 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
The self-association of therapeutic antibodies can result in elevated viscosity and create problems in manufacturing and formulation, as well as limit delivery by subcutaneous injection. The high concentration viscosity of some antibodies has been reduced by variable domain mutations or by the addition of formulation excipients. In contrast, the impact of Fc mutations on antibody viscosity has been minimally explored. Here, we studied the effect of a panel of common and clinically validated Fc mutations on the viscosity of two closely related humanized IgG1, κ antibodies, omalizumab (anti-IgE) and trastuzumab (anti-HER2). Data presented here suggest that both Fab-Fab and Fab-Fc interactions contribute to the high viscosity of omalizumab, in a four-contact model of self-association. Most strikingly, the high viscosity of omalizumab (176 cP) was reduced 10.7- and 2.2-fold by Fc modifications for half-life extension (M252Y:S254T:T256E) and aglycosylation (N297G), respectively. Related single mutations (S254T and T256E) each reduced the viscosity of omalizumab by ~6-fold. An alternative half-life extension Fc mutant (M428L:N434S) had the opposite effect in increasing the viscosity of omalizumab by 1.5-fold. The low viscosity of trastuzumab (8.6 cP) was unchanged or increased by 2-fold by the different Fc variants. Molecular dynamics simulations provided mechanistic insight into the impact of Fc mutations in modulating electrostatic and hydrophobic surface properties as well as conformational stability of the Fc. This study demonstrates that high viscosity of some IgG1 antibodies can be mitigated by Fc mutations, and thereby offers an additional tool to help design future antibody therapeutics potentially suitable for subcutaneous delivery.
Introduction
In the past several decades, antibody therapeutics have become increasingly prevalent for the treatment of a diverse array of serious human maladies, including many different cancers, as well as inflammatory, autoimmune, ophthalmologic, hematologic, infectious, and metabolic diseases.Citation1 Over 170 antibodies are currently approved as therapeutics, making antibodies one of the most clinically impactful class of drugs in the pharmaceutical armamentarium.Citation2
Antibody therapeutics are commonly administered to patients by intravenous infusion (IV),Citation1 but subcutaneous (SC) delivery is increasingly becoming an option, offering greater convenience for patients and healthcare professionals as well as potentially reducing healthcare costs.Citation3,Citation4 The typical injection volume for SC administration is 2.0 mL, commonly necessitating high antibody concentration (
100 mg/mL) to deliver the desired dose.Citation4 Such high antibody concentrations pose technical challenges that may raise the cost and delay the development of antibody therapeutics.Citation5,Citation6 Antibody high concentration properties, including viscosity, are typically first evaluated at a late stage in preclinical development when large quantities (>100 mg) of one or a few clinical candidates are available. High concentration problems identified at this stage can delay projects and be resource-intensive to fix through protein engineering, replacement of clinical lead candidates and/or formulation. Thus, earlier stage assessment of high concentration antibody properties is of high interest in the development of antibody therapeutics, particularly for SC delivery.
The primary method for directly measuring viscosity is by cone-and-plate rheometry that requires at least tens of milligrams of antibody protein.Citation7 Efforts to establish high-throughput low material methods to predict viscosity experimentally,Citation8–13 or through sequence-based computational methods,Citation14–20 have shown moderate correlations, but have yet to yield strong predictive power. Assessing self-association of antibody solutions can be measured, either directly or indirectly, in a variety of ways. Methods, including dynamic light scattering (DLS),Citation9–11,Citation21 self-interaction nanoparticle spectroscopy,Citation10,Citation22,Citation23 analytical ultracentrifugation,Citation9,Citation11 and fluorescence correlation spectroscopy,Citation13 have been used to investigate the self-association of antibodies and other proteins as proxies for the measurement of viscosity.
Antibody self-association can sometimes be mitigated through amino acid mutations in antibody variable domains.Citation21,Citation24–27 For example, single mutations of aromatic residues reduced the viscosity of a bispecific anti-IL-13/IL-17 IgG4 (13 centipoise (cP)) by up to 4-fold.Citation27 Multi-parameter optimization has recently been demonstrated for an anti-GCGR antibody.Citation21 Specifically, up to a 2-fold reduction in the viscosity of the parent antibody (36 cP) was achieved with single-point mutations in variable domains while maintaining comparable antigen-binding affinity, polyspecificity, predicted T cell epitopes, humanness score, computational developability score, and Fv isoelectric point (pI).Citation21 Additional focus on the Fv region electrostatics with PfAbNet generalization has highlighted how negative patches and corresponding neighboring positively charged residues contribute to high concentration viscosities.Citation28 Similar to critical residues, or hot spots, representing the dominant contributors to protein–protein binding free energy,Citation29 single hydrophobic and/or electrostatic residues may play prominent roles in high concentration antibody viscosity through either facilitating or blocking access to patches within their vicinity. Excipients used to reduce the viscosity of high concentration antibody therapeutics include L-arginine, L-proline, L-lysine, glycine, and sodium chloride.Citation3 There is an abundance of specific formulations based on a single antibody at a time or direct mutagenesis of the antigen-binding Fv region, but we are not aware of any extensive survey of modifications to the Fc region to reduce self-association.
Fc variants are commonly incorporated into antibody therapeutics to modulate functions, including increasing or decreasing serum half-life or secondary immune functions such as antibody-dependent cellular cytotoxicity (ADCC), antibody-dependent cellular phagocytosis (ADCP), and complement-dependent cytotoxicity (CDC). Additional applications of Fc variants include enabling site-specific conjugation, promoting heavy chain (HC) heterodimerization for making bispecific antibodies, and abrogating binding to staphylococcal protein A.Citation30,Citation31
Here, we investigated how Fc variants affect the viscosity of two humanized IgG1 antibodies with κ light chains (LC), namely, the anti-IgE antibody, omalizumabCitation32 and the anti-HER2 antibody, trastuzumab.Citation33 Omalizumab was chosen for its high viscosity, whereas trastuzumab was selected as a closely matched low viscosity antibody. Omalizumab and trastuzumab share the same human IgG1 HC and κ LC constant domains as well as the same consensus human framework region (FR) residues: variable heavy (VH) subgroup III and variable light (Vκ) subgroup I. These antibodies differ only in their antigen-binding complementarity-determining region (CDR) residues and a few FR residues known as Vernier zone residuesCitation34 (Figure S1). There are far too many known Fc variants to undertake a comprehensive rheology survey, so we selected representative Fc variants that are commonly used to modulate antibody functions ().Citation31 The formulation pH range for antibody therapeutics is pH 4.8 to pH 8.0, with most antibodies being formulated between pH 5.0 and pH 6.9.Citation4 Histidine is the most commonly used buffer in antibody therapeutics.Citation4 With these common formulation practices in mind, viscosity experiments in this study were performed at pH 5.5 in histidine acetate buffer. The use of histidine acetate, with its four ionizable groups, allowed the viscosity of omalizumab to be investigated over a broad pH range (pH 4.0 to pH 7.0) in a single buffer.
Table 1. CommonCitation9–11,Citation21 Fc variants evaluated in this study including their respective effects upon effector functions, plasma half-life and heavy-chain heterodimerization. ⇧, increased binding or function; ⇩, decreased binding or function. Residues are numbered using the Eu numbering scheme.Citation56,Citation57 Cited examples are antibody therapeutics that are approved or undergoing regulatory review that contain the listed mutations alone (*) or in combination with additional Fc mutations (**).2 Antibody examples include amubarvimab and romlusevimab (targeting SARS-CoV2) that are co-formulated.
