diversity VHH library and screening campaign for SARS-CoV-2 neutralizing VHHs. A) Library design using deposited sequences of functional VHHs from various sources. Data was mined to determine optimal CDR length and amino acid prevalence at each amino acid position within each respective CDR. B) Schematic for the biopanning approach used to identify neutralizing VHHs directed to the SARS-CoV-2 RBD. C) Polyclonal ELISA against multiple antigens show enrichment of a positive SARS-CoV-2 binding population of displayed VHH with each sequential round of panning. Data is from experimental conditions performed in triplicate; the error is the standard deviation from the mean. D) Neutralization of VSV-SARS-CoV-2 GFP viral infection with polyclonal VHH displaying phage from each round of panning. Data is from experimental conditions performed in triplicate; the error is the standard deviation from the mean (***, P < .001; **, P < .01; *, P < .05). The data represent one of three experiments with similar results
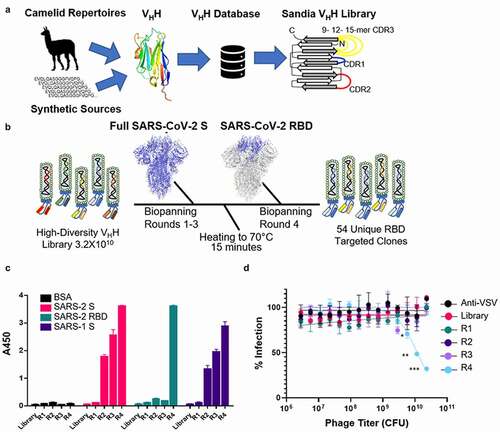
To obtain sufficient diversity coverage for the library (i.e., transformants), 150 electroporations were performed yielding approximately 3.38 × 1010 transformants. To determine the level of success for the ligation of the library into the vector backbone, colony PCR was performed. Of the 408 colonies selected, 395 contained the correct size amplified DNA fragment (95.9%). This value was used to adjust the calculated value for library diversity to 3.24 × 1010. Finally, library diversity, quality, and the distribution of CDR3 lengths were assessed by NGS from a total of 39,870,360 reads (Tables S1 and S2). The 9-amino acid CDR3 was the most prevalent at 40%, followed by 12-amino acid CDR3 at 34%, and lastly the 15-amino acid CDR3 at 25% of the observed diversity. Overall, there was good coverage of all represented CDR3s. Approximately 1% of sequences contained a stop codon and 99% of reads were unique sequences (38,592,027 reads). Roughly 1% of reads were duplicates, and 0.01% (1,095 sequences) were present in triplicate. With these corrections the adjusted diversity for this nanobody library is 3.18 × 1010.
Panning against SARS-COV-2 S and RBD
Four rounds of biopanning were used to identify clones that bind to the SARS-CoV-2 S protein RBD. For the first three rounds, full-length soluble purified SARS-CoV-2 S protein confirmed as a trimer was used, ensuring that conformational integrity of the RBD was maintained for initial selection. A 15-minute heat denaturing step at 70°C was used to remove unstable sequences and a final round of biopanning against SARS-CoV-2 RBD was conducted to identify therapeutically relevant VHH antibodies (). Enrichment of phage against SARS-CoV-2 S was observed over the initial three rounds of biopanning ( and Table S3) and there was a significant loss in phage recovered when the antigen was shifted to RBD (0.0005% compared to 0.004%).
Polyclonal ELISA showed that enrichment for SARS-CoV-2 S binders occurred over each round of biopanning. Interestingly, there was relatively little SARS-CoV-2 RBD binding until panning was conducted against RBD specifically, indicating preferred epitopes are outside the RBD (). Additionally, neutralization of SARS-CoV-2 pseudotyped vesicular stomatitis virus encoding enhanced green fluorescent protein (eGFP) (VSV-SARS2-GFP)Citation16 was only observed after RBD biopanning (R4), showing enrichment for RBD binders is required to remove clones binding to epitopes outside the RBD and to identify virus neutralizing clones (). Monoclonal characterization was performed using ELISA; 222 of the 384 clones were designated as hits. Clones that had OD450 ≥ 2.5 in the phage coat monoclonal ELISA against both SARS-CoV-2 S and RBD and that did not bind bovine serum albumin (BSA) were designated as hits (Figure S2). Of those designated as hits, 54 sequences were identified as unique.
Evaluation of VHH-huFc candidates
Humanized VHH-huFc antibodies were produced as a fusion to the hinge region and the Fc domain of human IgG1, which combines the advantages of the VHH with the improved half-life and effector functions of human IgG1 while reducing the overall size by half that of a conventional antibody, improving circulation and increasing their ability to penetrate target tissues in the body.Citation12 All 54 unique VHH sequences were commercially produced in a VHH-huFc format by GenScript for further characterization and triage (). Of the 54 VHH-huFc antibodies, 49 showed positive binding to both SARS-CoV-2 S and RBD, with little to no binding to BSA by ELISA (Figure S3). Those that showed positive binding by ELISA were then tested for their ability to compete with ACE2-huFc for binding to SARS-CoV-2 S by a competition ELISA (Table S4). Finally, 46 candidates were evaluated for their ability to neutralize VSV-SARS-CoV-2-GFP infection in vitro, with 34 VHH-huFc antibodies demonstrating full or partial neutralization of infection (Table S4).
Figure 2. Evaluation of VHH-huFc candidates. A) Triage flowchart for selection of top candidates, from clonal phage ELISA to selection of 16 candidates for reconfirmation. B) Reconfirmation ELISA of top three candidates binding to SARS-CoV-2 S and C) SARS-CoV-2 RBD. D) Top candidate VHH-huFcs compete with ACE2-huFc for binding to SARS-CoV-2 S by competition ELISA and E) neutralize VSV-SARS-CoV-2 GFP infection of Vero cells. The data for a–e are from experimental conditions performed in triplicate, the error is the standard deviation from the mean. F) All three VHH-huFcs also neutralize wt SARS-CoV-2 in a plaque reduction neutralization assay. The data from this is the mean of the plaque assay performed in duplicate, the error is the standard deviation from the mean. BLI sensorgrams show binding to SARS-CoV-2 RBD by SP1B4 (g), SP1D9 (h), and SP3H4 (i). At least five concentrations were used for global fit analysis. J) Table summarizing kinetic properties, neutralizing efficacy, and thermal stability properties for SP1B4, SP1D9, and SP3H4. Melting temperatures from DSF were determined from two independent experiments each performed in triplicate
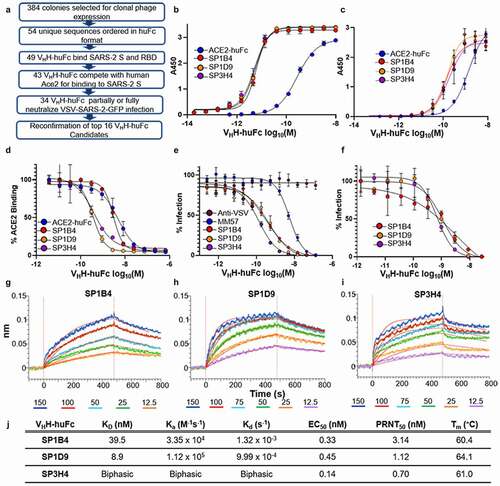
For reconfirmation of binding and neutralization, 16 VHH-huFc antibodies were selected based on low 50% effective inhibitory concentration (EC50) values and were produced and purified in-house (Figure S4). None of these candidates inhibited infection with pseudotyped, single-cycle VSV particles displaying the SARS-1 S protein (VSV-SARS-1-GFP), indicating specificity for SARS-CoV-2 S protein (Figure S4E). Three of the 16 VHH-huFc antibodies, SP1B4, SP1D9, and SP3H4, were identified as showing the greatest potency in viral neutralization with EC50s of 0.33, 0.45, and 0.14 nM, respectively, against VSV-SARS-CoV-2-GFP and EC50s of 3.14, 1.12, and 0.70 nM, respectively, against wtSARS-CoV-2 by a plaque neutralization assay (,)). The neutralization values were obtained with three independent virus-based assays (Table S5), suggesting that SP1B4, SP1D9, and SP3H4 are potent neutralizers of SARS-CoV-2, and this observed potency correlates with their strong competition with ACE2 for binding to SARS-CoV-2 S protein (). The thermal stability of VHH-huFc antibodies was evaluated by differential scanning fluorimetry (DSF). Melting temperatures for SP1B4, SP1D9, and SP3H4, 60.4°C, 64.0°C, and 61.0°C, respectively, are comparable to previously reported SARS-CoV-2 neutralizing VHH-huFc antibodies (; S5).Citation17
Dissociation constants were determined for the VHH-huFc antibodies using BLI, revealing high affinity for the SARS-CoV-2 RBD of 39.5 nM and 8.9 nM for SP1B4 and SP1D9, respectively (). SP3H4 exhibited biphasic association and dissociation kinetics that could not be reconciled with a 1:1 global fit analysis, and it was therefore not given kinetic parameters ().
