ABSTRACT
The crystallizable fragment (Fc) of immunoglobulin G (IgG) activates key immunological responses by interacting with Fc gamma receptors (FcɣR). FcɣRIIIb contributes to neutrophil activation and is involved in antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP). These processes present important mechanisms-of-actions of therapeutic antibodies. The very low affinity of IgG toward FcɣRIIIb (KD ~ 10 µM) is a technical challenge for interaction studies. Additionally, the interaction is strongly dependent on IgG glycosylation, a major contributor to proteoform heterogeneity. We developed an affinity chromatography–mass spectrometry (AC-MS) assay for analyzing IgG-FcɣRIIIb interactions in a proteoform-resolved manner. This proved to be well suited to study low-affinity interactions. The applicability and selectivity of the method were demonstrated on a panel of nine different IgG monoclonal antibodies (mAbs), including no-affinity, low-affinity and high-affinity Fc-engineered or glycoengineered mAbs. Thereby, we could reproduce reported affinity rankings of different IgG glycosylation features and IgG subclasses. Additional post-translational modifications (IgG1 Met252 oxidation, IgG3 hinge-region O-glycosylation) showed no effect on FcɣRIIIb binding. Interestingly, we observed indications of an effect of the variable domain sequence on the Fc-binding that deserves further attention. Our new AC-MS method is a powerful tool for expanding knowledge on structure–function relationships of the IgG-FcɣRIIIb interaction. Hence, this assay may substantially improve the efficiency of assessing critical quality attributes of therapeutic mAbs with respect to an important aspect of neutrophil activation.
Introduction
Immunoglobulin (Ig) G mediates key immunological responses by interacting with Fc gamma receptors (FcɣR).Citation1 FcɣRIII is found mainly on macrophages, natural killer cells and neutrophils, where it initiates various immune responses upon binding to opsonized IgG. The IgG-FcɣRIII interaction is strongly glycosylation-dependent.Citation2 This is attributed to unique glycan–glycan and glycan–protein interactions between the receptor and the crystallizable fragment (Fc) of IgGs.Citation3 Fcs with an afucosylated N-glycan show drastically increased FcɣRIII affinity.Citation3,Citation4 Increased affinity to activating FcɣRs, such as FcɣRIII, results in increased cytotoxicity.Citation5 Knowledge about the FcɣR-IgG interaction enabled the rational design of anti-cancer monoclonal antibodies (mAb), glycoengineered for increased cytotoxicity.Citation6
FcɣRIIIb is a particularly interesting receptor because it is uniquely expressed in humans. Neutrophils, the most abundant phagocytes in the circulation, show high levels of FcɣRIIIb expression; in fact, the highest of any FcɣR on any cell type.Citation7 Neutrophils exert antibody-dependent cellular cytotoxicity (ADCC), as well as antibody-dependent cellular phagocytosis (ADCP).Citation8 The neutrophil activation via FcɣRIIIb is considered an important mechanism of action of mAbs and is affected by the glycosylation, a critical quality attribute, of therapeutic mAbs.Citation8–11 The extracellular domain of FcɣRIIIb is highly homologous to FcɣRIIIa (>97% sequence homologyCitation12). However, FcɣRIIIb is the only FcɣR lacking a transmembrane and cytosolic signaling domain and is, instead, anchored by glycosylphosphatidylinositol. Further, the IgG1 affinity of FcɣRIIIb (KD ~ 10 µM) is up to ten-fold lower than for FcɣRIIIa (KD ~ 1 µM), which was attributed to a single amino acid difference.Citation9,Citation12 Of note, the KD values are highly dependent on the mAb glycoform. IgG subclass specificity of FcɣRIIIb interactions has been reported, with a higher affinity for IgG3 than IgG1 and no binding for IgG2 and IgG4.Citation5,Citation13 Three polymorphic variants of FcɣRIIIb, namely NA1, NA2 and SH, are known.Citation5 The two most common variants, NA1 and NA2, differ in four amino acids, leading to four (Asn38, Asn74, Asn162, Asn169) or six (Asn38, Asn45, Asn64 Asn74, Asn162, Asn169) glycosylation sites for NA1 and NA2, respectively. For IgG1 binding, only minor differences were observed for NA1 and NA2.Citation14
In vitro measurements of monovalent affinity have been acknowledged as important metrics in mAb optimization.Citation9 Of note, FcɣRIIIb affinity differences are difficult to measure with common techniques due to the low affinity and a high assay variability for FcɣRIIIb affinity assessments.Citation15,Citation16 Various studies have assessed the effect of mAb glycosylation on FcɣRIIIb affinity.Citation2,Citation9,Citation14,Citation15,Citation17 Besides fucosylation, galactosylation or bisecting N-acetylglucosamine (bisection) were found to modulate the interaction as well, but to a smaller extent. The naturally occurring heterogeneity of mAb glycosylation (i.e., glycan features and glycan pairing) is a major challenge for linking affinity differences to specific glyco- or proteoforms in most assays.
We recently developed an affinity chromatography – mass spectrometry (AC-MS) platform for a glycoform-resolved FcɣRIIIa binding assessment of mAbs.Citation18,Citation19 In contrast to AC-UV,Citation20 individual glycoforms within a complex mixture could be analyzed in a single run by AC-MS, omitting the need for glycoengineering. Furthermore, AC-MS enabled unpreceded insights into typically low-abundant glycoform pairings, which have not been addressed by previous studies. The molecular resolution obtained by AC-MS is an outstanding advantage over established physicochemical techniques such as surface plasmon resonance (SPR). In addition, retention time shifts in AC were previously linked to differences in ADCC activities.Citation21 This suggests that relevant approximations of the in vivo situation can be made by AC. Of note, physicochemical affinity assessment methods have high robustness and resolution. However, they do not fully reflect the in vivo complexity of biological interactions. Cell-based assays allow a better representation of the intricacies associated with immune complexes, at the expense of speed, robustness and resolution.Citation22 Therefore, these approaches are highly complementary and usually go hand in hand in drug discovery efforts.
This study reports, for the first time, the use of an FcɣRIIIb affinity column for the binding assessment of mAbs. We developed the AC conditions for compatibility with MS and showed the glycoform-resolved affinity profiles of several classical and glycoengineered mAbs with high amounts of bisected and hybrid-type glycans. Furthermore, we demonstrated the selectivity of the FcɣRIIIb AC-MS toward different IgG subclasses.