Results
Contribution of variable domains, Fc region and IgG1 format to the omalizumab and trastuzumab viscosity
Omalizumab and trastuzumab are closely matched humanized IgG1 antibodies with κ LC that differ only in their antigen-binding CDR loops plus two and seven FR residues in VL and VH, respectively (Figure S1). The viscosity of omalizumab (176 cP) at 180 mg/mL is much greater than for trastuzumab (8 cP) under the same conditions. These data suggest a major role for the variable domains, particularly CDRs, in the high viscosity of omalizumab. The viscosity of omalizumab was reduced by 9.1-fold in its corresponding F(ab′)2 fragment, at equivalent mass concentration, suggesting a significant contribution for the Fc region to the high viscosity of this antibody (). In contrast, the viscosity of the Fc fragment alone is very low (3 cP). Together, these data suggest that Fab-Fc may be important contributors to the high viscosity of omalizumab. Mixing equimolar amounts of omalizumab F(ab′)2 (117 mg/mL) and Fc (63 mg/mL) did not restore the high viscosity of omalizumab IgG1 (180 mg/mL) (). Thus, the high viscosity of omalizumab is also dependent on the intactness of this IgG1.
Figure 1. Contribution of Fab arms and Fc region to the viscosity of omalizumab and trastuzumab IgG1 antibodies. Cartoon representation of antibody formats highlighting corresponding protein concentrations used in viscosity experiments including the equimolar fragmented mixture of F(ab’)2 and Fc. Viscosity data were obtained by rheometry at a total protein concentration of 180 mg/mL in 20 mM histidine acetate, pH 5.5 at 25.0°C for omalizumab and trastuzumab. Data shown are the mean viscosity values (n = 2–5) ± SD analyzed using one-way ANOVA with *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 versus the corresponding parent antibody.
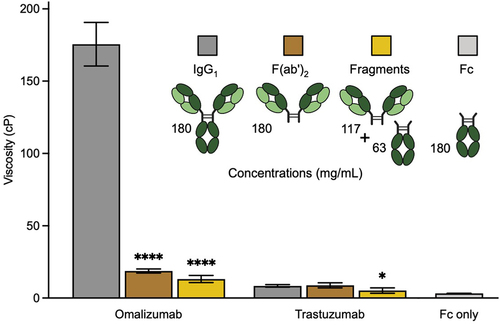
In contrast to omalizumab, the viscosity of trastuzumab IgG1 and corresponding F(ab′)2 fragment is comparable, consistent with no major role for the Fc regions in viscosity (). However, the low viscosity of trastuzumab IgG1 is moderately reduced when converted to equimolar mixtures of corresponding F(ab′)2 and Fc fragments. Thus, there is apparently some dependence on intact IgG1 format for the viscosity of trastuzumab.
Effect of Fc variants on omalizumab and trastuzumab viscosity
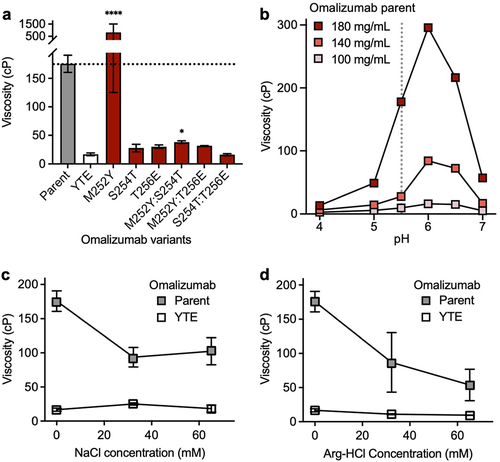
Next, the effect of some commonly used Fc variants () on the high concentration viscosity of omalizumab and trastuzumab () was studied. The most striking results for omalizumab were for the YTE (plasma half-life extension),Citation50 NG (aglycosylation)Citation51 and LPLIL (increased effector function)Citation47 variants, which decreased the viscosity of the parent antibody (176 cP) by 10.7-, 2.2-, and 1.6-fold, respectively (). In contrast, an alternative plasma half-life extension variant, LS,Citation49 had the opposite effect in increasing the viscosity of omalizumab by 1.5-fold (). Other Fc variants tested for omalizumab had comparable or slightly decreased viscosity compared to the parent antibody ().
Figure 2. Survey of some commonly used Fc variants () on the high concentration viscosity of (a) omalizumab and (b) trastuzumab. Viscosity measurements were made by rheometry with 180 mg/mL IgG1 antibody variant solutions in 20 mM histidine acetate, pH 5.5 at 25.0°C. Data shown are mean viscosity values (n = 1–5) ± SD analyzed using one-way ANOVA with *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 versus the corresponding parent antibody.
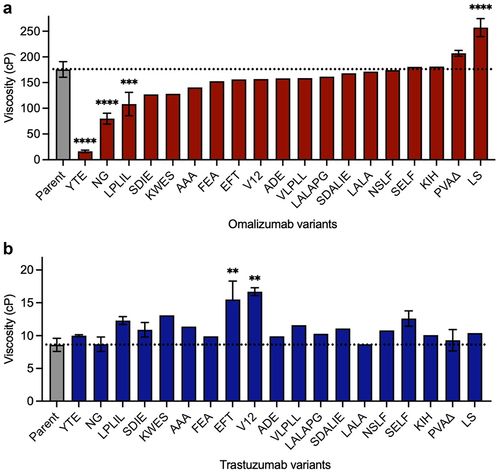
In contrast to omalizumab, none of the Fc variants significantly reduced the already low viscosity of trastuzumab (8.6 cP, ). However, several Fc variants increased the viscosity of trastuzumab. The largest increases were seen for EFT,Citation46 and V12Citation38 which increased the viscosity of trastuzumab by 2.0-fold and 1.9-fold, respectively (). These data demonstrate that unwanted self-association can sometimes result from Fc modifications that are commonly used in clinical-stage antibodies.Citation31
Spatial localization of Fc variant mutations with significant viscosity effects
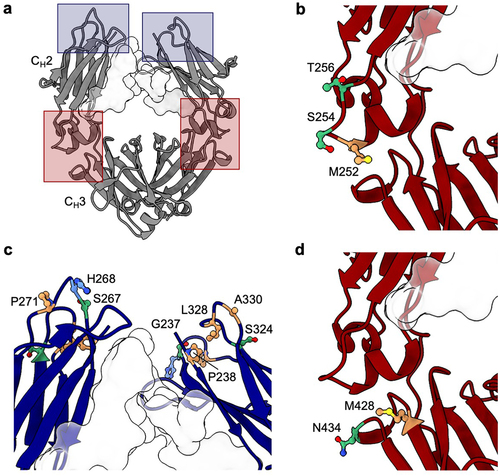
The large collection of different mutations analyzed in multiple antibodies offers an opportunity to identify topological areas linked to self-association in the Fc (). For omalizumab, viscosity-reducing variants NG and LPLIL have solvent-accessible surface residues in the upper CH2 domain, as well as glycan-facing residues in the CH2 and CH3 domains. The most significant viscosity reducing variant for omalizumab, YTE, is found within the CH2/CH3 elbow region (). This region also contains the location of the LS mutations, the only omalizumab Fc variant identified with a statistically significant elevation in viscosity (). The striking contrast in viscosity between the YTE and LS Fc variants of omalizumab (17 cP and 240 cP, respectively), in close proximity to one another, suggests a single self-association interface unique to omalizumab. As a whole, the spatial orientation of viscosity-affecting mutations in omalizumab was significantly different from that of trastuzumab (). All variants identified that substantially increase the viscosity of trastuzumab involve replacement of residues that are located in the upper CH2 domain ().
Figure 3. Location of Fc residues where mutations modulate the viscosity of omalizumab and trastuzumab. The Fc structure was visualized using UCSF ChimeraX software,Citation53 with the Fc glycan at N297 highlighted as a light gray surface. (a) Molecular structure of Fc region (PDB structure 7LBL) highlighting areas where mutations resulted in the largest changes in viscosity for omalizumab (red boxes) and trastuzumab (blue boxes). CH2/CH3 elbow region indicating the location of residues (b) M252, S254, and T256 mutated in the YTE variant that decreases the viscosity of omalizumab and (d) residues M428 and N434 mutated in the LS variant that increases the viscosity of omalizumab. (c) Upper CH2 residues that when mutated resulted in the largest increases in viscosity for trastuzumab. Polar (green), nonpolar (orange), and positively charged (blue) side chains are highlighted.