Epitope mapping by escape mutant formation and BLI
Epitopes were mapped by sequencing VSV-SARS-CoV-2-GFP virus containing escape mutants generated in the presence of SP1B4, SP1D9, or SP3H4, followed by confirmation with BLI binding studies of escape mutant SARS-CoV-2 RBDs (). Within 48–60 hours of the first passage, viral escape was apparent from all VHH-huFc antibodies at the highest concentration tested, with all cells expressing GFP. The viral supernatant used for the second passage was completely resistant to neutralization from a 15-fold excess of the corresponding VHH-huFc (). Escape mutations were characterized by RNAseq for each of the VSV-SARS-CoV-2-GFP supernatants (). Interestingly, for SP1B4 and SP1D9, E484K (the mutation found in the Beta variant) seems to be selected against in the second passage and Q493R and S494P seem to stabilize as the predominant mutations observed (). This may indicate these two VHH-huFc antibodies have similar binding modes. For SP3H4, the predominant escape mutation observed was L452R, and no shared mutations with SP1B4 and SP1D9 were detected.
Figure 3. Genereation and Characterization of VHH-huFc Neutralization Escape Mutants. A) Subdomains of the SARS-CoV-2 S gene are color coded with correspoding structural context (PDB: 7CAK). Loci of mutations on S gene from NGS evaluation of virus supernatant from virus infected cells passaged in the presence of VHH-huFc antibodies. In this structure the RBD is in the “up” position. B) The RBD is designated with the dominant mutations characterized in this study (magenta). C) VSV-SARS-CoV-2-GFP neutralization escape occurs in a single passage. VSV-SARS-CoV-2-GFP neutralization is recorded 12 hpi. The black arrow designated the well from which the virus-containing supernatant was taken and diluted for the second passage. Data for this is from a single replicate. D) NGS and kinetic summary of each of the neutralization escape mutant profiles for SP1B4, SP1D9, and SP3H4. Dissociation constants for each individual RBD mutation with ACE2-huFc and VHH-huFc antibodies. N.B. = no binding observed at 150 nM SARS-CoV-2 RBD by BLI (Supplementary Figure 7–9). Sensorgrams were designated biphasic if they did not agree with a global 1:1 fit analysis. The prevalence of each escape mutation is presented as a percentage at that amino acid postion and is conditionally colored with increasing intesity of red
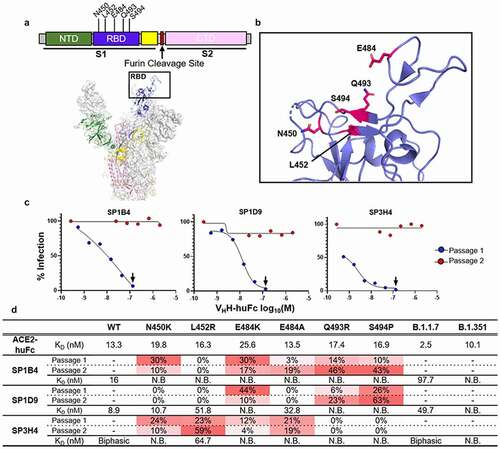
SARS-CoV-2 RBD variants with single mutations were produced and their affinity for human ACE2 and the three VHH-huFc antibodies was determined by BLI (Figure S6-S9 and Table S6). In all cases, individual mutations were shown to have little to no impact on ACE2 binding, suggesting minimal impact on viral infection. Mutations in the RBD significantly affected VHH-huFc binding. Interestingly, SP3H4 maintained the ability to bind to L452R, despite this being the predominant mutation observed in the escape mutant VSV-SARS-CoV-2-GFP virus generated. Additionally, BLI results show 1:1 binding kinetics for SP3H4 binding to the L452R mutant RBD. The biphasic kinetics observed with SP3H4 and wt RBD combined with the 1:1 binding kinetics of SP3H4 for RBD with the L452R mutation may indicate that SP3H4 has multiple binding sites. SP1D9 maintained the ability to bind many of the point mutations generated, though binding was abrogated by Q493R and S494P () as expected based on the escape mutant analysis. All mutations generated in RBD abrogated binding by SP1B4.
Next, SARS-CoV-2 RBDs from the Alpha and Beta variants were characterized for affinity to the human ACE2 receptor and top VHH-huFc antibodies identified in this study (Figure S6-S9 and Table S6). The Alpha variant RBD showed an ~5-fold increased affinity to the ACE2 receptor (KD = 2.5 nM vs 13.3 nM for WT). While SP1B4 (KD = 97.7 nM) and SP1D9 (KD = 49.7 nM) maintained binding affinity for the Alpha RBD, there was a 2.5 -fold and 5.5 -fold decrease in affinity in comparison to the wt or Wuhan RBD (KD = 39.5 nM, and 8.9 nM, respectively, (Figure S7-S8) and a visible difference in binding to this RBD for SP3H4 (Figure S9). For the SARS-CoV-2 RBD from the Beta variant, the KD for ACE2 was similar to the KD for the Wuhan strain, but there was no detectable binding for SP1B4, SP1D9, or SP3H4 (Figure S7-S9).
Preclinical evaluation of VHH-huFc antibodies:
Top performing VHH-huFc antibodies SP1B4, SP1D9 and SP3H4 were assessed for in vivo prophylactic (−24 hours) and therapeutic (+24 hours) efficacy against fully virulent wt SARS-CoV-2 using the recently described K18-hACE2 transgenic mouse model of SARS-CoV-2 infection.Citation18 Two of the three VHH-huFc antibodies tested, SP1D9 and SP3H4, provided very significant protection (p < .0001) over the isotype control in prophylactically dosed mice. Although SP1B4 was a potent neutralizer in vitro, it did not demonstrate efficacy against challenge with SARS-CoV-2 in vivo (,) and was not pursued in further in vivo studies. The SP1D9 group showed 94% survival (n = 16), and the SP3H4 group showed 87.5% survival (n = 16) at 10 d post infection (dpi), indicating effective neutralizing activity in vivo (). While 81.3% (n = 16) of control-treated mice lost more than 20% of their starting weight by 6 dpi, requiring euthanasia, those pretreated with SP1D9 lost an average of only 3.9% (±4.8%) of their starting weight ( and Figure S10). Similarly, the SP3H4-dosed group exhibited a 5.4% average (±9.3%) weight loss. Animals dosed prophylactically with the poorest performing VHH-huFc, SP1B4, lost an average of 17.7% (±8.3%) of their starting weight, with 50% of animals succumbing to infection by 6 dpi ( & Figure S10).