Materials and methods
Chemicals
All chemicals were at least analytical grade and were purchased from Sigma-Aldrich (Steinheim, Germany), if not stated otherwise. A Purelab Ultra system (Veolia Water Technologies Netherlands B.V., Ede, Netherlands) system was used for deionized water. Mobile phases were prepared using an ammonium acetate solution (7.5 M) and glacial acetic acid (Fluka-Honeywell). First, a 1 M stock solution was prepared and further diluted to the target concentration. Proteases (GluC and chymotrypsin) were obtained from Worthington Biochemical Corp. (Lakewood, USA).
Antibodies
Nine different monoclonal antibodies (mAbs) were used in this study. mAb1, mAb3 and mAb4, as well as a Pro329Gly, Leu234Ala and Leu235Ala mutant (PGLALA) of mAb1, were provided by Roche (Penzberg, Germany and Zurich, Switzerland). mAb2 (anti-RhD, reported as +B)Citation2 and anti-TNP subclasses (reported as IgG1*03, IgG2*01, IgG3*01, IgG4*03)Citation16 were provided by Sanquin (Amsterdam, The Netherlands).
Bottom-up liquid chromatography-MS/MS analysis
FcɣRIIIb was subjected to in-gel digestion using GluC and chymotrypsin as reported previously.Citation23 Digested FcɣRIIIb was subjected to bottom-up analysis by reversed-phase liquid chromatography (LC)-MS/MS. The separation of (glyco)peptides was performed on an Easy nLC 1200 system (Thermo Fisher Scientific) using a precolumn (15 mm × 100 μm; Reprosil-Pur C18-AQ 3 μm, Dr. Maisch, Ammerbuch, Germany) and an analytical nanoLC column (150 mm × 75 μm; Reprosil-Pur C18-AQ 3 μm). Mobile phase A was 0.1% formic acid in MQ. A gradient from 10% to 40% mobile phase B (0.1% formic acid/80% acetonitrile) was used for elution of (glyco-)peptides. The LC was hyphenated to an Orbitrap Exploris 480 mass spectrometer. MS1 scans were acquired in an m/z range of 400– 3,500. The MS1 resolution was set to 120,000 (FcɣRIIIb). Data-dependent higher-energy C-trap dissociation (HCD) was used for MS/MS fragmentation. An isolation window of 1.2 Th and a resolution of 30,000 was applied. The charge states 2–7 (FcɣRIIIb) were included for fragmentation. For FcɣRIIIb analysis, HCD with normalized collision energy (NCE) of 30% was performed. In addition, triggered MS/MS (HexNAc loss (204.087)) was used applying stepped NCE of 20%, 30% and 50% combined to one spectrum in an m/z range 110– 3,500.
FcɣRIIIb glycoproteomic data analysis
The obtained (glyco-)peptide cleavage products were verified by automated MS/MS identification using Byonic (v. 3.7.13 Protein Metrics). Next, glycopeptide compositions for each glycosylation site were analyzed based on mass accuracies and retention time differences (MSCitation1 information) by GlycopeptideGraphMS.Citation24 A glycan list covering all glycans from the different glycosylation sites was generated, then applied to each glycopeptide portion, integrated and manually checked based on retention time, mass accuracy (<10 ppm) and isotopic pattern quality (idotp > .85) in Skyline.Citation25 Relative abundances were calculated based on the total area normalization for each glycosylation site.
FcɣRIIIb column preparation
The FcɣRIIIb affinity column was prepared as reported for FcɣRIIIa AC.Citation18,Citation21 In short, human FcɣRIIIb_NA1 was produced in HEK cells as a construct with C-terminal AviTag, IgA protease cleavage site (no cleavage was performed) and IgG Fc part (PGLALA mutant) as reported.Citation26 The material was then biotinylated and 3 mg receptor was immobilized on streptavidin sepharose beads and packed in a Tricorn column housing (5 mm x 50 mm, GE Healthcare). The column volume was 1 mL.
FcɣRIIIb affinity liquid chromatography–mass spectrometry
FcɣRIIIb affinity chromatography was performed using a biocompatible Thermo Ultimate3000 instrument (Thermo Fisher Scientific). For method development, UV detection at 280 nm and online pH monitoring (PCM-3000) were used. The system was operated at 20°C and with a flow rate of 0.5 mL/min. Mobile phase A (30 mM ammonium acetate, pH 6.8) and mobile phase B (50 mM acetic acid, pH 3.0) were used. All samples were buffer-exchanged to mobile phase A (30 kDa molecular weight cutoff filter, Merck, Darmstadt, Germany). 50 µg of the sample were injected for each run. Prior to injection, the column was conditioned for 30 min with mobile phase A. Upon injection, a washing step of 10 min mobile phase A was applied. Then, a linear gradient of 30 min to 80% mobile phase B was used for elution. Next, an additional washing step with 80% mobile phase B for 10 min was used before returning to the starting conditions. The flow was split, diverting approximately 30 µL/min to the mass spectrometer. Online native MS detection was performed using a 15 T solariX Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker Daltonics, Bremen, Germany). MS settings were as described previously.Citation18 Acquired spectra were manually inspected and visualized using DataAnalysis 5.0 (Bruker Daltonics). Deconvolution of mass spectra was performed using the Maximum Entropy tool with an instrument resolving power of 3,000. The deconvolution mass range was set from 145,000 to 155,000 Da. Proteoform assignment was performed manually on deconvoluted mass spectra using a mass tolerance of 50 ppm. The web-based Protein Tool (https://protpi.ch) was used for calculation of average masses considering the mAb sequence, C-terminal lysine clipping, glycosylation, disulfide bonds and N-terminal pyro-glutamine (if applicable). Extracted ion chromatograms were generated for charge states 22+ to 29+ in a window of ±0.4 Th.