Effect of YTE and LS variants on additional high viscosity antibodies
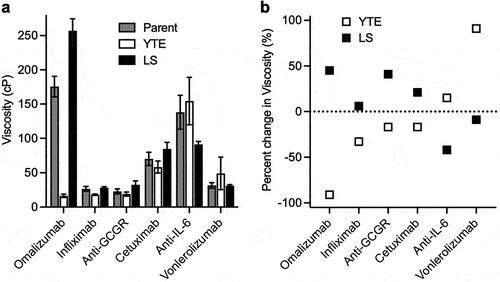
The large decrease and increase in the viscosity of omalizumab from YTE and LS variants, respectively (), motivated us to evaluate these Fc variants in the context of different antibodies. Using the same human HC and κ LC constant domains (Table S1), five additional IgG1 antibodies were selected with moderate-to-high parent viscosity, namely: anti-GCGR (22.9 cP), infliximab (anti-TNF, 26.6 cP), vonlerolizumab (anti-OX40, 34.4 cP), cetuximab (anti-EGFR, 70.1 cP), and anti-IL-6 (156 cP). The effect of the YTE and LS mutations on the viscosity of these additional antibodies was dependent on the variable domain context (). Amongst this set of antibodies, any viscosity reductions were much less pronounced than for the YTE variant of omalizumab. Indeed, the largest viscosity reductions observed were for the YTE variant of infliximab (1.5-fold decrease) and the LS variant of anti-IL-6 (1.7-fold decrease) (). Incorporation of the YTE mutations into vonlerolizumab had the opposite effect, increasing the viscosity by 1.9-fold, which further demonstrates the antibody-specific effect of this Fc variant on self-association. For each of the five antibodies tested, either YTE or the LS half-life extension mutations gave a small to moderate reduction (1.1- to 1.7-fold) in the viscosity compared to the corresponding parent IgG1 antibody ().
Figure 4. Viscosity of different IgG1 antibodies as parent, YTE, and LS variants. Viscosity measurements were made by rheometry with 180 mg/mL IgG1 antibody solutions in 20 mM histidine acetate, pH 5.5 at 25.0°C presented as (a) raw viscosity values. Data shown are the mean values (n = 2–5) ± SD. (b) percent-change in viscosity from antibodies containing the parent Fc.
High viscosity of omalizumab mitigated by individual YTE mutations
The large reduction in viscosity of omalizumab from the YTE triple mutation (176 cP to 16.4 cP) was further investigated by evaluating all possible single (M252Y, S254T, and T256E) and double (M252Y:S254T, M252Y:T256E, and S254T:T256E) component mutations of the YTE triple mutation. The large decrease in viscosity for omalizumab by the YTE triple mutation (10.7-fold) can be partially recapitulated with individual S254T (6.1-fold decrease) or T256E mutations (5.8-fold decrease) (). In contrast, the single mutant M252Y has the opposite effect, increasing the viscosity of omalizumab by 4.6-fold. Strikingly, the viscosity reducing effects of either the S254T or T256E single mutations are not significantly attenuated by combining them with the M252Y mutation.
Figure 5. Viscosity of omalizumab and YTE-related variants in the presence or absence formulation excipients. Rheometry measurements were obtained at 180 mg/mL IgG1 antibody solutions in 20 mM histidine acetate, pH 5.5 at 25.0°C. Data shown are mean viscosity values (n = 2–5) ± SD. (a) The viscosity of omalizumab parent IgG1 (gray bar) was compared to the YTE triple mutant (open bar) and component single and double mutants (red bars). Data were analyzed using one-way ANOVA with *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 versus the YTE (M252Y:S254T:T256E) variant. (b) Viscosity measurements for omalizumab parent IgG1 at different concentrations and pH values. Viscosity measurements of omalizumab parent IgG1 and YTE mutant at 180 mg/mL IgG1 in the presence of either (c) NaCl or (d) Arg-HCl.
Solution pH, electrostatics, and hydrophobicity influence omalizumab viscosity
Next, the solution pH effect on omalizumab viscosity was investigated in 20 mM histidine acetate (). The maximal measured viscosity of omalizumab at 180 mg/mL was 296 cP at pH 6.0. The viscosity of omalizumab decreased sharply under more acidic conditions with the lowest measured viscosity of 13.4 cP at pH 4.0. The viscosity of omalizumab was also reduced with increasing pH: 57.1 cP at pH 7.0. As for omalizumab concentration, the viscosity was highest at 180 mg/ml and reduced at 140 mg/mL with a qualitatively similar pH-dependence. The viscosity of omalizumab was lowest at 100 mg/mL with only a small pH dependence. This strong pH dependence of omalizumab appears consistent with ionizable groups such as histidines contributing to omalizumab viscosity.
The type of noncovalent interactions contributing to the high viscosity of omalizumab were studied by the addition of the excipients, NaCl and arginine-hydrochloride (Arg-HCl). The viscosity of omalizumab was reduced 1.7- to 1.9-fold by NaCl (), suggesting shielding of electrostatic interactions. In contrast, the viscosity of the YTE variant of omalizumab was slightly increased by addition of NaCl (), implying Fab-Fab self-associations are not primarily driven by electrostatics. The addition of Arg-HCl reduced the viscosity for the omalizumab parent antibody (2.0- to 3.3-fold) and also its YTE variant (1.5- to 1.7-fold) (), suggesting acidic and aromatic groups contribute to high viscosity of omalizumab through both Fab-Fc and Fab-Fab self-associations. Additional analysis of omalizumab YTE variant compared to the parent revealed decreased temperature effects, reduced shear thinning at high shear rates and a significant delay in concentration-dependent pseudo-exponential growth in viscosity (Figure S2).
Molecular dynamics of parent and YTE Fc regions
To gain deeper insights into how Fc variants affect the surface patch properties of the Fc region, we conducted microsecond-scale molecular dynamics (MD) simulations on glycosylated Fc regions, including both parent and various YTE mutations. We calculated the spatial aggregation propensity (SAP), a descriptor shown to modulate antibody viscosity,Citation21,Citation54,Citation55 and the electrostatic potential at the residue level across the entire Fc averaged over the MD trajectory (Figures S3 and S4). Notable average shift in surface patch properties of Fc mutants relative to the parent Fc were observed, particularly around the loop containing point mutations, residues 246–258, Eu numbering.Citation56,Citation57 Therefore, we focused our comparative analysis specifically on this loop (). The M252Y single mutant showed a large increase in the SAP score, with smaller increases in SAP score for corresponding double (M252Y:S254T and M252Y:T256E) and triple (YTE) mutants (). This observation aligns with the notable viscosity increase observed in M252Y, implying the key role of tyrosine in enhancing the aromaticity or hydrophobicity of the Fc region. This enhancement could potentially foster various intermolecular interactions, such as cation-π, anion-π or π–π interactions. However, the reduction in viscosity observed in the M252Y:S254T variant contradicts the anticipated increase in SAP scores, suggesting additional factors at play.
Figure 6. Impact of mutations on surface properties and conformational stability of the elbow loop in the Fc (a) distribution of SAP scores summed over the elbow loop (residues 246–258) during MD simulations. (b) Distribution of the electrostatic potential calculated using APBS summed over the residues in the elbow (residues 246–258) during MD simulations. (c) The value of SAP and APBS values calculated for the elbow loop averaged over all the MD frames. The green region highlights lowered hydrophobicity and more negative Fc, thus reducing viscosity. The red region shows increased hydrophobicity and more positive Fc, increasing the risk of viscosity. (d) Distribution of RMSD of heavy atoms of the elbow residues relative to the starting structure during MD simulations.