Figure 4. Top VHH-huFc candidates provide protection from lethal SARS-CoV-2 infection in vivo. Kaplan–Meier curve illustrating percentage survival of K18-hACE2 mice infected intranasally with 2.5 × 104 PFU SARS-CoV-2 and dosed with 10 mg/kg VHH-huFc via intraperitoneal injection prophylactically (a) at 24 hours prior to infection or therapeutically (c) at 24 hpi. A log rank test was performed with Bonferroni multiple comparison correction applied. B) Percentage weight loss for mice in (A). D) Percentage weight loss for mice in (C). For (b) and (d), statistical analyses were performed at time points when all mice were alive to avoid survivor bias. Data displayed are the mean ± standard error of the mean. A & B) Isotype control n = 16, SP1B4 (−24 hpi) n = 8, SP1D9 (−24 hpi) n = 16, SP3H4 (−24 hpi) n = 16. C & D) Isotype control n = 8, SP1D9 (+24 hpi) n = 8, SP3H4 (+24 hpi) n = 8. Data represent two experiments (**** p < .0001; ** p < .01; ns = not significant)
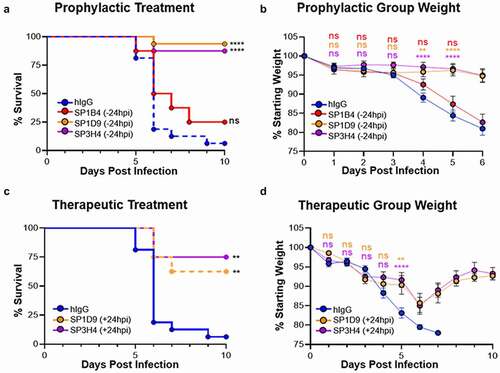
Since SARS-CoV-2 viral titers peak as early as 48 hours post-infection (hpi) in vivo, allowing the virus to establish infection for 24 hours prior to therapeutic administration should demonstrate the versatility of these VHH-huFc antibodies in disease treatment.Citation18 To evaluate the efficacy of SP1D9 and SP3H4 VHH-huFc antibodies post-exposure, mice were infected intranasally with 2.5 × 104 plaque forming units (PFU) SARS-CoV-2, then administered 10 mg/kg of VHH-huFc antibodies 24 hpi. Impressively, both SP1D9 and SP3H4 demonstrated significant therapeutic protection (p = .0033, p = .0013, respectively) over that of the isotype control. The SP1D9 group showed 58% survival (n = 8), and the SP3H4 group showed 75% survival (n = 8) at 10 dpi (). Of note, several surviving individuals in each of the post-exposure VHH-huFc-treated groups exhibited 10–15% weight loss up to 5 dpi, but then rebounded from 6 to 10 dpi, gaining back 90–95% of their starting weight ( and Figure S10). Taken together, these data demonstrate protection from lethal infection after a single 10 mg/kg dose, and further suggest that neutralization of SARS-CoV-2 by VHH-huFc antibodies in vivo can promote recovery from an ongoing infection.
Discussion
The COVID-19 pandemic caused by SARS-CoV-2 has resulted in 184 million cases and over 3.9 million deaths and an estimated global economic cost of over 10.3 trillion dollars in forgone output for 2020 and 2021.Citation19 As variants of SARS-CoV-2 emerge, it is essential to possess a diverse set of prophylactic and therapeutic tools, in order to maintain the progress made toward ending this pandemic. While vaccines have been a great success, therapeutic biologics are emerging as a critical tool in preventing progression to severe disease in those who become infected.Citation20
Several therapeutic antibody candidates with efficacy against SARS-CoV-2 have been recently identified, including three with emergency use authorization, but there are a number of caveats associated with conventional antibodies as therapeutics, such as reliance on immunized or patient sera, expensive and labor-intensive production, and the large doses required to achieve clinical efficacy.Citation9,Citation21–24 High stability, solubility, and the ability to be multimerized are a few of the many reasons why VHH-based antibody therapeutics (VHH-Fc) represent a highly promising method for treatment.Citation12 One VHH-based therapeutic is approved by the FDA for treatment of a rare blood clotting disorder and many more are in late stage clinical trials.Citation12,Citation25 The small size of VHH-huFc antibodies and the wider distribution of CDR3 loop lengths compared to human IgGs expands the type of epitope that can be effectively targeted.Citation12 Finally, preliminary evidence suggests that VHH-Fc antibodies are transported more efficiently into the blood and lung parenchyma following intraperitoneal administration compared to conventional IgG1 antibodies.Citation26,Citation27 Several groups have recently used in vitro screening techniques to identify VHH domains with high affinity for the SARS-CoV-2 S protein and neutralization of SARS-CoV-2 with EC50s ranging from 0.02 nM to 2 μM in pseudotyped virus and in wt SARS-CoV-2 virus assays.Citation25,Citation26,Citation28–34 However, in addition to neutralization, there are several antibody and immune functions that contribute to prophylactic and therapeutic effectiveness in vivo, and only one of these studies characterized the in vivo efficacy of a single VHH-Fc construct (VHH-Fc ab8) at reducing lung viral titers at 2 or 5 d post infection.Citation26 Importantly, the mouse-adapted SARS-CoV-2 and the golden hamster SARS-CoV-2 infection models used in this study are not lethal, rather, they inform efficacy for more mild disease cases.Citation18,Citation26,Citation35,Citation36 In this regard, the K18-hACE2 SARS-CoV-2 infection model is particularly valuable for testing the protective and therapeutic properties of novel antibodies in the context of severe infection and disease.Citation37,Citation38
In this study, we identified and characterized several prospective therapeutic neutralizing VHH-huFc antibodies both in vitro and in vivo from a high diversity synthetic library. Three candidate VHH-huFc antibodies are highlighted in this study because of their potent sub-nanomolar EC50s in preventing viral infection. For the SP1B4, SP1D9, and SP3H4 VHH-huFc constructs, affinity maturation was not required to achieve potent neutralization against SARS-CoV-2 pseudovirus (EC50s of 0.33, 0.45, and 0.14 nM, respectively) or wt SARS-CoV-2 virus (EC50s of 3.14, 1.12 and 0.70 nM, respectively).
Two of the VHH-huFc antibodies provided protection of K18-hACE2 transgenic mice when administered 24 hours before infection with wt SARS-CoV-2. A single dose of SP1D9 or SP3H4 exhibited significant therapeutic value when administered a full 24 hpi, which is closer to the peak of infection than was used to evaluate therapeutic efficacy of VHH-Fc ab8 (6 hpi).Citation26,Citation37 In this study, the onset of weight loss at 2–4 dpi, indicative of effective disease progression, and subsequent improvement after treatment establishes the potential for efficacy in the presence of an established wt SARS-CoV-2 infection ( and Figure S10). Although SP1B4 was a potent neutralizer in vitro, it failed to protect mice from infection with wt SARS-CoV-2, highlighting the importance of animal validation studies to triage top candidates for clinical evaluation.
Selection for escape mutants by individual VHH-huFc antibodies demonstrate the high propensity for SARS-CoV-2 to escape neutralization in vitro. Several studies, including this one, have generated mutations in vitro that exist in circulating SARS-CoV-2 variants and revealed that these mutations partially or completely block binding of top anti-SARS-CoV-2 therapeutic antibodies.Citation39 The potential for continued mutation acquisition and escape in SARS-CoV-2 highlights the critical importance of having multiple options for antibody cocktails.Citation29 Furthermore, sequencing analysis of the latest strains’ mutation status would allow patient stratification to identify which therapeutic regimen will be most effective in combating the latest emerging variant.Citation29
In this study, viral escape was readily apparent in the presence of our top candidates, with several mutations in the RBD. Interestingly, many of the initial mutations were selected against in the second passage (), and none of these individual mutations in the RBD significantly affected binding to ACE2 ( and Figure S6). This suggests that these positions of the S gene are highly susceptible to single-nucleotide polymorphisms and should be considered when identifying future preclinical antibody candidates because they may be present in future variant strains. Escape mutations observed for our two most promising candidates are all found in proximity to each other, likely indicating overlapping epitopes, though they may engage this epitope with different geometries (). SP3H4 did not share any of the mutations observed for SP1B4 or SP1D9, indicating that SP3H4 may engage the RBD region differently.