Results
Recombinant Fc gamma receptor glycosylation
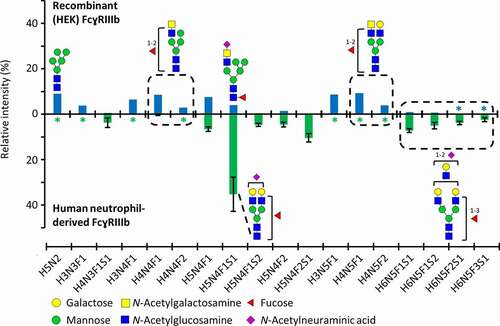
A human embryonic kidney (HEK) cell-produced FcɣRIIIb (NA1) was used in this study. The NA1 variant of FcɣRIIIb has four glycosylation sites (Asn38, Asn74, Asn162, Asn169). At least one peptide moiety for each glycosylation site was verified by MS/MS analysis (Figure S1–S4). A comprehensive site-specific analysis revealed 43 (Asn38), 56 (Asn74), 81 (Asn162) and 25 (Asn169) compositions at the individual sites, amounting to 95 different glycan compositions in total (Figure S5, Table S1). Asn162 glycosylation is visualized in more detail due to its functional importance (). The relative abundance of aglycosylated Asn162 glycopeptide was negligible (0.1%). Furthermore, glycan compositions and structures of highly abundant glycopeptides from recombinant (HEK) and previously reported human neutrophil-derived FcɣRIIIb were compared ().Citation23
Figure 1. Asn162 glycosylation profiles of FcɣRIIIb, comparing recombinant (HEK) and human neutrophil-derived receptor (the latter extracted from Wojcik et al.Citation23). The data was normalized to the sum of all quantified compositions. For recombinant (HEK) FcɣRIIIb, these are listed in Table S1. Glycan structures of selected compositions are suggested based on MS/MS experiments and previous findings on HEK cell-derived or human-derived neutrophil FcɣRIII glycosylation.Citation23,Citation27 Asterisks (*) indicate that a glycopeptide was not detected
Fc gamma receptor IIIb affinity chromatography–mass spectrometry
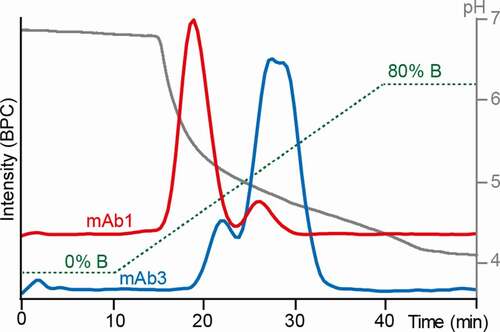
As mAbs have significantly lower affinity for FcɣRIIIb than for FcɣRIIIa, the separation was optimized starting from the previously reported conditions.Citation18,Citation20 Two mAbs were used for gradient optimization, representing either low afucosylation/low-affinity glycoforms (mAb1) or glycoengineered high afucosylation/high-affinity glycoforms (mAb3), respectively. A glycoengineered mAb with high levels of afucosylation showed up to seven-fold increased FcɣRIIIb affinity compared to a mAb with a classical CHO-cell glycosylation profile, considering the average of all glycoforms.Citation8 Therefore, the glycoengineered mAb used for method development (mAb3) was expected to show higher affinity glycoforms compared to mAb1. The fucosylated species showed no retention using reported FcɣRIIIa AC-MS and AC-UV conditions (Figure S6). Sufficient binding of fucosylated species was achieved by lowering the ammonium acetate concentration to 30 mM (mobile phase A). Further, the pH was increased to 6.8 and the column temperature decreased to 20°C, facilitating retention of fucosylated species. A complete elution within the gradient was obtained by an acetic acid concentration of 50 mM in mobile phase B, which resulted in a final pH above 4 (). For both mAbs, doubly fucosylated species could be chromatographically separated from singly and doubly afucosylated species. The charge state distribution of native MS spectra ([M + 22 H]22+ to [M + 29 H]29+) did not change within the gradient. This indicates that the mAbs do not undergo large conformational changes under the applied separation conditions (Figure S7). An Fc-engineered version of mAb1 was used as a negative control to check for nonspecific interactions. This mAb1 version contained the Pro329Gly, Leu234Ala and Leu235Ala mutations, which abolish Fc receptor binding.Citation28 None of the glycoforms showed any retention (Figure S8).
Glycoform-resolved FcɣRIIIb AC-MS of IgG1 mAbs
Four different IgG1 mAbs, produced in Chinese hamster ovary (CHO) cells, were analyzed in a glycoform-resolved manner by FcɣRIIIb AC-MS (Table S2, S3, ). Of note, the constant part of the heavy chains of mAb1, mAb3 and mAb4 is based on the G1m17 allotype (mAb1 = G1m17, mAb3/4 = G1m17,1), whereas mAb2 is based on the G1m3 allotype. The constant part of the light chains of all four mAbs is based on the Km3 allotype.
Figure 3. FcɣRIIIb AC-MS analysis of four IgG1 mAbs. A – D represent mAb1 to mAb4. Extracted ion chromatograms of glycoforms are displayed (Figure S9, S11 – 13, Table S3). Zoom in A visualizes low-abundant afucosylated glycoforms of mAb1. Some assigned minor glycoforms (< 5% relative to the main peak) of mAb2 – mAb4 are not displayed for visibility reasons
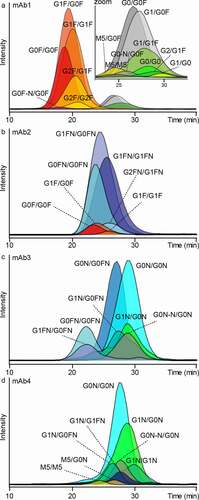
Doubly fucosylated (GxF/GxF), singly afucosylated (GxF/Gx) and doubly afucosylated (M5/M5, Gx/Gx) species were resolved by FcɣRIIIb AC-MS. The assigned glycoforms of mAb1, affinity ranking and inter-day retention time variability showed high similarity to previously reported FcɣRIIIa AC-MS results (Table S3, Figure S9, Figure S10).Citation18
Next, the applicability of FcɣRIIIb AC-MS to different glycoengineered mAbs (mAb2 – mAb4) was tested. Glycoforms of mAb2 were assigned mainly to fucosylated structures with varying levels of galactosylation and bisection (Figure S11, Table S3). Both galactosylation and bisection of mAb2 slightly increased retention in FcɣRIIIb AC-MS. When comparing different IgG1 mAbs (), the amino acid sequence apparently contributes to the retention. For example, the doubly fucosylated glycoforms (G0F/G0F, G1F/G0F, G1F/G1F) of mAb2 had increased retention compared to mAb1. In addition, the bisected fucosylated glycoforms of mAb2 (GxFN/GxFN) had increased retention compared to mAb3. Of note, the assigned glycoforms showed a preferential pairing of bisected glycoforms (GxFN/GxFN) or non-bisected glycoforms (GxF/GxF), whereas hemi-bisected glycoforms (GxFN/GxF) were not assigned. A minor amount of M5/M5 was detected and showed the highest retention time in mAb2 (Figure S11, Table S3).