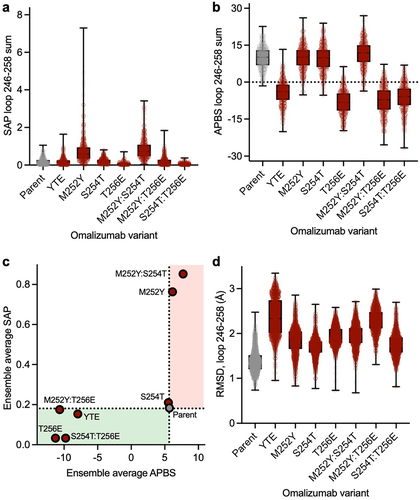
The electrostatic surface potential Adaptive Poisson-Boltzmann Solver (APBS) score surrounding the point mutations is notably influenced by the presence or absence of a glutamic acid residue. Specifically, single (T256E), double (M252Y:T256E and S254T:T256E), and triple (YTE) mutants all exhibit a shift toward a negative electrostatic potential within the specified loop, compared to parent and other variants which maintain a positive electrostatic potential on the Fc loop (). Given that the variable domain of omalizumab features a strong electrostatic negative patch,Citation58 a potential explanation is that the presence of a strong positive electrostatic patch on the Fc would enhance the favorability of Fab-Fc interaction through electrostatic attraction. Consequently, the decreased viscosity observed in all variants featuring a glutamic acid mutation (T256E, M252Y:T256E, S254T:T256E, and YTE) may reflect the reduction in electrostatic attraction resulting from the reduced positivity of the Fc.
Next, we considered changes in ensemble average electrostatic and hydrophobic (SAP) surface properties together across different variants (). All variants featuring a glutamic acid mutation exhibit a shift toward a negative surface potential in the local Fc region, consistent with the observed viscosity reduction (). The M252Y variant shows notably higher SAP score on the Fc compared to the parent, which appears consistent with the increased viscosity observed in this variant. However, the M252Y:S254T variant has a similarly high SAP score and much lower viscosity than the omalizumab parent (). Thus, surface properties reflected in SAP scores alone are not sufficient to account for the differences in viscosity of M252Y and M252Y:S254T variants compared to the omalizumab parent antibody.
Destabilization of the Fc may contribute to the reduction in viscosity of omalizumab
We analyzed the root mean square deviation (RMSD) of the Fc region for each variant relative to the initial parental crystal structure across different MD frames to better understand how Fc point mutations affect the structure and conformational stability of the Fc region. The parent Fc exhibited the lowest RMSD of the loop relative to the crystal structure (). However, the YTE triple mutant showed the most significant conformational change and instability, followed by the M252Y:T256E double mutant. Other mutants, incorporating either a glutamic or tyrosine substitution, also displayed higher RMSD instability within the corresponding loop relative to the parent Fc. The S254T variant also showed enhanced instability relative to the parent Fc, even though it is lower than the other variants. The destabilization of the Fc could potentially disrupt the intermolecular interactions through an increase in the entropic cost of Fab-Fc interaction.
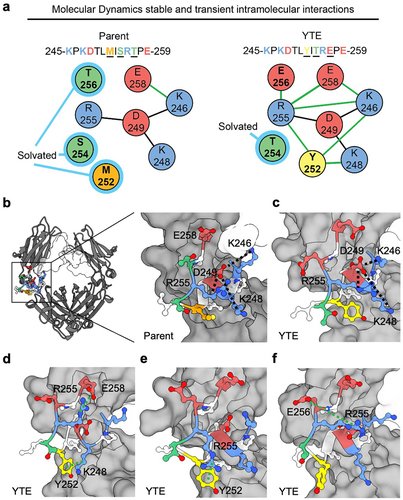
MD analysis revealed a potential mechanism underlying the destabilization of the Fc by the YTE mutations (). MD simulations indicate the presence of a stable intramolecular salt-bridge network involving positively charged residues R255, K246, and K248, which encapsulate the negatively charge D249 at their core. E258 is the only competitor to D249 in interacting with its neighboring positively charged residues. The M252, S254, and T256 residues in the parent Fc remain solvated by water rather than engaging in significant intramolecular interactions. However, the introduction of M252Y and/or T256E mutations, disrupts this stability by introducing a competing set of transient intramolecular interactions. For example, Y252 interacts with both K248 and R255 via cation-π interactions, and sometimes, it extends its reach to interact with E258 or K246 (). Similarly, E256 exhibits favorable interactions with R255, and to a lesser extent with other basic residues through salt-bridge formations. Unlike the parental structure, which predominantly adopts a single meta-stable conformation, the YTE mutations display several favorable conformations. The transitions between these conformations contribute to the observed conformational instability in YTE, as evident by its large RMSD ().
Figure 7. Modulations in intramolecular interaction network in the parent Fc and YTE variant. (a) Representative comparison of intramolecular interaction networks for parent Fc and YTE variant. A stable salt-bridge network (black line between residues) primarily involving residues R255, K248, K246, and D249 is shown for each while residues M252, S254, and T256 remain predominantly solvated (light blue highlight) in the parental Fc. In contrast, the introduction of M252Y and/or T256E mutations in the YTE variant disrupts this stability, leading to the formation of transient intramolecular interactions (green lines between residues). (b) Molecular structure of Fc with focus on CH2/CH3 elbow region depicting intramolecular salt-bridge network. (c-f) selected frames of MD simulation highlighting (c) retained R255, K248, K246, and D249 salt-bridge, (d) Y252 cation-π interaction with K248, R255 salt-bridge with E258, (e) additional Y252 cation-π interaction with R255 and (f) E256 forms favorable interactions primarily with R255, with lesser involvement with other basic residues through salt-bridge formations. An occasional interaction between E258 and K246 was observed but not shown explicitly in the selected frames. Polar (green), nonpolar (orange), aromatic (yellow), negatively charged (red), and positively charged (blue) side chains are highlighted.
Discussion
Fc mutations can affect the viscosity for some IgG1 antibodies
SC delivery has recently emerged as a desirable and increasingly common route of administration for antibody therapeutics, including for the most common current format, i.e., IgG1.Citation1 Desirable attributes of antibodies for SC clinical applications include favorable high concentration properties, such as low viscosity and high solubility plus low propensity for self-aggregation, opalescence, and gelation.Citation5 The impact of antibody variable domain sequences on IgG1 high concentration properties has been extensively explored.Citation24–27,Citation54,Citation59–62 Additionally, the viscosity contribution of different IgG isotypes has also been investigated.Citation9,Citation63 For example, Lai et al. compared the viscosity of 14 matched pairs of IgG1 and hinge-stabilized (S228P) IgG4 antibodies with identical variable domains.Citation63 Similar or higher viscosity was observed for all of the hinge-stabilized IgG4 antibodies.Citation63
The effect of Fc mutations on antibody high concentration properties is largely unknown. This is an important knowledge gap to address, as such mutations are very widely used with antibody therapeutics to modulate Fc functions.Citation31 To this end, we investigated the effect of a panel of commonly used Fc variants () on the high concentration viscosity properties of two closely related humanized IgG1 antibodies, omalizumab, and trastuzumab. These antibodies differ mainly in their CDR residues, with two and seven FR differences in VL and VH, respectively (Figure S1), whereas the constant domains are identical.