All three VHH-huFc antibodies neutralize viral infection by targeting the RBD of the wt SARS-CoV-2 S protein and compete directly with ACE2 binding to trimeric SARS-CoV-2 S, which indicates they are class I nanobodies.Citation25 Although the VHH-huFc antibodies described in this study bind to a single epitope, other groups have recently identified VHH antibodies that target a distinct epitope and could be used in combination with the VHH antibodies identified in this study to develop highly potent therapeutic cocktails that prevent the emergence of escape mutants.Citation29,Citation31,Citation34
We also evaluated our VHH-huFc antibodies’ ability to engage RBDs with mutations found in clinically relevant SARS-CoV-2 variants, including the L452R mutation found in the Epsilon (B.1.427/B.1.429), Kappa (B.1.617.1) and Delta (B.1.617.2) variants, and the mutations found in the Alpha (B.1.1.7) and Beta (B.1.351) variants, which pose a significant hurdle to therapeutic efficacy and vaccine-derived immunity. Interestingly, we observed a ~5-fold increase in affinity of the Alpha variant RBD for the ACE2 receptor, which may account for the increased infectivity observed with this strain of SARS-CoV-2. While none of our most promising candidates bind to the Beta variant, SP1B4, SP1D9, and SP3H4 still bind the Alpha RBD variant, albeit with reduced affinity in comparison to wt SARS-CoV-2 RBD (Figure S7-S9). The E484K mutation, observed in the Beta variant and now a new Iota (B.1.526) variant, confers total abrogation of VHH-huFc binding. The L452R mutation, found in circulating Epsilon, Kappa, and Delta variant strains, was selected for with SP3H4.Citation40–42 Interestingly, SP3H4 retains affinity for SARS-CoV-2 RBD with the L452R mutations. Furthermore, the kinetics appear to shift from biphasic to 1:1 with the L452R mutation, which may indicate that SP3H4 has two non-overlapping binding sites.
While many of the mutations in clinically relevant SARS-CoV-2 variants emerged in this study, several mutations that arose in this study have yet to be highlighted. Using these VHH-huFc antibodies to rapidly generate escape mutants could help to preemptively identify and develop countermeasures for future variants before they arise and circulate in the population.
In this work, we reported the design of a large synthetic VHH library (3.18 x 1010) and described the library’s utility in identifying over 50 VHH candidates that bind to wt SARS-CoV-2. We triaged top candidates based on neutralization efficiency in vitro, and evaluated efficacy in vivo using a mouse model of severe SARS-CoV-2 disease. To our knowledge, this is the first study to examine survival up to 10 dpi with a single dose of VHH-Fc given 24 hpi in the context of a lethal SARS-CoV-2 infection. While in vitro neutralization assays and short-range in vivo titer reduction studies are informative, they do not always recapitulate likelihood of survival. There are several reasons why in vitro potency may not correlate with in vivo protection, including an antibody’s pharmacokinetic properties and/or a broad range of immunomodulatory functions required for viral clearance.Citation38,Citation43–45 Studies are currently underway to evaluate the efficacy of VHH-huFc combinations administered at later time points post-infection and to investigate the role of Fc effector function in protection against SARS-CoV-2. The demonstrated ability of this virus to generate escape mutants emphasizes the need for multiple therapeutic options, with demonstrated in vivo activity in a post-infection context, to rapidly react to the emergence of resistant variants as expanded vaccine and antibody therapy use creates selection pressures on the viral population.
Materials and methods
DNA manipulation and production of proteins
pSF-CMV-SARS-CoV-2-S was constructed as follows. DNA fragments encoding amino acids 1–1208 of the SARS-CoV-2 S gene (GenBank:MN908947.3) were amplified as several independent fragments to introduce several mutations described in Wrapp et al.Citation46 from a vector containing the full-length codon-optimized S gene. Additionally, a double-stranded DNA fragment encoding a C-terminal T4 fibritin trimerization domain, TEV cleavage site, Twin-Step-Tag, and octahistadine was commercially obtained. pSF-SARS-CoV-2-RBD was produced by commercial production of double-stranded DNA encoding a N-terminal Kozak, a signal peptide (MDWTWRFLFVVAAATGVQS), 319–577 of the SARS-CoV-2 S gene (GenBank:MN908947.3), a TEV site, and a C-terminal, decahistadine tag. Mutant SARS-CoV-2 RBDs were produced by site-directed mutagenesis. All fragments were synthesized commercially (IDT) and terminal fragments had overlapping sequence with the downstream digested vector. All DNA fragments and pSF-CMV vector restriction digested with EcoRI and BamHI were assembled using NEBuilder HiFi DNA master (NEB, E2621S).
Constructs for expression of ACE2-Fc (human Fc domain) fusion proteins were produced as follows. pAce2-huFc was produced by subcloning the ectodomain of human ACE2 (Sino Biological) into pCR-Fc using the NotI and BamHI restriction sites. To produce Ace2-rbFc (rabbit Fc domain), a gBlock (IDT) of this same region of Ace2 was subcloned into pFUSE rIgG-Fc2 (Invivogen, pfuse-rfc2).
Soluble SARS-CoV-2 S and RBD were both produced by transient expression in Expi293F suspension cells (Thermo, A14527). Cells were transfected with 1 μg/mL plasmid at a density of 3.0 × 106 cells/mL using Expifectamine (Thermo, A14635). Cells were supplemented as instructed by the manufacturer and grown at 37°C with 8% CO2. Cell supernatant was harvested on day 4 by centrifugation at 4,000 x g for 30 min at 4°C. Clarified supernatant was passed through a 0.22 μm filter and then applied to a 5 mL HisTrap Excel column (Cytiva, 17,371,206) pre-equilibrated with 10 mM Tris (pH 8.0), 300 mM NaCl for SARS-CoV-2 S and 20 mM phosphate (pH 7.4), 300 mM NaCl for SARS-CoV-2 RBD. The column was washed with 10 column volumes equilibration buffer followed by 5 column volumes equilibration buffer with 20 mM imidazole. The S and RBD proteins were eluted with a step gradient to 500 mM imidazole. Fractions containing SARS-CoV-2 S protein were pooled and dialyzed in 20 mM HEPES (pH 8.0), 200 mM NaCl, prior to concentration. Protein was then concentrated and filtered prior to application to an Enrich 650 SEC column (Biorad, 7,801,650) equilibrated with dialysis buffer for SARS-CoV-2 S and phosphate-buffered saline) PBS for SARS-CoV-2 RBD. Purified SARS-CoV-2 proteins were stored at −80°C.
ACE2-huFc, ACE2-rbFc, and VHH-huFc antibodies were produced by transient expression in CHO-S cells using the ExpiCHO expression system (Thermo, A29133). In brief, cells were transfected at a density of 6 × 106 cells/mL and grown for 18 hours at 37°C with 8% CO2. Cells were then supplemented as per the manufacture’s guidelines and transferred to 32°C with 5% CO2. Cell supernatant containing soluble protein was harvested on day 10 by centrifugation at 4,000 x g for 30 min at 4°C. Clarified supernatant was passed through a 0.22 μm filter and the applied to a 1 mL HighTrap MabSelect PrismA column (Cytiva, 17,549,851). The column was washed with 20 mM sodium phosphate (pH 7.4), 150 mM NaCl. ACE2-Fc fusion proteins were eluted with 100 mM sodium citrate (pH 3.0) and immediately neutralized with 1 M Tris (pH 9.0). Protein was then concentrated and filtered before application to an Enrich 650 SEC column equilibrated with 10 mM sodium phosphate (pH 7.2), 140 mM NaCl. Purified ACE2-huFc, ACE2-rbFc, and VHH-huFc were stored at −80°C. VHH-huFc antibodies identified from screening were commercially obtained from GenScript.