mAbs glycoengineered via GlycoMab technologyCitation29 (mAb3 and mAb4) showed predominantly bisected, afucosylated glycoforms (Figure S12, S13, Table S3). mAb3 showed bisected glycoforms with different fucosylation levels (2x, 1x, 0x). For mAb4, no doubly fucosylated glycoforms and overall higher afucosylation were observed. In addition, glycoforms consisting only of the conserved pentasaccharide core and a bisecting N-acetylglucosamine (G0N-N) were assigned, based on glycan data of the GlycoMab technology.Citation29 The influence of galactosylation was more pronounced for mAb4 (e.g., ∆G0N/G0N vs. G1N/G0N) compared to mAb3. Low abundant high mannose (M5/M5) species showed the lowest retention of afucosylated glycoforms for both mAbs.
In addition, a singly oxidized (Met252) variant of mAb4 was subjected to FcɣRIIIb AC-MS (Figure S14). Enrichment was achieved by neonatal Fc receptor (FcRn) chromatography as described previously.Citation30 The glycoform masses of the oxidized version were in agreement with one additional oxidation. No notable influence of the oxidation on retention in FcɣRIIIb AC-MS was observed (Figure S14).
IgG subclass specificity of FcɣRIIIb visualized by AC-MS
We demonstrated that the reported IgG subclass specificity (IgG3 > IgG1, no retention of IgG2 and IgG4) of FcɣRIIIb binding was reflected in the AC-MS approach. For this, a representative allotype of each of the four subclasses was analyzed. The different subclasses are referred to as IgG1 (G1m3; Km3), IgG2 (G2m(.); Km3), IgG3 (G3m(b*); Km3) and IgG4 (G4m(a); Km3). Similar glycosylation profiles and the same antigen-binding fragment (Fab) sequence (anti-trinitrophenol (TNP)) allowed direct comparison of glycoforms (). Mainly, fucosylated complex-type glycans with varying levels of galactosylation (GxF/GxF) were observed. As expected, IgG2 and IgG4 did not show retention on FcɣRIIIb AC ().
Figure 4. Anti-TNP IgG subclass comparison by FcɣRIIIb AC-MS. Extracted ion chromatograms of the major glycoforms are displayed for IgG1-4 (in panels A-D, respectively)
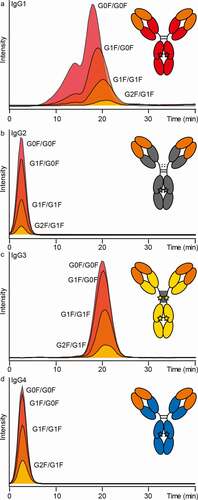
The doubly fucosylated glycoforms of IgG1 (G1m3; Km3) showed comparable retention to the same glycoforms in mAb1 (G1m17; Km3). In addition to this main peak (16–25 min; peak 2), a partially separated peak with reduced retention (7–16 min; peak 1) was observed (). The two peaks did not show a difference in the intact mass profiles (Figure S15). Both peaks were fractionated in triplicates, analyzed by tryptic bottom-up analysis and checked for deamidation levels in the Fc (data not shown). However, the bottom-up data were inconclusive to link the partial separation to a potential Fc deamidation site.
IgG3 showed a slightly higher affinity compared to IgG1, but a less pronounced increase in retention time for galactosylation (, c). In addition to the main IgG3 glycoforms (GxF/GxF), glycoforms with O-glycosylation (+1x H1N1S2 or +2x H1N1S2) were assigned (Figure S16). However, no noticeable influence on the FcɣRIIIb affinity was observed for the IgG3 hinge O-glycosylation (Figure S17).
Discussion
Receptor glycosylation
FcɣRIIIb glycosylation changes at Asn162 were reported to impact the interaction of FcɣRIIIb with an IgG1 Fc fragment.Citation17 Therefore, it is imperative to report both the antibody and the receptor glycosylation, in order to facilitate the comparison of results between different interaction studies.Citation5,Citation31 The recombinant FcɣRIIIb glycan profile of Asn162 showed differences compared to recently reported natural FcɣRIIIb glycosylation (). Recombinant vs. natural FcɣRIIIb showed higher levels of oligomannose forms (9.9% vs. 1.7%), lower levels of sialylation (average number of sialic acids per glycan 0.2 vs. 0.9) and LacDiNAc structures were exclusively present on recombinant FcɣRIIIb.Citation31 Furthermore, the amount of LacNAc units was less abundant in recombinant FcɣRIIIb (). FcɣRIIIb from both sources showed antennary fucosylation (, average number of fucoses per glycan 1.0 vs. 1.3). Interpreting the complex interplay between FcɣRIIIb and antibody glycosylation remains challenging due to the scarcity of receptor glycosylation studies. Though we found glycosylation differences between recombinant and primary human FcɣRs, there is as yet no indication that this would affect the affinity ranking of different antibody glycoforms. Nonetheless, the observed differences in oligomannose forms may well affect the absolute affinity of the antibodies.Citation17,Citation32 In conclusion, as we only report affinity rankings of IgG glycoforms, the recombinant FcɣRIIIb is expected to provide state-of-the-art information. If, in the future, the use of recombinant FcɣR with more human-like glycosylation profiles would be attainable and desirable, our method offers the possibility to quickly adapt to the potential changes in affinity.