Many Fc variants resulted in only small perturbations in viscosity, whereas large increases or decreases in viscosity are seen in some cases (). Strikingly, the YTE variant,Citation50 clinically validated for antibody plasma half-life extension ()Citation2 reduced the high viscosity of omalizumab (176 cP) by 10.7-fold. In contrast, an alternative clinically validated half-life extension variant, LS,Citation49 had the opposite effect and increased the viscosity of omalizumab by 1.5-fold. Investigation of YTE and LS variants for five additional antibodies revealed diverse effects that were dependent upon the variable domain context () and sometimes opposite to the effects on omalizumab. For example, the viscosity of vonlerolizumab (34.4 cP) was increased 1.9-fold by the YTE variant. In contrast, the viscosity of the anti-IL-6 antibody (156 cP) was decreased 1.7-fold by the LS variant. Thus, for future antibody therapeutics intended for SC delivery, it may be helpful to empirically screen high concentration properties of alternative Fc variants for achieving the desired modulation of Fc function. Half-life extension variants YTE and LS have both been successfully used in approved antibody therapeutics () and could potentially be used interchangeably if one offered better biophysical properties for a specific antibody therapeutic candidate. The YTE Fc variant has also previously been incorporated by Liu et al. into a next-generation omalizumab alongside variable domain mutations.Citation64
Four interaction site model for omalizumab viscosity
Alternative possible molecular mechanisms underlying the high IgG1 viscosity were considered focusing on omalizumab and the domain location of interaction sites. A priori, IgG self-association sites may include Fab-Fab, Fab-Fc, and Fc-Fc interactions (). Interactions between Fab-Fab,Citation24–27 are consistently reported in the literature while there are a few examples of Fab-Fc interactions.Citation65,Citation66 In contrast, there is no clear evidence, to our knowledge, to support significant Fc-Fc interactions contributing to antibody high concentration viscosity. The data reported here suggest that the high viscosity of omalizumab requires the variable domains, Fc region, and an intact IgG1 format (). In toto, these observations suggest that Fab-Fab and Fab-Fc, but not Fc-Fc interactions, make significant contributions to the high viscosity of omalizumab ().
Figure 8. Model for potential interaction sites for (a) IgG and (b) omalizumab including its corresponding F(ab′)2 fragment plus Fc mixture and the omalizumab YTE variant.
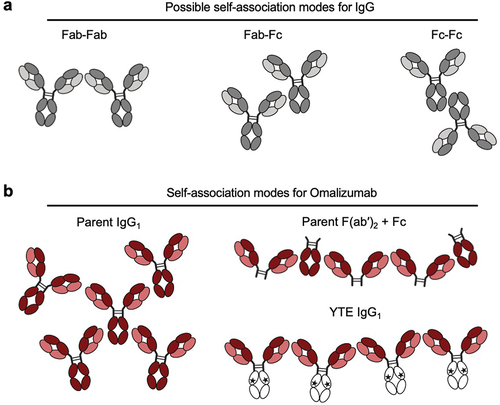
Fragmentation of omalizumab into F(ab′)2 and Fc components greatly reduces viscosity (). This may reflect lowering of the number of potential self-association sites per molecule from four for omalizumab IgG1 to two for the corresponding F(ab′)2 plus Fc mixture (). Fc mutations such as YTE appear to attenuate the Fab-Fc interactions for omalizumab. Strikingly similar reductions of omalizumab viscosity (176 cP) were achieved by fragmentation to F(ab′)2 plus Fc (13.2 cP) and the YTE variant (16.4 cP). Beyond the number of possible different sites for interaction of omalizumab IgG1, the pH and different types of non-covalent interaction that might contribute to its high viscosity were considered. Modulation of the solution pH resulted in omalizumab peak viscosity at pH 6.0 (), suggesting histidine side chains, with known pKa of ~ 6,Citation67 in the VH (H97, H100a, and H100c) or Fc (H310, H433, and H435) are likely contributing to Fab-Fc self-association in some capacity. This strong pH-dependence of the viscosity of omalizumab is consistent with a pioneering earlier publication from the laboratory of our late colleague, Dr. Steven Shire.Citation68
Charge patches and also hydrophobic patches are known to contribute to the high viscosity of some proteins, including antibodies.Citation69 The high viscosity of omalizumab was reduced by the addition of NaCl and Arg-HCl (), consistent with shielding of electrostatic interactions from charge patches and aromatic groups, respectively.Citation70
High viscosity antibody solutions may result from a network of transient and weak interactions, including through electrostatics.Citation20 We attempted to measure the affinity of the proposed omalizumab Fab-Fc and Fab-Fab interactions through surface plasmon resonance. Our initial attempts using full-length omalizumab IgG1 and fragmented omalizumab components in conjunction with multiple capture approaches and variable concentrations (5 to 250 nM) did not yield an interpretable response because of high background. Ongoing efforts to characterize these putative omalizumab Fab-Fab and Fab-Fc interactions include hydrogen-deuterium exchange mass spectrometry, fluorescence polarization, analytical ultracentrifugation, and isothermal titration calorimetry.
Molecular mechanisms for Fc variants that reduce omalizumab viscosity
Next, possible mechanisms by which the YTE variant might reduce the viscosity of omalizumab were considered. Initially, the impact of YTE variants on the surface patch properties directly linked to viscosity were assessed (). The introduction of a glutamic acid in the YTE variants reduced the overall positivity of the Fc and resulted in a negative electrostatic potential around the elbow. This charge mutation could disrupt the full-length molecular charge asymmetry, consequently reducing the electrostatic attraction between the Fab and Fc regions. Further, the presence of tyrosine in the YTE mutations, particularly in the absence of glutamate, increased the strength of the hydrophobic patch on the elbow, which is consistent with the increase of viscosity in the M252Y mutation. However, the contribution of S254T mutations to the viscosity reduction of omalizumab could not be explained solely by surface patch properties.
Beyond the surface patch properties, YTE variants could affect the conformational landscape of the Fc region, potentially modulating the intermolecular interactions. The X-ray crystallographic structure of the YTE Fc region (PDB structure 3FJT)Citation71 is highly similar to the parent Fc region (PDB structure 3AVE),Citation72 as previously reported by Dall’Acqua and colleaguesCitation71 (Figure S5). Nevertheless, the YTE mutations reduce the thermal melting temperature (Tm) of the IgG1 CH2 domain by ~ 7°C,Citation73,Citation74 consistent with some structural destabilization of the CH2 domain. Hydrogen/deuterium exchange experiments with matched parental and YTE variant antibodies revealed greater flexibility of the 241–251 segment (Eu numbering)Citation56,Citation57 of the CH2 domain in the YTE variant.Citation73 The other Fc variant identified that greatly reduces the viscosity of omalizumab, namely, NG (2.2-fold), also lowers the thermal stability through destabilization of the CH2 domain.Citation75 This reduction in viscosity was abrogated in the PVANG variant (substitution of IgG1 lower hinge sequence with that from IgG2 plus removal of glycosylation site), which may restabilize CH2 through deletion of residue G236, known to alter alignment of the lower hinge.Citation44 The high viscosity of omalizumab (176 cP) was reduced in the NG (74 cP) and YTE (17 cP) variants (). Combining these Fc variants (i.e., YTENG) resulted in an intermediate viscosity (25 cP) (Figure S6), suggesting these Fc variants reduce viscosity through distinct mechanisms.
The increased conformational flexibility of the YTE variant is consistent with increased transient intramolecular interactions present in MD simulations (). The tyrosine and glutamate residues within the YTE mutations seem to form additional intramolecular interactions (cation-π and salt-bridge), which disrupt the stable network of intramolecular interactions in the parental Fc.
Destabilization of the Fc region may exert a dual effect on the Fab-Fc intermolecular interactions. Firstly, Fc instability may increase the entropic cost associated with protein–protein interaction, making a stable Fab-Fc interaction less favorable. Consequently, this could lead to facilitated dissociation of intermolecular interactions, potentially reducing the viscosity of all YTE variants. This hypothesis aligns with the observed viscosity reduction in the YTE variant and related single and double mutants for omalizumab. Secondly, Fc destabilization may increase exposure of certain amino acids, changing the surface properties of the Fc. For example, in the YTE mutations, the interaction of Y252 with either K248 or R255 can release D249 for intermolecular interaction. The exposure of D249, coupled with the addition of E256, can form a local negative charge patch on the elbow of the Fc, which is typically positively charged in the parent. The change in electrostatic surface properties of the Fc has the potential to either increase or decrease viscosity, depending on the properties of the variable domains.