VHH phage library construction and production
Synthetic VHH library was designed as follows. A high diversity library was designed by incorporating the natural prevalence of amino acids at positions in CDR1 and CDR2 based off 670 functional VHH antibodies deposited on sdAb-DB (www.sdab-db.ca).Citation14 Amino acids cysteine and methionine were omitted from all CDR loops and full diversity was used for CDR3. Asparagine was also omitted from CDR1 and CDR2. The library was also designed such that there would be 3 different lengths of CDR3 (9-, 12-, and 15-amino acids). Linear double-stranded DNA fragments incorporating the specifications mentioned above, as well as terminal flanking BglI restriction sites, were synthetically produced commercially by Twist Bioscience. A humanized VHH framework characterized in Moutel et al.Citation15 was used to house the designer CDRs.
Library was assembled as follows. The linear library DNA fragment was amplified by PCR. PCR reaction was divided into a 384-well format to minimize potential amplification bias. Five cycles of PCR were performed (Step 1: 98°C, 3 minutes, 1 cycle; Step 2: 98°C, 10 seconds; 68°C, 10 seconds; 72°C, 15 seconds, 5 cycles; Step 3: 72°C, 10 minutes, 1 cycle; Step 4: 12°C, hold). Amplified DNA was pooled and purified by Monarch Nucleic Acid Purification Kit (NEB, T1020S). The library was digested with 5 U/μg DNA SfiI at 50°C for 16 hours. Digest reactions were again column purified as previously mentioned. Ligations were set up as follows. pADL20c (205 μg) previously digested with BglI and treated with rSAP was added to a 40 mL reaction containing the linear digested library (42 μg) at a vector to insert ratio of 1:2, 1 mM ATP and 1,100,000 units of T4 Ligase (Antibody Design Laboratories, PD0109) . The reaction was allowed to proceed at 16°C for 16 hours, then at 37°C for 1 hour, and lastly was heat inactivated at 70°C for 20 minutes. DNA was purified and concentrated by a modified ethanol precipitation protocol utilizing tRNA carrier at 15 μg/mL.
Transformation of the library into TG1 E. coli was performed as follows. A total of 9 liters of fresh electrocompetent TG1 E. coli cells were produced and 150 electroporations performed. To 350 μL cells, 15 μL of ligated DNA was added and electroporation performed. After 1-hour recovery with the addition of 650 μL SOC at 37°C, cells were plated on 2xYT agar plates supplemented with 100 mM glucose and 100 μg/mL carbenicillin (2xYT-GA). E. coli was harvested after growth at 37°C for 16 hours, supplemented with glycerol to 10%, and stored at −80°C in aliquots for further use. Diversity of the nanobody library was determined by both NGS and colony PCR.
The VHH library was added to 2xYT-GA to a final OD600 of 0.08 and allowed to grow at 37°C until an OD600 of 0.5 was reached. Superinfection with CM13 (Antibody Design Laboratories, PH020L) was performed with 2.0 × 1012 helper phage per liter of E. coli culture for 15 minutes without shaking followed by rigorous shaking for 30 minutes. Cell were collected by centrifugation at 5,500 × g for 10 minutes and resuspended in the previous volume used of 2xYT supplemented with 100 μg/mL carbenicillin and 50 μg/mL kanamycin (2xYT-AK). E. coli were grown overnight with shaking at 28°C.
Supernatant containing packaged phagemid was clarified by centrifugation at 6,000 × g for 15 minutes and supplemented with one-fourth volume 20% PEG-8000 and 2.5 M NaCl. Phage were precipitated overnight at 4°C and were collected by centrifugation at 5,500 × g for 60 minutes at 4°C. The pellet containing phage was resuspended in PBS and centrifugated for 15 minutes at 5,500 × g to remove E. coli particulates. Clarified phage was again precipitated as mentioned above and incubated on ice for 60 minutes. The phage was finally centrifuged at 17,900 × g for 10 minutes at 4°C, the supernatant was removed, and the pellet was again centrifuged at 17,900 × g for 1 minute to remove trace amounts of PEG-8000. The pellet was resuspended in PBS and passed through a 0.22 μm filter before determining the colony forming units (CFU).
Next-generation sequencing
All samples for NGS were processed as follows. The minimum region containing all 3 CDR domains, approximately 300 bps, was excised from the pADL20c backbone by two-step restriction digests, BglI followed by DdeI/BstEII double digests on the gel-purified small fragment from the BglI restriction reaction. The sequencing library was prepared using NEBNext® Ultra™ II DNA Library Prep Kit for Illumina®(NEB, E7645S) and xGen™ UDI-UMI Adapters (IDT, 10,006,914) as instructed in the manufacture’s manuals and sequenced on Illumina NextSeq 500/550 platform with High Output v2.5 300-cycles (Illumina, 20,024,908), paired-end mode. BCL files were converted to FASTQ and demutiplexed using the bcl2fastq script from MyIllumina (https://my.illumina.com). The quality filtering and adaptor trimming were performed using fastp (https://github.com/OpenGene/fastp) with following parameters, -q 30 -l 100 -x 7. R2 read was reformatted to be reverse complemented and merged with R1 read using BBTools (BBMap, https://sourceforge.net/projects/bbmap/).Citation47 Three CDR domains with correct sequence lengths were extracted, concatenated and translated and counted unique amino acid sequences using a custom python script.
Affinity selection for SARS-CoV-2 S clones by panning
A total of four rounds of affinity selection were conducted, three rounds against soluble full-length SARS-CoV-2 S and a final round against SARS-CoV-2 RBD. Panning was performed by immobilizing antigen in 96-well Immulon® HBX microplates (Thermo, 3855). For the first round of panning, 3 μg SARS-CoV-2 S was immobilized in 50 μL coating buffer (100 mM NaHCO3 (pH 8.3), 150 mM NaCl) overnight at 4°C. Wells containing antigen were washed five times with 300 μL PBS-t (phosphate buffered saline supplemented with 0.05% Tween-20). Wells were blocked with 300 μL Pierce PBS Protein Free Blocking Solution (Thermo, 37,572) for 2 hours. After another round of washing, 200 μL of 2.5 × 1012 CFU/mL phage were added to each well in blocking buffer and allowed to incubate with vigorous shaking at 25°C for 2 hours. Wells were extensively washed with increasing amounts of Tween-20, with a peak concentration of 0.5% Tween-20. Phage were eluted with the addition of 50 μL of elution buffer (10 mM Tris pH 7.4, 137 mM NaCl, 1 mM CaCl2 and 100 µg/mL Trypsin) by incubating for 30 minutes at 37°C. Eluate was removed and pooled and a second 50 μL of elution buffer was added for 20 minutes. This was collected and pooled with the first round of elution buffer. Eluted phage was added to an equal volume of log phage TG1 E. coli for reamplification and quantification.
For all subsequent rounds of panning, 2 μg of antigen was used to coat wells. For round 2, a total of 1012 CFU phage were used for panning. For rounds 3 and 4, panning was conducted with an input of 1011 CFU phage. With each round of panning, washing was progressively increased from 12 washes in the first round to 15, then to 21 for the final two rounds. For all rounds after the first, a maximum of 1% Tween-20 was used for washing. The plates were then extensively washed with PBS to remove excess Tween-20. Additionally, the blocking solution for each round of panning was cycled. For the second round, PBS-t supplemented with 1% bovine serum albumin (BSA) (w/v) was used, for the third round PBS-t supplemented with 2% nonfat milk, and for the final round BSA was again used. A denaturing step was conducted before the last round of panning against the SARS-CoV-2 RBD to remove unstable clones by heating the phage to 70°C for 15 minutes before panning. All other elution, reamplification, and tittering steps were conducted as mentioned above.