Specific advantages of affinity chromatography for low-affinity interactions
FcɣRIIIb shows a very low affinity toward IgG1 (KD ~ 10 µM) even when compared to other low-affinity FcɣRs (FcɣRIIIa, FcɣRIIa).Citation14,Citation16 For IgG1, a 10-fold lower affinity toward FcɣRIIIb compared to FcɣRIIIa was reported.Citation13,Citation33 The low affinity of the receptor–antibody interaction was reported to lead to assay-to-assay variation for FcɣRIIIb binding.Citation15 A recent study applying SPR could not even determine binding affinities for different mAbs because the FcɣRIIIb affinity was too weak.Citation16 In our study, AC conditions reported for FcɣRIIIa showed insufficient retention for some glycoforms of the tested mAbs in FcɣRIIIb AC. However, a generic FcɣRIIIb AC gradient was developed with which both low- and high-affinity mAb glycoforms could be analyzed. A pH gradient alone was not sufficient to achieve this broad coverage. Because we had observed a stronger susceptibility of the FcɣRIIIb interaction to the buffer concentration, compared to FcɣRIIIa, we successfully supplemented the pH gradient with a minor increase in buffer concentration. Of note, the pH gradient itself leads to a decrease in ionic strength. This likely counteracts the elution of antibodies, which is mitigated by increasing the buffer concentration with the gradient. This demonstrates one advantage of an AC-based assay for receptor antibody interaction studies. The chromatographic conditions offer much design space, while at the same time being tightly controlled. For example, different gradient parameters (ionic strength, pH) can be easily varied, and the temperature is more easily controlled in a flow than in a batch setup. Consequently, AC-MS assays can be both highly adaptable and robust. A significant presence of nonspecific binding, for example, through ionic interactions with the column material, was excluded using negative controls (mAb1 PGLALA variant, anti-TNP IgG2, anti-TNP IgG4). For different IgG1 mAbs and IgG subclasses, we could demonstrate that FcɣRIIIb AC is a powerful and generic assay for assessing weak antibody–receptor interactions. It should be noted that the weak binding toward FcɣRIIIb is expected to be less critical when studying multivalent interactions (avidity), i.e., of immune complexes, as opposed to monomeric mAb interactions (affinity). The affinity ranking of individual proteoforms is more insightful than solely KD determination of proteoform mixtures by classical binding assays, such as SPR.
Advantages of AC-MS for glycoform resolution
The online hyphenation of FcɣRIIIb AC to native MS allows comprehensive proteoform assessment of heterogeneous mAbs and can resolve subtle affinity differences. We demonstrated recently the advantages of FcɣRIIIa AC-MS.Citation18 In this and the previous study, mAb1 was comprehensively analyzed in a glycoform-resolved manner. The FcɣRIIIb AC-MS glycoform affinity ranking and interday retention time stability of mAb1 showed high similarity compared to FcɣRIIIa AC-MS (Figure S10).
Of note, the data are not directly comparable, due to the need to adapt the chromatographic conditions to the lower FcɣRIIIb affinity. The level of fucosylation had the biggest effect on the FcɣRIIIb affinity, which is attributed to a unique glycan–glycan interaction between antibody and receptor.Citation3 This is in line with other reports on FcɣRIIIb-IgG interactions.Citation2,Citation8,Citation9,Citation12,Citation15
We observed differences in the effect of galactosylation in the studied mAbs. Of note, galactosylation has recently been found to affect the hexamerization potential of IgG1, which may possibly increase avidity contributions to FcɣR interactions.Citation34,Citation35 We found no indication that other multivalent interactions, such as aggregate formation exist in our analytical setup. Interestingly, the effect of galactosylation in glycoengineered mAbs was more pronounced for mAb4 than for mAb3 (G0N/G0N vs G1N/G0N). This may be related to different linkages of galactosylation (presence of terminal galactose on the 1,3-arm or the 1,6-arm), which show a positive (1,6) or no effect (1,3) on FcɣRIIIa binding.Citation19,Citation36
mAb2 showed the highest affinity of all doubly fucosylated and afucosylated (M5/M5) glycoforms (Table S3). A positive impact of the allotype can be excluded when comparing G0F/G0F of mAb2 with IgG1 (both G1m3; Km3). mAb2 was previously analyzed by FcɣRIIIa AC-MS and the glycoforms showed increased RT as well when comparing to mAb1.Citation18 However, the difference in FcɣRIIIa affinity was not emphasized in the previous work. As similar FcɣRIIIa binding profiles were reported, independent of the IgG1 allotype,Citation16 the data hints to an influence of the variable domains on FcɣRIII binding. This is in line with a recently demonstrated influence of the Fab on IgG1-FcɣRIII receptor interaction.Citation37,Citation38 Although the Fab interaction sites were mainly attributed to the CH1 domain, amino acids within the variable domain were also found to contribute to the interaction.Citation37 However, further studies using orthogonal techniques are needed to validate the observed inter-mAb FcɣRIIIb differences. If a strong impact of the variable Fab domain on FcɣRIII would be confirmed, this may have very important implications for modulating mAb effector functions.
The glycoengineered mAbs featured several unstudied pairings, for example, the combination of highly abundant G0N with oligomannose (G0N/M5) or bisected mono-antennary (G0N/G0N-N) glycoforms. Their investigation would require laborious glycoengineering, reducing, but not even fully eliminating, the heterogeneity of the mAb glycoform profiles. This highlights the advantage of FcɣRIIIb AC-MS for affinity ranking of individual proteoforms. Here, we described the analysis of (therapeutic) mAbs produced in CHO or HEK cells, which are widely used production systems.Citation39 Potential glycosylation differences in human IgG antibodies are extensively discussed in the literature.Citation10 The developed method is expected to apply to the majority of therapeutic mAbs. However, it should be noted that the proteoform resolution may be highly impaired for mAbs with additional complexity besides Fc glycosylation, e.g., Fab glycosylation. These may require additional method development as shown for the analysis of cetuximab by FcɣRIIIa AC-MS.Citation19 Finally, it should be stressed that the developed assay targets qualitative differences between glycoforms. More precise insights may be obtained by comparing retention time differences in AC-MS to quantitative data of highly homogeneous glycoengineered mAbs or fractions obtained by AC-MS.
Affinity impact of additional post-translational modifications
Moreover, the glycoform-resolution obtained by FcɣRIIIb AC-MS allowed study of the affinity impact of post-translational modifications (PTMs), other than Fc N-glycosylation.
We concluded that oxidation at Met252 is not critical for FcɣRIIIb binding (mAb4, Figure S14). In contrast, FcRn and FcɣRIIa binding have been reported to be decreased in the presence of Met252 oxidation.Citation30,Citation40 These differences with respect to Fc receptor binding are in line with the proximity to the different binding sites. In conclusion, we could demonstrate that AC-MS allowed the analysis of specifically enriched oxidized mAb variants for FcɣRIIIb binding differences. Of note, PTMs cannot be located by intact mass analysis alone, and the co-occurrence of multiple PTMs may hamper the data interpretation.