Figure S7 illustrates the impact of YTE and LS mutations on antibody viscosity as a function of three molecular descriptors of the Fv known to influence viscosity: 1) negative electrostatic potential on the Fv (Fv APBS negative),Citation76 2) charge asymmetry on the Fv (Fv CAP),Citation77 and 3) SAP score on the Fv (Fv SAP).Citation21 Overall, our data suggest that the YTE mutations benefit antibodies with extreme charge asymmetry and a large negative charge patch (e.g., omalizumab), while potentially increasing viscosity in antibodies with either positive charge or high hydrophobicity on their Fv (e.g., trastuzumab, anti-IL-6). The introduction of a local negative patch by YTE mutations can increase Fv-Fc repulsion when the Fv is negatively charged. In contrast, the addition of tyrosine and glutamate residues in YTE variants may enhance interactions with aromatic-rich Fv regions (e.g., anti-IL-6), due to strengthened Fab-Fc interactions driven by hydrophobic, cation-π, anion-π, or π-π interactions. Furthermore, the results shown in Figure S7b demonstrate that the LS mutations tend to reduce viscosity driven by hydrophobic interactions (e.g., anti-IL-6 antibody) while potentially increasing electrostatic-driven viscosity (e.g., omalizumab). It is important to note that the correlation observed is imperfect and that our current dataset is small in size. Indeed, the complex interplay involving conformational changes, protein–protein interactions, and solvent effects necessitates further investigation to fully elucidate the underlying mechanisms and their broader implications across different antibodies.
We demonstrate for the first time, to our knowledge, that some mutations that are commonly used to modulate Fc functions for antibody therapeutics () can decrease or increase the viscosity of some antibodies. For example, the YTE triple mutation (M252Y:S254T:T256E)Citation50 commonly used for plasma half-life extensionCitation31 reduced the viscosity of omalizumab from 176 cP to 16.4 cP (10.7-fold). Indeed, even single Fc mutants can sometimes result in large changes in viscosity as illustrated by the S254T variant of omalizumab variants: 6.1-fold viscosity decrease. Further work is needed to verify that IgG1 viscosity can be reduced by such Fc variants without modulating Fc function when this is needed. For some antibodies, it may be possible to mitigate unfavorable high concentration properties, such as high viscosity, by selecting from amongst alternative available Fc variants. For example, for six different antibodies tested here, either the YTE or LS half-life extension variant, but never both variants for the same antibody, decreased viscosity to some extent (). For future antibody therapeutics designed for SC administration, it is desirable to evaluate clinical candidates as early as possible for viscosity, as well as for other high concentration properties.
Materials and methods
Antibody mutagenesis, expression, purification, and preparation
The amino acid sequences for all parental antibodies with unmodified Fc regions used in this study are listed in Table S1. Gene synthesis was employed to produce DNA fragments encoding for modified HC. Mutations to amino acids and all references to amino acid positions in the text follow Eu numbering.Citation56,Citation57 Gene fragments were inserted into a mammalian expression vector and transient transfection was carried out in high titer Chinese hamster ovary (HT CHO) cells followed by two-step purification by protein A chromatography and size exclusion chromatography (SEC), as previously described.Citation78 For F(ab′)2 generation, intact IgG1 antibodies were IdeS digested and purified with CH1-XL Affinity Matrix, as previously described.Citation27 Samples were buffer-exchanged against 20 mM histidine acetate, pH 5.5, or some other specified pH (), using Slide-A-Lyzer 10 kDa MWCO Dialysis Cassettes (Thermo Fisher Scientific). Dialyzed antibody samples were concentrated to greater than 180 mg/mL using Amicon Ultra-15 10 kDa MWCO centrifugal filter units (MilliporeSigma) by centrifugation at 4,000 g. Concentration determinations were made by gravimetric analysis in duplicate with 2–3 µL samples diluted ~ 500-fold before A280 measurement on 8453 UV/Vis Spectrometer (Agilent). The A280 was then multiplied by the dilution factor and the product was divided by the antibody extinction coefficient, individually calculated based upon corresponding primary sequence, to yield sample concentration. Extinction coefficients and isoelectric points (pI) for corresponding IgG1 antibodies determined with internal software and confirmed with ExPASy ProtParam tool.Citation79 Positive displacement pipettes were used for dispensing high concentration antibody samples to enable accurate volume transfers.
Rheometry
For viscosity determinations, samples were analyzed on a Discovery-HR-30 cone-and-plate rheometer (TA Instruments) using 20 mm diameter stainless steel 1° cone angle Peltier plate geometry. Concentrated antibody samples were prepared at 180 mg/mL by dilution in 20 mM histidine acetate, pH 5.5, or other specified pH (), prior to loading 40 µL onto the rheometer plate. The cone was lowered and samples were measured at temperature-controlled 25°C with fixed shear-rate of 1,000 s−1. Each reported value represents the mean of 12 measurements collected over 1 min. The rheometer was calibrated with 8 cP (s6) and 30 cP (s20) viscosity standards (Cannon Instrument Company) to ±5% at the start of each day. Viscosity determination for parental IgG1 antibodies with unmodified Fc regions was subject to repeat measurements (n = 5). Given the high IgG1 protein needs for rheometry, the initial survey of Fc variants was done with a single viscosity measurement. Variants with viscosity values that were substantially different from the corresponding parental antibodies (reduced or increased by > 15%) were subject to repeat measurements (n = 2–5). All viscosity data from this study, including the number of replicates are tabulated in Table S2.
Molecular dynamics simulations
The software Molecular Operating Environment (MOE)Citation80 from Chemical Computing Group was used to prepare a full length glycosylated IgG1 crystal structure (PDB:1HZH). The Fab regions were removed by truncating the hinge region just above the disulfide bonds (CPPC in the HC). The parent, YTE variant, and related single and double mutants were generated by applying point mutations to the parental crystal structure. The prepareforleap tool in CPPTRAJCitation81 was used to rename the glycans and reformat the pdb for tleap. FF14SB force field was used for proteinsCitation82 and the Glycam06 force field for carbohydrates.Citation83 The system was solvated using TIP3P water moleculesCitation84 in a rectangular box extending 10 Å from protein edges. Sodium and chloride counter ions at a 0.15 M were added to neutralize the system using parameters derived by Joung and Cheatham.Citation85
The GPU implementation of Amber 2019 MD software packageCitation86 with the SPFP precision modelCitation87 was used for the MD simulation using the following protocol. First, the structure was relaxed with 2,000 steps of conjugate-gradient energy minimization, using harmonic restraining potential with the force constant of 10 kcal mol−1 Å−2 to restrain the solute to the initial structure. Then, the solvent molecules were allowed to move using NPT ensemble with a temperature of 300 K. Another step of conjugate-gradient energy minimization was performed with 2,000 steps while removing all the restraints. Next, the pressure was maintained at 1 atmosphere and the thermostat temperature increased to 300 K over the course of 500 picoseconds, while Harmonic positional restraints of strength 10 kcal mol−1 Å−2 was applied to the solute. The system was then equilibrated for 1 ns with a restraint force constant of 1 kcal mol−1 Å−2. All restraints were removed for the production stage. The hydrogen mass repartition option of Amber was used allowing a time step of 4 femtoseconds.Citation88 The production simulation was carried out using NPT conditions. Langevin dynamicsCitation89 was used to maintain the temperature at 300 K with a collision frequency of 3 ps−1. The production stage of the MD simulation was performed for 350 ns. The protocol described above was repeated to generate three independent replicates of 350-ns trajectories, adding up to ~ 1.0 microsecond trajectories for each structure.