Polyclonal and monoclonal phage ELISA
Polyclonal phage ELISA was used to characterize antigenic enrichment and specificity for each round of panning. Multiple antigens were tested, including BSA, SARS-CoV-1 S, SARS-CoV-2 S, and SARS-CoV-2 RBD. Immulon HBX microtiter 384-well plates were coated with at 0.05 μg/well of each antigen overnight at 4°C. Plates were washed 5 times with PBS-t and blocked with 50 μL of PBS with 2% nonfat milk and 0.2% Tween-20 (MPBS-t). Plates were washed and 109 phage from the unenriched library and each round of panning was added to each well for all of the antigens in blocking solution. After 2 hours of shaking at 25°C, the plates were again washed 5 times and mouse anti-M13 coat protein (Thermo, MA529950) was added 1:2000 in MPBS-t for 1 hour. The plates were again washed, and rabbit anti-mouse IgG horseradish peroxidase (HRP; Thermo, PA1-28,568) was added 1:1000 in MPBS-t for 1 hour with shaking. The plates were finally washed 5 times with PBS-t and developed with TMB Ultra substrate (Thermo, 34,028) and quenched with 2 M H2SO4 (Sigma, 258,105) before reading the absorbance at 450 nm. Monoclonal phage ELISAs were performed in a similar fashion to the polyclonal ELISA mentioned above with homogeneous preparations of VHH-phage to validate individual clones.
VHH-huFc ELISA
Indirect ELISAs were performed as follows. Full-length soluble SARS-CoV-2 S or RBD were immobilized on Immulon 384-well microtiter plates (0.05 μg/well) overnight at 4°C in coating buffer. After washing, plates were blocked for 2 hours with Pierce PBS Protein Free Blocking buffer. Serial dilutions of VHH-huFc antibodies in blocking buffer were added to plates for 2 hours after washing. Secondary antibody, goat anti-human H + L IgG HRP (Thermo, 31,410) was added for an additional hour before washing. Plates were developed with TMB Ultra and the reaction stopped by addition of equal volume 2 M H2SO4 after washing. Absorbance was read at 450 nm.
Competition of soluble ACE2-huFc by VHH-huFc candidates identified from display screening was performed as follows. SARS-CoV-2 antigen was immobilized and blocked as described above. After blocking, serial dilutions of VHH-huFc antibodies were prepared in blocking solution and allowed to incubate for at least 2 hours, after which ACE2-rbFc was added to a final concentration of 0.1 μg/mL (SARS-CoV-1 S) or 0.06 μg/mL (SARS-CoV-2 S) and allowed to incubate for 1 hour with shaking at 25°C. Plates were washed and 1:10,000 HRP-conjugated goat anti-rabbit IgG (Thermo, 31,466) was added. Plates were developed as described above.
Differential scanning fluorimetry
Thermostability for the VHH-huFc antibodies was determined by DSF. Each reaction contained 10 μg of VHH-huFc in PBS with 10 μM SYPRO Orange (Thermo, S6651). Reactions were heated from 10–95°C at a heating rate of 1°C/min and monitored in the FRET channel of a BioRad CFX96. The melting point for each of the VHH-huFc antibodies was calculated by the first derivative method.Citation48
Viruses and cell culture
Vero and Vero E6 cells (African green monkey kidney, ATCC CCL-81 and CRL-1586, respectively) were maintained in culture medium supplemented with 10% fetal bovine serum (FBS), 100 units/ml penicillin, and 100 μg/ml streptomycin (Invitrogen, 15,070,063), at 37°C in 5% CO2. For risk group (RG) two experiments and viral stocks, Vero cells were cultured in supplemented minimum essential medium alpha (alpha MEM). For RG 3 experiments and viral stocks, Vero E6 cells were cultured in supplemented DMEM. A pseudotyped, replication-competent vesicular stomatitis virus (VSV) expressing eGFP as a marker of infection and the SARS-CoV-2 spike gene (VSV-SARS-CoV-2) in place of its own VSV-G gene was provided by Dr. Sean Whelan.Citation16, Pseudotyped, single cycle VSV particles displaying the SARS-1 spike protein (VSV-SARS-1-GFP) were derived from recombinant VSV-DG-GFP in which the VSV-G envelope protein has been replaced with GFP. VSV-SARS-1-GFP single cycle particles were produced in Expi293 cells through transfection per manufacturer’s instructions with the SARS-CoV-1 spike expression plasmid (Sino Biological, VG40150-G-N) containing a 19-aa deletion in the cytoplasmic tail. The Expi293 cells were subsequently infected with VSV-DG-GFP, itself pseudotyped with VSV-G, at 72 h post-transfection using a multiplicity of infection (MOI) of 3. The resulting VSV-SARS-1-GFP pseudotyped viruses were collected at 24 h post-infection. WT SARS-CoV-2 was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52,281. An infectious clone of SARS-CoV-2 expressing a NeonGreen reporter gene (icSARS-CoV-2-mNG, or SARS-CoV-2-NG) was provided by Dr. Pei Yong Shi.Citation49 Viral stocks were amplified in Vero E6 cells, supernatants were harvested upon extensive cytopathic effect, 48 (VSV-SARS-CoV-2-GFP) or 72 (SARS-CoV-2) hpi. Cellular debris was removed by centrifugation, and aliquots were stored at −80°C. Titers of viral stocks were determined by plaque assay. All work with WT SARS-CoV-2 was performed in Institutional Biosafety Committee approved BSL-3 and ABSL-3 facilities at Lawrence Livermore National Laboratory using appropriate personal protective equipment and protective measures.
SARS-CoV-2 fluorescent reporter neutralization assays
Serial dilutions of VHH-huFc were prepared in supplemented media at 2× the desired final concentration. SARS-CoV-2-NG, VSV-SARS-CoV-2-GFP, or VSV-SARS-1-GFP was added to the serially diluted VHH-huFc antibodies for a final MOI of 0.2 (RG 2) or 0.1 (RG 3) and allowed to incubate at 37°C for 30 minutes to 1 hour with shaking prior to transfer of virus-VHH-huFc mixture to cells seeded in 96- (RG 3) or 384-well plates (RG 2). VHH-huFc-virus complexes were incubated with Vero cells at 37°C with 5% CO2 for 12–16 h. For RG 2 experiments, cells were subsequently fixed in 4% paraformaldehyde (Millipore Sigma, 818,715) containing 10 mg/mL Hoechst 33,342 nuclear stain (Invitrogen, 62,249) for 30 min at room temperature, when fixative was replaced with PBS, and images were acquired with the Tecan Spark® Cyto multi-mode plate reader and image cytometer in both the DAPI and FITC channels to visualize nuclei and infected cells (i.e., eGFP-positive cells), respectively (4X objective, covering the entire well). Images were analyzed using the Spark Control Image Analyzer Software. For RG 3 experiments with, cells were lysed in RIPA buffer plus Halt protease inhibitor cocktail (Thermo, 87,786) and fluorescence was measured using 485 nm excitation and 528 emission wavelengths. Fluorescent values were background subtracted using no-infection controls and normalized to no-treatment infection values. The dose response curves and 50% effective inhibitory concentrations (EC50) were generated using Graphpad Prism 9.
Plaque neutralization assay
VHH-huFc-WT SARS-CoV-2 virus complexes were preincubated at 37°C with 5% CO2 for 1 hour prior to addition to subconfluent Vero E6 cells in 12-well plates. After a 30-minute incubation of VHH-huFc-virus complexes with Vero E6 cells at 37°C with 5% CO2, overlays of 2 mL per well of 0.6% microcrystalline cellulose (MCC, Sigma, 435,244), in 8% FBS, 1% P/S complemented 2X MEM were added. To stain, the MCC was aspirated, wells rinsed with PBS, and 0.4% crystal violet in 100% methanol was added for 10 minutes and then removed. Wells were then washed twice with water, and titers were recorded in PFU/mL.
Biolayer interferometry
Affinity measurements for VHH-huFc antibodies were performed using BLI using an Octet 384 Red system (Sartorius). Measurements were conducted in 10 mM phosphate (pH 7.4), 300 mM NaCl, 1 mg/mL BSA, 0.1% NP-40 (Thermo, 28,324). VHH-huFc ligands were immobilized on human Fc capturing sensors. WT and mutant SARS-CoV-2 RBD were used as the analyte and sensorgrams were fit to a global 1:1 fit. Sensorgrams that deviated from a 1:1 global fit or fit well to a heterogeneous ligand model were designated as biphasic.