The partial separation of IgG1 glycoforms () was hypothesized to be related to Asn deamidation in the Fc. Increased Asn325 deamidation levels were previously reported to reduce FcɣRIIIa binding.Citation41,Citation42 However, additional experiments were inconclusive in supporting this interpretation. Partially reduced or scrambled disulfide bonds are another potential PTM, which is not resolved by intact mass analysis. This modification was also shown to affect FcɣRIIIa binding.Citation43 Additional experiments would be needed to pinpoint the modification responsible for the additional separation of IgG1 proteoforms.
In contrast to the other IgG subclasses, some IgG3 allotypes additionally contain O-glycans. We could demonstrate the feasibility of measuring intact IgG3 with FcɣRIIIb AC-MS. For the first time, we showed that the hinge O-glycosylation of IgG3 does not significantly influence the FcɣRIIIb affinity. Moreover, the affinity of IgG3 was found to be slightly higher compared to IgG1, which is in line with recent findings.Citation16 IgG3-based biopharmaceuticals are not yet on the market, but the first clinical trials were recently started.Citation33 Despite its disadvantages, IgG3 has several benefits and may be reconsidered for biopharmaceutical applications in the future. This makes a proteoform-resolved method for Fc receptor affinity assessments highly desirable.Citation33
The combination of FcɣRIIIb AC and MS is very powerful to unravel complex glycosylated antibodies. It allows unpreceded molecular insights into the relative affinity ranking of unstudied glycoform pairings. We demonstrated the applicability to different (glycoengineered) mAbs. In addition, our data show the potential of AC-MS-based affinity assessments to simultaneously study the impact of different forms of post-translational modifications. All of this makes the method attractive for supporting the assessment of critical quality attributes.
Importance of understanding FcɣRIIIb-mediated effector functions
The role of FcɣRIIIb binding is less studied and understood compared to FcɣRIIIa. Whereas the FcɣRIIIa affinity of mAbs positively correlates with natural killer cell-mediated ADCC, recent studies demonstrated that glycoengineered/high-affinity mAbs increased ADCP, whereas ADCC was impaired.Citation8,Citation44,Citation45 In this context, it is debated whether FcɣRIIIb supports or hinders the FcɣRIIa mediated activation of neutrophils.Citation7,Citation44 There is also evidence that FcɣRIIIb by itself can lead to neutrophil activation.Citation46 Therefore, it is important to be aware of the relative expression levels and affinity of mAb proteoforms toward FcɣRIIIb and FcɣRIIa.Citation47Citation48 This could be crucial in understanding which neutrophil effector functions are activated by a therapeutic mAb. In addition, the use of high FcɣRIII-affinity mAbs has been linked to safety concerns due to first infusion reactions, mediated by neutrophil FcɣRIIIb.48 Since FcɣRIIIb is uniquely present in humans, these effects cannot be assessed by animal-based preclinical studies. In conclusion, an analytical platform for differentiating FcɣRIIIb affinities of mAb proteoforms, as presented in this study, is of high relevance to better understand and predict the safety and efficacy of therapeutic mAbs.
Abbreviations
ACAffinity chromatography
ADCCAntibody-dependent cellular cytotoxicity
ADCPAntibody-dependent cellular phagocytosis
CHConstant domain heavy chain
CHOChinese hamster ovary
FabFragment antigen binding
FcFragment crystallizable
FcɣRFc gamma receptor
FcRnNeonatal Fc receptor
FT-ICRFourier transform ion cyclotron resonance
HCDHigher-energy C-trap dissociation
HEKHuman embryonic kidney
IgGImmunoglobulin G
LCLiquid chromatography
mAbMonoclonal antibody
MSMass spectrometry
NCENormalized collision energy
PTMPost-translational modification
SPRSurface-plasmon resonance
UVUltraviolet
Supplemental Material
Download MS Word (1.9 MB)Acknowledgments
This research was supported by the Roche/Genentech Global Strategy Team (GST). David Falck received funding from the Dutch Research Council (NWO) in the framework of the ENW PPP Fund for the top sectors (project Proteoform-resolved pharmacokinetics of biopharmaceuticals, no. 019.012). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No[765502], A4B project.
Disclosure statement
Alexander Knaupp, Erwin van Puijenbroek, Dietmar Reusch and Tilman Schlothauer are employees of Roche/Genentech, the manufacturer of the column and the therapeutic antibodies used in this research.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Nimmerjahn F, Ravetch JV. Fcγ receptors as regulators of immune responses. Nat Rev Immunol. 2008;8(1):34–10. doi:10.1038/nri2206. PMID: 18064051.
- Dekkers G, Treffers L, Plomp R, Bentlage AEH, De Boer M, Koeleman CAM, Lissenberg-Thunnissen SN, Visser R, Brouwer M, Mok JY. Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of fc-receptor- and complement-mediated-effector activities. Front Immunol. 2017;8(877). doi:10.3389/fimmu.2017.00877. PMID: 28824618.
- Ferrara C, Grau S, Jäger C, Sondermann P, Brünker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M. Unique carbohydrate–carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc Natl Acad Sci U S A. 2011;108(31):12669–74. doi:10.1073/pnas.1108455108. PMID: 21768335.
- Radaev S, Motyka S, Fridman W-H, Sautes-Fridman C, Sun PD. The structure of a human type III Fcγ receptor in complex with Fc. J Biol Chem. 2001;276(19):16469–77. doi:10.1074/jbc.M100350200. PMID: 11297532.
- Barb AW. Fc γ receptor compositional heterogeneity: considerations for immunotherapy development. J Biol Chem. 2021;296:100057. PMID: 33172893. doi:10.1074/jbc.REV120.013168.
- Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat Rev Drug Discov. 2009;8(3):226–34. doi:10.1038/nrd2804. PMID: 19247305.
- Kerntke C, Nimmerjahn F, Biburger M. There is (scientific) strength in numbers: a comprehensive quantitation of Fc gamma receptor numbers on human and murine peripheral blood leukocytes. Front Immunol. 2020;11(118). doi:10.3389/fimmu.2020.00118. PMID: 32117269.
- Golay J, Da Roit F, Bologna L, Ferrara C, Leusen JH, Rambaldi A, Klein C, Introna M. Glycoengineered CD20 antibody obinutuzumab activates neutrophils and mediates phagocytosis through CD16B more efficiently than rituximab. Blood. 2013;122(20):3482–91. doi:10.1182/blood-2013-05-504043. PMID: 24106207.