From each MD trajectory, we extracted 438 frames evenly distributed along the trajectory. For each frame, we calculated the SAP and the electrostatic potential around the loop containing point mutations (residues 246–258), employing the APBS tool (). The per-residue APBS surface potentials were calculated using the Moldesk surface analyzer.Citation76 The CPPTRAJ toolCitation81 available in AmberTools 2019 was used to calculate RMSD from the starting structure.
Abbreviations
| ADCC | = | Antibody-dependent cellular cytotoxicity |
| ADCP | = | Antibody-dependent cellular phagocytosis |
| APBS | = | Adaptive Poisson-Boltzmann Solver |
| Arg-HCl | = | Arginine-hydrochloride |
| CDC | = | Complement-dependent cytotoxicity |
| CDR | = | Complementarity-determining region |
| CHO | = | Chinese hamster ovary |
| cP | = | Centipoise |
| DLS | = | Dynamic light scattering |
| FR | = | Framework region |
| HC | = | Heavy chain |
| IV | = | Intravenous |
| LC | = | Light chain |
| MD | = | Molecular dynamics |
| pI | = | Isoelectric point |
| RMSD | = | Root mean square deviation |
| SAP | = | Spatial aggregation propensity |
| SC | = | Subcutaneous |
| SEC | = | Size-exclusion chromatography |
| Tm | = | Thermal melting temperature |
| VH | = | Variable heavy |
| VL | = | Variable light |
Heisler Rev Table S2.docx
Download MS Word (13.6 KB)Heisler Rev Fig S7.jpeg
Download JPEG Image (431.3 KB)Heisler Rev Fig S3.jpeg
Download JPEG Image (355.8 KB)Heisler Rev Fig S4.jpeg
Download JPEG Image (372.6 KB)Heisler Rev Fig S5.jpeg
Download JPEG Image (442.9 KB)Heisler Rev Fig S6.jpeg
Download JPEG Image (211.4 KB)Heisler Rev Fig S2.jpeg
Download JPEG Image (307 KB)Heisler Rev Fig S1.jpeg
Download JPEG Image (294.7 KB)Heisler Rev Table S1.docx
Download MS Word (10.5 KB)Acknowledgments
We thank Kristen Nailor and Sowmya Balasubramanian and their respective teams at Research Materials Group and Protein Chemistry Department within Genentech for antibody product acquisition, expression and purification. We also thank Stuart McNelles and Jing Dai for helpful input on this research including this manuscript.
Disclosure statement
All authors are current employees of Genentech, Inc., which develops and commercializes therapeutics including antibodies.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/19420862.2024.2379560.
Additional information
Funding
References
- Carter PJ, Rajpal A. Designing antibodies as therapeutics. Cell. 2022;185(15):2789–15. doi:10.1016/j.cell.2022.05.029.
- The Antibody Society. Therapeutic monoclonal antibodies approved or in regulatory review. accessed 2024 May 3]. www.antibodysociety.org/antibody-therapeutics-product-data.
- Ghosh I, Gutka H, Krause ME, Clemens R, Kashi RS. A systematic review of commercial high concentration antibody drug products approved in the US: formulation composition, dosage form design and primary packaging considerations. MAbs. 2023;15(1):2205540. doi:10.1080/19420862.2023.2205540.
- Strickley RG, Lambert WJ. A review of formulations of commercially available antibodies. J Pharm Sci. 2021;110(7):2590–608 e2556. doi:10.1016/j.xphs.2021.03.017.
- Shire SJ, Shahrokh Z, Liu J. Challenges in the development of high protein concentration formulations. J Pharm Sci. 2004;93(6):1390–402. doi:10.1002/jps.20079.
- Narasimhan C, Mach H, Shameem M. High-dose monoclonal antibodies via the subcutaneous route: challenges and technical solutions, an industry perspective. Ther Deliv. 2012;3(7):889–900. doi:10.4155/tde.12.68.
- Zarraga IE, Taing R, Zarzar J, Luoma J, Hsiung J, Patel A, Lim FJ. High shear rheology and anisotropy in concentrated solutions of monoclonal antibodies. J Pharm Sci. 2013;102(8):2538–49. doi:10.1002/jps.23647.
- Jain T, Sun T, Durand S, Hall A, Houston NR, Nett JH, Sharkey B, Bobrowicz B, Caffry I, Yu Y. et al. Biophysical properties of the clinical-stage antibody landscape. Proc Natl Acad Sci USA. 2017;114(5):944–49. doi:10.1073/pnas.1616408114.
- Kingsbury JS, Saini A, Auclair SM, Fu L, Lantz MM, Halloran KT, Calero-Rubio C, Schwenger W, Airiau CY, Zhang J. et al. A single molecular descriptor to predict solution behavior of therapeutic antibodies. Sci Adv. 2020;6(32):eabb0372. doi:10.1126/sciadv.abb0372.
- Starr CG, Makowski EK, Wu L, Berg B, Kingsbury JS, Gokarn YR, Tessier PM. Ultradilute measurements of self-association for the identification of antibodies with favorable high-concentration solution properties. Mol Pharm. 2021;18(7):2744–53. doi:10.1021/acs.molpharmaceut.1c00280.
- Connolly BD, Petry C, Yadav S, Demeule B, Ciaccio N, Moore JM, Shire SJ, Gokarn YR. Weak interactions govern the viscosity of concentrated antibody solutions: high-throughput analysis using the diffusion interaction parameter. Biophys J. 2012;103(1):69–78. doi:10.1016/j.bpj.2012.04.047.
- Roche A, Gentiluomo L, Sibanda N, Roessner D, Friess W, Trainoff SP, Curtis R. Towards an improved prediction of concentrated antibody solution viscosity using the Huggins coefficient. J Colloid Interface Sci. 2022;607(Pt 2):1813–24. doi:10.1016/j.jcis.2021.08.191.
- Hung JJ, Zeno WF, Chowdhury AA, Dear BJ, Ramachandran K, Nieto MP, Shay TY, Karouta CA, Hayden CC, Cheung JK. et al. Self-diffusion of a highly concentrated monoclonal antibody by fluorescence correlation spectroscopy: insight into protein-protein interactions and self-association. Soft Matter. 2019;15(33):6660–76. doi:10.1039/c9sm01071h.
- Lai P-K, Swan JW, Trout BL. Calculation of therapeutic antibody viscosity with coarse-grained models, hydrodynamic calculations and machine learning-based parameters. MAbs. 2021;13(1):1907882. doi:10.1080/19420862.2021.1907882.
- Lai PK, Fernando A, Cloutier TK, Gokarn Y, Zhang J, Schwenger W, Chari R, Calero-Rubio C, Trout BL. Machine learning applied to determine the molecular descriptors responsible for the viscosity behavior of concentrated therapeutic antibodies. Mol Pharm. 2021;18(3):1167–75. doi:10.1021/acs.molpharmaceut.0c01073.
- Izadi S, Patapoff TW, Walters BT. Multiscale coarse-grained approach to investigate self-association of antibodies. Biophys J. 2020;118(11):2741–54. doi:10.1016/j.bpj.2020.04.022.
- Tomar DS, Li L, Broulidakis MP, Luksha NG, Burns CT, Singh SK, Kumar S. In-silico prediction of concentration-dependent viscosity curves for monoclonal antibody solutions. MAbs. 2017;9(3):476–89. doi:10.1080/19420862.2017.1285479.
- Agrawal NJ, Helk B, Kumar S, Mody N, Sathish HA, Samra HS, Buck PM, Li L, Trout BL. Computational tool for the early screening of monoclonal antibodies for their viscosities. MAbs. 2016;8(1):43–48. doi:10.1080/19420862.2015.1099773.
- Kuroda D, Tsumoto K. Engineering stability, viscosity, and immunogenicity of antibodies by computational design. J Pharm Sci. 2020;109(5):1631–51. doi:10.1016/j.xphs.2020.01.011.