Generation and analysis of escape mutants
Mutant VSV-SARS-CoV-2-GFP virus with the ability to escape VHH-huFc neutralization was generated in a similar fashion to that described in Baum et al.Citation21 In brief, a series of 500 μL dilutions of VHH-huFc antibodies were incubated with 500 μL of 1.5 × 106 PFU VSV-SARS-CoV-2-GFP virus for 1 hour at 37°C. The VHH-huFc and virus mixture was added to Vero cells and allowed to incubate for up to 72 hours until apparent cytopathic effect (CPE) or widespread viral replication could be observed by GFP fluorescence. Supernatant from wells containing the highest concentrations of VHH-huFc was collected and viral RNA isolated from total RNA using TRIzol. A portion of this supernatant was diluted 500-fold and exposed to a series of higher concentrations of VHH-huFc antibodies for 1 hour at 37°C prior to addition to Vero cells. Cells were monitored for 60 hours for CPE and GFP fluorescence. Supernatants at the highest concentration of VHH-huFc with clear viral replication were collected and processed as mentioned above and viral RNA analyzed as described below.
VSV-SARS-CoV-2 genomic RNA was isolated for NGS. For library preparation, 25–100 ng of total RNA was used to deplete rRNA using DNA probes and RNase H provided in RiboErase (HMR) kit (Roche). The depleted RNA was fragmented prior to cDNA synthesis followed by Illumina adaptor ligation using KAPA RNA HyperPrep kit (Roche). The library was analyzed using High Sensitivity DNA ScreenTape 1000 (Agilent) for quality and quantification and pulled together for the final library denaturation. Sequencing was performed on Illumina NextSeq 500/550 platform with High Output v2.5 75-cycle mode (Illumina, 20,024,906). BCL files were converted to FASTQ and demutiplexed using the bcl2fastq script from MyIllumina (https://my.illumina.com). The quality filtering and adaptor trimming were performed using fastp with -q 30 option. The filtered reads were mapped to Spike protein coding region of the SARS-CoV-2 Wuhan Hu-1 isolate (GenBank:MN908947.3) using Bowtie 2.Citation50 The variant calling on the mapped reads was performed using mpileup from Bcftools (https://samtools.github.io/bcftools/) and inspected using Integrative Genomics Viewer (IGV).Citation51
Animal studies
All animal work was performed in accordance with protocols approved by the Lawrence Livermore National Laboratory Institutional Animal Care and Use Committee. Groups of male and female K18-hACE2 C57BL/6 J transgenic mice (Jackson Laboratory) ranging in age from 12 to 16 weeks were inoculated intranasally with 2.5 × 104 PFU SARS-CoV-2 (USA-WA 01/2020) while under anesthesia (4–5% isoflurane in 100% oxygen). Animals were dosed prophylactically (−24 hours pre-infection) or therapeutically (+24 hpi) by intraperitoneal injection with 10 mg/kg of VHH-huFc or isotype control antibody. Body weight was measured daily and any animal falling below 80% of their starting weight was humanely euthanized in accordance with animal welfare guidelines.
Statistical analysis
Raw data for infection assays measured by reporter fluorescence were compared using a two-tailed t test for each individual experiment. For plate assays, untreated, infected controls and uninfected controls were included on every plate. Antibody-treated cells that were infected were compared to control samples infected with the same virus. Kaplan–Meier survival curves were generated based on two independent experiments and log rank tests were performed with Bonferroni multiple comparison correction applied (GraphPad Prism). P values were considered significant when they were <0.05 (*) and very significant when they were <0.01 (**), <0.001 (***), or <0.0001 (****).
Abbreviations
Supplemental Material
Download Zip (3.4 MB)Acknowledgments
We would like to thank Prof. Sean Whelan from Washington University School of Medicine St. Louis for graciously providing access to the pseudotyped replication competent VSV-SARS-CoV-2-GFP virus used in this study. We would also like to thank Dr. Pei Yong Shi from the World Reference Center for Emerging Viruses and Arboviruses at University of Texas, Medical Branch for generously providing the infectious clone of SARS-CoV-2 expressing a NeonGreen reporter gene. We would also like to thank Robert Meager for critically reviewing the paper. This paper describes objective technical results and analysis. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the U.S. Department of Energy or the United States Government. All work performed at Lawrence Livermore National Laboratory is performed under the auspices of the U.S. Department of Energy under Contract DE-AC52-07NA27344.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020 May;20(5):533–13. doi:10.1016/S1473-3099(20)30120-1.
- Yimer B, Ashebir W, Wolde A, Teshome M. COVID-19 and Global Health Security: overview of the Global Health Security Alliance, COVID-19 response, African countries’ approaches, and ethics. Disaster Med Public Health Prep. 2020 Oct;2:1–5. doi:10.1017/dmp.2020.360.
- Katz LM. A little clarity on convalescent plasma for Covid-19. N Engl J Med. 2021 Feb 18;384(7):666–68. doi:10.1056/NEJMe2035678.
- Ali MG, Zhang Z, Gao Q, Pan M, Rowan EG, Zhang J. Recent advances in therapeutic applications of neutralizing antibodies for virus infections: an overview. Immunol Res. 2020 Dec;68(6):325–39. doi:10.1007/s12026-020-09159-z.
- Jiang S, Zhang X, Yang Y, Hotez PJ, Du L. Neutralizing antibodies for the treatment of COVID-19. Nat Biomed Eng. 2020 Dec;4(12):1134–39. doi:10.1038/s41551-020-00660-2.
- Ryman JT, Meibohm B. Pharmacokinetics of monoclonal antibodies. CPT Pharmacometrics Syst Pharmacol. 2017 Sep;6(9):576–88. doi:10.1002/psp4.12224.
- Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020 2020 Aug 1;5847821:443–49. doi:10.1038/s41586-020-2548-6.
- Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med. 2021 Jan 21;384(3):238–51. doi:10.1056/NEJMoa2035002.
- Chen P, Nirula A, Heller B, Gottlieb RL, Boscia J, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021 Jan 21;384(3):229–37. doi:10.1056/NEJMoa2029849.
- Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins. 2018 Jul;86(7):697–706. doi:10.1002/prot.25497.
- Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol. 2007 Nov;77(1):13–22. doi:10.1007/s00253-007-1142-2.
- Bannas P, Hambach J, Nanobodies K-NF. Nanobody-based human heavy chain antibodies as antitumor therapeutics. Front Immunol. 2017;8:1603–1603. doi:10.3389/fimmu.2017.01603.
- Grant OC, Montgomery D, Ito K, Woods RJ. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci Rep. 2020 Sep 14;10(1):14991. doi:10.1038/s41598-020-71748-7.
- Wilton EE, Opyr MP, Kailasam S, Kothe RF, Wieden H-J. sdAb-DB: the Single Domain Antibody Database. ACS Synth Biol. 2018 Nov 16;7(11):2480–84. doi:10.1021/acssynbio.8b00407.
- Moutel S, Bery N, Bernard V, Keller L, Lemesre E, de Marco A, Ligat L, Rain JC, Favre G, Olichon A, et al. NaLi-H1: a universal synthetic library of humanized nanobodies providing highly functional antibodies and intrabodies. Elife. 2016;19;5:e16228.
- Case JB, Rothlauf PW, Chen RE, Liu Z, Zhao H, Kim AS, Bloyet LM, Zeng Q, Tahan S, Droit L, et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host & Microbe. 2020 2020/09/09/;28(3):475-485.e5.
- Dong J, Huang B, Wang B, Titong A, Gallolu Kankanamalage S, Jia Z, Wright M, Parthasarathy P, Liu Y. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci Rep. 2020 Oct 20;10(1):17806. doi:10.1038/s41598-020-74761-y.
- Winkler ES, Bailey AL, Kafai NM, Nair S, McCune BT, Yu J, Fox JM, Chen RE, Earnest JT, Keeler SP, et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020 Nov;21(11):1327–35. doi:10.1038/s41590-020-0778-2.
- What is the economic cost of covid-19? The Economist. 2021 Jan 7, 2021.