- Subedi GP, Barb AW. The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc γ receptor. mAbs. 2016;8(8):1512–24. doi:10.1080/19420862.2016.1218586. PMID: 27492264.
- Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes. Glycobiology. 2015;25(12):1325–34. doi:10.1093/glycob/cwv065. PMID: 26263923.
- Alt N, Zhang TY, Motchnik P, Taticek R, Quarmby V, Schlothauer T, Beck H, Emrich T, Harris RJ. Determination of critical quality attributes for monoclonal antibodies using quality by design principles. Biologicals. 2016;44(5):291–305. doi:10.1016/j.biologicals.2016.06.005. PMID: 27461239.
- Roberts JT, Barb AW. A single amino acid distorts the Fc γ receptor IIIb/CD16b structure upon binding immunoglobulin G1 and reduces affinity relative to CD16a. J Biol Chem. 2018;293(51):19899–908. doi:10.1074/jbc.RA118.005273. PMID: 30361439.
- Caaveiro JM, Kiyoshi M, Tsumoto K. Structural analysis of Fc/FcγR complexes: a blueprint for antibody design. Immunol Rev. 2015;268(1):201–21. doi:10.1111/imr.12365. PMID: 26497522.
- Bruhns P, Iannascoli B, England P, Mancardi DA, Fernandez N, Jorieux S, Daëron M. Specificity and affinity of human Fcγ receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113(16):3716–25. doi:10.1182/blood-2008-09-179754. PMID: 19018092.
- Peipp M, Lammerts van Bueren JJ, Schneider-Merck T, Bleeker WWK, Dechant M, Beyer T, Repp R, van Berkel PHC, Vink T, van de Winkel JGJ. Antibody fucosylation differentially impacts cytotoxicity mediated by NK and PMN effector cells. Blood. 2008;112(6):2390–99. doi:10.1182/blood-2008-03-144600. PMID: 18566325.
- De Taeye SW, Bentlage AEH, Mebius MM, Meesters JI, Lissenberg-Thunnissen S, Falck D, Sénard T, Salehi N, Wuhrer M, Schuurman J. FcγR binding and ADCC activity of human IgG allotypes. Front Immunol. 2020;11(740). doi:10.3389/fimmu.2020.00740. PMID: 32435243.
- Subedi GP, Barb AW. CD16a with oligomannose-type N-glycans is the only “low-affinity” Fc γ receptor that binds the IgG crystallizable fragment with high affinity in vitro. J Biol Chem. 2018;293(43):16842–50. doi:10.1074/jbc.RA118.004998. PMID: 30213862.
- Lippold S, Nicolardi S, Domínguez-Vega E, Heidenreich A-K, Vidarsson G, Reusch D, Haberger M, Wuhrer M, Falck D. Glycoform-resolved FcɣRIIIa affinity chromatography–mass spectrometry. mAbs. 2019;11(7):1191–96. doi:10.1080/19420862.2019.1636602. PMID: 31276431.
- Lippold S, Nicolardi S, Wuhrer M, Falck D. Proteoform-resolved FcɤRIIIa binding assay for fab glycosylated monoclonal antibodies achieved by affinity chromatography mass spectrometry of Fc moieties. Front Chem. 2019;7(698). doi:10.3389/fchem.2019.00698. PMID: 31709228.
- Dashivets T, Thomann M, Rueger P, Knaupp A, Buchner J, Schlothauer T. Multi-angle effector function analysis of human monoclonal IgG glycovariants. PLoS One. 2015;10(12):e0143520. doi:10.1371/journal.pone.0143520. PMID: 26657484.
- Thomann M, Schlothauer T, Dashivets T, Malik S, Avenal C, Bulau P, Rüger P, Reusch D. In vitro glycoengineering of IgG1 and its effect on Fc receptor binding and ADCC activity. PLoS One. 2015;10(8):e0134949. doi:10.1371/journal.pone.0134949. PMID: 26266936.
- Cymer F, Beck H, Rohde A, Reusch D. Therapeutic monoclonal antibody N-glycosylation – structure, function and therapeutic potential. Biologicals. 2018;52:1–11. PMID: 29239840. doi:10.1016/j.biologicals.2017.11.001.
- Wojcik I, Sénard T, de Graaf EL, Janssen GMC, De Ru AH, Mohammed Y, Van Veelen PA, Vidarsson G, Wuhrer M, Falck D. Site-specific glycosylation mapping of Fc gamma receptor IIIb from neutrophils of individual healthy donors. Anal Chem. 2020;92(19):13172–81. doi:10.1021/acs.analchem.0c02342. PMID: 32886488.
- Choo MS, Wan C, Rudd PM, Nguyen-Khuong T. GlycopeptideGraphMS: improved glycopeptide detection and identification by exploiting graph theoretical patterns in mass and retention time. Anal Chem. 2019;91(11):7236–44. doi:10.1021/acs.analchem.9b00594. PMID: 31079452.
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–68. doi:10.1093/bioinformatics/btq054. PMID: 20147306.
- Freimoser–Grundschober A, Rueger P, Fingas F, Sondermann P, Herter S, Schlothauer T, Umana P, Neumann C. FcγRIIIa chromatography to enrich a-fucosylated glycoforms and assess the potency of glycoengineered therapeutic antibodies. J Chromatogr A. 2020;1610:460554. PMID: 31597603. doi:10.1016/j.chroma.2019.460554.
- Zeck A, Pohlentz G, Schlothauer T, Peter-Katalinić J, JT R. Cell type-specific and site directed N-glycosylation pattern of FcγRIIIa. J Proteome Res. 2011;10(7):3031–39. doi:10.1021/pr1012653. PMID: 21561106.
- Schlothauer T, Herter S, Koller CF, Grau-Richards S, Steinhart V, Spick C, Kubbies M, Klein C, Umaña P, Mössner E. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng Des Sel. 2016;29(10):457–66. doi:10.1093/protein/gzw040. PMID: 27578889.
- Umaña P, Jean–Mairet J, Moudry R, Amstutz H, Bailey JE. Engineered glycoforms of an antineuroblastoma IgG1 with optimized antibody-dependent cellular cytotoxic activity. Nat Biotechnol. 1999;17(2):176–80. doi:10.1038/6179. PMID: 10052355.
- Cymer F, Thomann M, Wegele H, Avenal C, Schlothauer T, Gygax D, Beck H. Oxidation of M252 but not M428 in hu-IgG1 is responsible for decreased binding to and activation of hu-FcγRIIa (His131). Biologicals. 2017;50:125–28. PMID: 28988621. doi:10.1016/j.biologicals.2017.09.006.