- FDA authorizes revisions to fact sheets to address SARS-CoV-2 variants for monoclonal antibody products under emergency use authorization [Internet]. U.S. Food & Drug Administration; 2021; Mar 10, 2021;[1]. Available from: https://www.fda.gov/drugs/drug-safety-and-availability/fda-authorizes-revisions-fact-sheets-address-sars-cov-2-variants-monoclonal-antibody-products-under
- Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020 Aug 21;369(6506):1014–18. doi:10.1126/science.abd0831.
- Chames P, Van Regenmortel M, Weiss E, Baty D, Therapeutic antibodies: successes, limitations and hopes for the future. Br J Pharmacol. 2009 Jun 30; received Sep 1; revised 01/12/accepted;1572:220–33. doi:10.1111/j.1476-5381.2009.00190.x.
- Dhama K, Sharun K, Tiwari R, Dadar M, Malik YS, Singh KP, Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020 Jun 2;16(6):1232–38. doi:10.1080/21645515.2020.1735227.
- Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, Yu J, Shan S, Zhou B, Song S, et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature.2020/08/01;584(7819):115-119.
- Schoof M, Faust B, Saunders RA, Sangwan S, Rezelji V, Hoppe N, Boone M, Billesbolle CB, Puchades C, Azumaya CM, et al. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020 Dec 18;370(6523):1473–79.
- Li W, Schäfer A, Kulkarni SS, Liu X, Martinez DR, Chen C, Sun Z, Leist SR, Drelich A, Zhang L, et al. High potency of a bivalent human V(H) domain in SARS-CoV-2 animal models. Cell. 2020 Oct 15;183(2):429–41. e16. doi:10.1016/j.cell.2020.09.007.
- Wesolowski J, Alzogaray V, Reyelt J, Unger M, Juarez K, Urrutia M, Cauerhff A, Danquah W, Rissiek B, Scheuplein F, et al. Single domain antibodies: promising experimental and therapeutic tools in infection and immunity. Med Microbiol Immunol. 2009 Aug;198(3):157–74. doi:10.1007/s00430-009-0116-7.
- Huo J, Le Bas A, Ruza RR, Duyvesteyn HME, Mikolajek H, Malinauskas T, Tan TK, Rijal P, Dumoux M, Ward PN, et al. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat Struct Mol Biol. 2020 2020 Sep 1;279:846–54. doi:10.1038/s41594-020-0469-6.
- Koenig P-A, Das H, Liu H, Kümmerer BM, Gohr FN, Jenster L-M, Schiffelers LDJ, Tesfamariam YM, Uchima M, Wuerth JD, et al. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371(6530):eabe6230. doi:10.1126/science.abe6230.
- Wrapp D, De Vlieger D, Corbett KS, Torres GM, Wang N, Van Breedam W, Roose K, van Schie L, Hoffmann M, Pöhlmann S, et al. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181(5):1004–1015.e15. doi:10.1016/j.cell.2020.04.031.
- Bracken CJ, Lim SA, Solomon P, Rettko NJ, Nguyen DP, Zha BS, Schaefer K, Byrnes JR, Zhou J, Lui I, et al. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat Chem Biol. 2021 Jan;17(1):113–21. doi:10.1038/s41589-020-00679-1.
- Sun Z, Chen C, Li W, Martinez DR, Drelich A, Baek D-S, Liu X, Mellors JW, Tseng C-T, Baric RS et al. Potent neutralization of SARS-CoV-2 by human antibody heavy-chain variable domains isolated from a large library with a new stable scaffold. MAbs. 2020 Jan 1;12(1):1778435. doi:10.1080/19420862.2020.1778435.
- Lu Q, Zhang Z, Li H, Zhong K, Zhao Q, Wang Z, Wu Z, Yang D, Sun S, Yang N, et al. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J Nanobiotechnology. 2021 2021 Jan 29;191:33. doi:10.1186/s12951-021-00768-w.
- Xiang Y, Nambulli S, Xiao Z, Liu H, Sang Z, Duprex PW, Schneidman-Duhovny D, Zhang C, Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Science. 2020;370(6523):1479–84.
- Sia SF, Yan L-M, Chin AWH, Fung K, Choy K-T, Wong AYL, Kaewpreedee P, Perera RAPM, Poon LLM, Nicholls JM, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. 2020 2020 Jul 1;5837818:834–38. doi:10.1038/s41586-020-2342-5.
- Dinnon KH 3rd, Leist SR, Schäfer A, Edwards CE, Martinez DR, Montgomery SA, West A, Yount BL, Hou YJ, Adams LE, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020 Oct;586(7830):560–66. doi:10.1038/s41586-020-2708-8.
- Rosenfeld R, Noy-Porat T, Mechaly A, Makdasi E, Levy Y, Alcalay R, Falach R, Aftalion M, Epstein E, Gur D, et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat Commun. 2021 2021 Feb 11;121:944. doi:10.1038/s41467-021-21239-8.
- Winkler ES, Gilchuk P, Yu J, Bailey AL, Chen RE, Chong Z, Zost SJ, Jang H, Huang Y, Allen JD, et al. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021 2021 Apr 1. 1847:1804–20. e16. doi:10.1016/j.cell.2021.02.026
- Zhou D, Dejnirattisai W, Supasa P, Liu C, Mentzer AJ, Ginn HM, Zhao Y, Duyvesteyn HME, Tuekprakhon A, Nutalai R, et al. 2021. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine induced sera. Cell. 2021 Feb 23. 184(9):2348-2361.e6
- Tchesnokova V, Kulakesara H, Larson L, Bowers V, Rechkina E, Kisiela D, Sledneva Y, Choudhury D, Maslova I, Deng K, et al. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. bioRxiv. 2021 2021 Feb 22. 432189.
- Zhang W, Davis BD, Chen SS, Sincuir Martinez JM, Plummer JT, Vail E. Emergence of a Novel SARS-CoV-2 Variant in Southern California. JAMA. 2021;325(13):1324. doi:10.1001/jama.2021.1612.
- Deng X, Garcia-Knight MA, Khalid MM, Servellita V, Wang C, Morris MK, Sotomayor-Gonzalez A, Glasner DR, Reyes KR, Gliwa AS, Reddy NP, et al. Transmission, infectivity, and antibody neutralization of an emerging SARS-CoV-2 variant in California carrying a L452R spike protein mutation. medRxiv. 2021Mar 7;21252647.
- Schäfer A, Muecksch F, Lorenzi JCC, Leist SR, Cipolla M, Bournazos S, Schmidt F, Maison RM, Gazumyan A, Martinez DR, et al. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J Exp Med. 2020;218:3.
- Liu R, Oldham RJ, Teal E, Beers S, Cragg M. Fc-engineering for modulated effector functions—Improving antibodies for cancer treatment. Antibodies. 2020;9(4):64. doi:10.3390/antib9040064.
- van Erp EA, Luytjes W, Ferwerda G, van Kasteren PB. Fc-mediated antibody effector functions during respiratory syncytial virus infection and disease [Review]. Front Immunol. 2019 2019 March 22;10(548). doi:10.3389/fimmu.2019.00548.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Mar 13;367(6483):1260–63. doi:10.1126/science.abb2507.
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018 Sep 1;34(17):i884–i890. doi:10.1093/bioinformatics/bty560.
- Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2(9):2212–21. doi:10.1038/nprot.2007.321.
- Xie X, Muruato A, Lokugamage KG, Narayanan K, Zhang X, Zou J, Liu J, Schindewolf C, Bopp NE, Aguilar PV, et al. An Infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27(5):841–848.e3. doi:10.1016/j.chom.2020.04.004.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012 Mar 4;9(4):357–59. doi:10.1038/nmeth.1923.
- Robinson JT, Thorvaldsdóttir H, Wenger AM, Zehir A, Mesirov JP. Variant review with the integrative genomics viewer. Cancer Res. 2017 Nov 1;77(21):e31–e34. doi:10.1158/0008-5472.CAN-17-0337.