- Hayes JM, Frostell A, Karlsson R, Müller S, Martín SM, Pauers M, Reuss F, Cosgrave EF, Anneren C, Davey GP. Identification of Fc gamma receptor glycoforms that produce differential binding kinetics for rituximab. Mol Cell Proteomics. 2017;16(10):1770–88. doi:10.1074/mcp.M117.066944. PMID: 28576848.
- Cambay F, Forest-Nault C, Dumoulin L, Seguin A, Henry O, Durocher Y, De Crescenzo G. Glycosylation of Fcγ receptors influences their interaction with various IgG1 glycoforms. Mol Immunol. 2020;121:144–58. PMID: 32222585. doi:10.1016/j.molimm.2020.03.010.
- Chu TH, Patz EF, Ackerman ME. Coming together at the hinges: therapeutic prospects of IgG3. mAbs. 2021;13(1):1882028. doi:10.1080/19420862.2021.1882028. PMID: 33602056.
- Wei B, Gao X, Cadang L, Izadi S, Liu P, Zhang H-M, Hecht E, Shim J, Magill G, Pabon JR. Fc galactosylation follows consecutive reaction kinetics and enhances immunoglobulin G hexamerization for complement activation. mAbs. 2021;13(1):1893427. doi:10.1080/19420862.2021.1893427. PMID: 33682619.
- Thijs LJ, Van Osch JN, Derksen NIL, van Mierlo G, Ellen van der Schoot C, Wuhrer M, Rispens T, Vidarsson G. Fc galactosylation promotes hexamerization of human IgG1, leading to enhanced classical complement activation. Journal of Immunology (Baltimore, Md: 1950). 2021. doi:10.4049/jimmunol.2100399. PMID: 34408013.
- Aoyama M, Hashii N, Tsukimura W, Osumi K, Harazono A, Tada M, Kiyoshi M, Matsuda A, Ishii-Watabe A. Effects of terminal galactose residues in mannose α1-6 arm of Fc-glycan on the effector functions of therapeutic monoclonal antibodies. mAbs. 2019;11(5):826–36. doi:10.1080/19420862.2019.1608143. PMID: 30990348.
- Yogo R, Yamaguchi Y, Watanabe H, Yagi H, Satoh T, Nakanishi M, Onitsuka M, Omasa T, Shimada M, Maruno T. The fab portion of immunoglobulin G contributes to its binding to Fcγ receptor III. Sci Rep. 2019;9(1):11957. doi:10.1038/s41598-019-48323-w. PMID: 31420591.
- Sun Y, Izadi S, Callahan M, Deperalta G, Wecksler AT. Antibody–receptor interactions mediate antibody-dependent cellular cytotoxicity. J Biol Chem. 2021;297(1):100826. doi:10.1016/j.jbc.2021.100826. PMID: 34044019.
- de Haan N, Falck D, Wuhrer M. Monitoring of immunoglobulin N- and O-glycosylation in health and disease. Glycobiology. 2019;30(4):226–40. doi:10.1093/glycob/cwz048. PMID: 31281930.
- Schlothauer T, Rueger P, Stracke JO, Hertenberger H, Fingas F, Kling L, Emrich T, Drabner G, Seeber S, Auer J. Analytical FcRn affinity chromatography for functional characterization of monoclonal antibodies. mAbs. 2013;5(4):576–86. doi:10.4161/mabs.24981. PMID: 23765230.
- Evans AR, Capaldi MT, Goparaju G, Colter D, Shi FF, Aubert S, Li L-C, Mo J, Lewis MJ, Hu P. Using bispecific antibodies in forced degradation studies to analyze the structure–function relationships of symmetrically and asymmetrically modified antibodies. mAbs. 2019;11(6):1101–12. doi:10.1080/19420862.2019.1618675. PMID: 31161859.
- Lu X, Machiesky LA, De Mel N, Du Q, Xu W, Washabaugh M, Jiang X-R, Wang J. Characterization of IgG1 Fc deamidation at asparagine 325 and its impact on antibody-dependent cell-mediated cytotoxicity and FcγRIIIa binding. Sci Rep. 2020;10(1):383. doi:10.1038/s41598-019-57184-2. PMID: 31941950.
- Bahou C, Love EA, Leonard S, Spears RJ, Maruani A, Armour K, Baker JR, Chudasama V. Disulfide modified IgG1: an investigation of biophysical profile and clinically relevant Fc interactions. Bioconjug Chem. 2019;30(4):1048–54. doi:10.1021/acs.bioconjchem.9b00174. PMID: 30855134.
- Derer S, Glorius P, Schlaeth M, Lohse S, Klausz K, Muchhal U, Desjarlais JR, Humpe A, Valerius T, Peipp M. Increasing FcγRIIa affinity of an FcγRIII-optimized anti-EGFR antibody restores neutrophil-mediated cytotoxicity. mAbs. 2014;6(2):409–21. doi:10.4161/mabs.27457. PMID: 24492248.
- Treffers LW, van Houdt M, Bruggeman CW, Heineke MH, Zhao XW, van der Heijden J, Nagelkerke SQ, Verkuijlen PJJH, Geissler J, Lissenberg-Thunnissen S. FcγRIIIb restricts antibody-dependent destruction of cancer cells by human neutrophils. Front Immunol. 2019;9:3124–3124. PMID: 30761158. doi:10.3389/fimmu.2018.03124.
- García-García E, Nieto-Castañeda G, Ruiz-Saldaña M, Mora N, Rosales C. FcγRIIA and FcγRIIIB mediate nuclear factor activation through separate signaling pathways in human neutrophils. Journal of Immunology (Baltimore, Md: 1950). 2009;182(8):4547–56. doi:10.4049/jimmunol.0801468. PMID: 19342628.
- Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14(2):94–108. doi:10.1038/nri3582. PMID: 24445665.
- Weber F, Breustedt D, Schlicht S, Meyer CA, Niewoehner J, Ebeling M, Freskgard PO, Bruenker P, Singer T, Reth M. First infusion reactions are mediated by FcγRIIIb and neutrophils. Pharm Res. 2018 Jun 27;35(9):169. doi:10.1007/s11095-018-2448-8. PMID: 29951887; PubMed Central PMCID: PMCPMC6021477.