- Buck PM, Chaudhri A, Kumar S, Singh SK. Highly viscous antibody solutions are a consequence of network formation caused by domain-domain electrostatic complementarities: insights from coarse-grained simulations. Mol Pharm. 2015;12(1):127–39. doi:10.1021/mp500485w.
- Dai J, Izadi S, Zarzar J, Wu P, Oh A, Carter PJ. Variable domain mutational analysis to probe the molecular mechanisms of high viscosity of an IgG1 antibody. MAbs. 2024;16(1):2304282. doi:10.1080/19420862.2024.2304282.
- Phan S, Walmer A, Shaw EW, Chai Q. High-throughput profiling of antibody self-association in multiple formulation conditions by PEG stabilized self-interaction nanoparticle spectroscopy. MAbs. 2022;14(1):2094750. doi:10.1080/19420862.2022.2094750.
- Sule SV, Dickinson CD, Lu J, Chow CK, Tessier PM. Rapid analysis of antibody self-association in complex mixtures using immunogold conjugates. Mol Pharm. 2013;10(4):1322–31. doi:10.1021/mp300524x.
- Makowski EK, Chen H, Lambert M, Bennett EM, Eschmann NS, Zhang Y, Zupancic JM, Desai AA, Smith MD, Lou W. et al. Reduction of therapeutic antibody self-association using yeast-display selections and machine learning. MAbs. 2022;14(1):2146629. doi:10.1080/19420862.2022.2146629.
- Nichols P, Li L, Kumar S, Buck PM, Singh SK, Goswami S, Balthazor B, Conley TR, Sek D, Allen MJ. Rational design of viscosity reducing mutants of a monoclonal antibody: hydrophobic versus electrostatic inter-molecular interactions. MAbs. 2015;7(1):212–30. doi:10.4161/19420862.2014.985504.
- Yadav S, Sreedhara A, Kanai S, Liu J, Lien S, Lowman H, Kalonia DS, Shire SJ. Establishing a link between amino acid sequences and self-associating and viscoelastic behavior of two closely related monoclonal antibodies. Pharm Res. 2011;28(7):1750–64. doi:10.1007/s11095-011-0410-0.
- Tilegenova C, Izadi S, Yin J, Huang CS, Wu J, Ellerman D, Hymowitz SG, Walters B, Salisbury C, Carter PJ. Dissecting the molecular basis of high viscosity of monospecific and bispecific IgG antibodies. MAbs. 2020;12(1):1692764. doi:10.1080/19420862.2019.1692764.
- Rai BK, Apgar JR, Bennett EM. Low-data interpretable deep learning prediction of antibody viscosity using a biophysically meaningful representation. Sci Rep. 2023;13(1). doi:10.1038/s41598-023-28841-4.
- Keskin O, Ma B, Nussinov R. Hot regions in protein-protein interactions: the organization and contribution of structurally conserved hot spot residues. J Mol Biol. 2005;345(5):1281–94. doi:10.1016/j.jmb.2004.10.077.
- Chiu ML, Goulet DR, Teplyakov A, Gilliland GL. Antibody structure and function: the basis for engineering therapeutics. Antibodies (Basel). 2019;8(4):55. doi:10.3390/antib8040055.
- Wilkinson I, Hale G. Systematic analysis of the varied designs of 819 therapeutic antibodies and Fc fusion proteins assigned international nonproprietary names. MAbs. 2022;14(1):2123299. doi:10.1080/19420862.2022.2123299.
- Presta LG, Lahr SJ, Shields RL, Porter JP, Gorman CM, Fendly BM, Jardieu PM. Humanization of an antibody directed against IgE. J Immunol. 1993;151(5):2623–32. doi:10.4049/jimmunol.151.5.2623.
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci USA. 1992;89(10):4285–89. doi:10.1073/pnas.89.10.4285.
- Foote J, Winter G. Antibody framework residues affecting the conformation of the hypervariable loops. J Mol Biol. 1992;224(2):487–99. doi:10.1016/0022-2836(92)91010-M.
- Chu SY, Vostiar I, Karki S, Moore GL, Lazar GA, Pong E, Joyce PF, Szymkowski DE, Desjarlais JR. Inhibition of B cell receptor-mediated activation of primary human B cells by coengagement of CD19 and Fc gamma RIIb with Fc-engineered antibodies. Mol Immunol. 2008;45(15):3926–33. doi:10.1016/j.molimm.2008.06.027.
- Lazar GA, Dang W, Karki S, Vafa O, Peng JS, Hyun L, Chan C, Chung HS, Eivazi A, Yoder SC. et al. Engineered antibody Fc variants with enhanced effector function. Proc Natl Acad Sci USA. 2006;103(11):4005–10. doi:10.1073/pnas.0508123103.
- Kellner C, Otte A, Cappuzzello E, Klausz K, Peipp M. Modulating cytotoxic effector functions by Fc engineering to improve cancer therapy. Transfus Med Hemother. 2017;44(5):327–36. doi:10.1159/000479980.
- Mimoto F, Katada H, Kadono S, Igawa T, Kuramochi T, Muraoka M, Wada Y, Haraya K, Miyazaki T, Hattori K. Engineered antibody Fc variant with selectively enhanced FcγRIIb binding over both FcγRIIaR131 and FcγRIIaH131. Protein Eng Des Sel. 2013;26(10):589–98. doi:10.1093/protein/gzt022.
- Richards JO, Karki S, Lazar GA, Chen H, Dang W, Desjarlais JR. Optimization of antibody binding to FcgammaRIIa enhances macrophage phagocytosis of tumor cells. Mol Cancer Ther. 2008;7(8):2517–27. doi:10.1158/1535-7163.MCT-08-0201.
- Shang LM, Daubeuf B, Triantafilou M, Olden R, Depis F, Raby AC, Herren S, Dos Santos A, Malinge P, Dunn-Siegrist I. et al. Selective antibody intervention of toll-like receptor 4 activation through Fc gamma receptor tethering. J Biol Chem. 2014;289(22):15309–18. doi:10.1074/jbc.M113.537936.
- Ellwanger K, Reusch U, Fucek I, Wingert S, Ross T, Muller T, Schniegler-Mattox U, Haneke T, Rajkovic E, Koch J. et al. Redirected optimized cell killing (ROCK(R)): a highly versatile multispecific fit-for-purpose antibody platform for engaging innate immunity. MAbs. 2019;11(5):899–918. doi:10.1080/19420862.2019.1616506.
- Xu DL, Alegre ML, Varga SS, Rothermel AL, Collins AM, Pulito VL, Hanna LS, Dolan KP, Parren PWHI, Bluestone JA. et al. In vitro characterization of five humanized OKT3 effector function variant antibodies. Cell Immunol. 2000;200(1):16–26. doi:10.1006/cimm.2000.1617.
- Lo M, Kim HS, Tong RK, Bainbridge TW, Vernes JM, Zhang Y, Lin YL, Chung S, Dennis MS, Zuchero YJY. et al. Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J Biol Chem. 2017;292(9):3900–08. doi:10.1074/jbc.M116.767749.
- Armour KL, Clark MR, Hadley AG, Williamson LM. Recombinant human IgG molecules lacking Fc gamma receptor I binding and monocyte triggering activities. Eur J Immunol. 1999;29(8):2613–24. doi:10.1002/(SICI)1521-4141(199908)29:08<2613:AID-IMMU2613>3.0.CO;2-J.
- Idusogie EE, Wong PY, Presta LG, Gazzano-Santoro H, Totpal K, Ultsch M, Mulkerrin MG. Engineered antibodies with increased activity to recruit complement. J Immunol. 2001;166(4):2571–75. doi:10.4049/jimmunol.166.4.2571.
- Moore GL, Chen H, Karki S, Lazar GA. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs. 2010;2(2):181–89. doi:10.4161/mabs.2.2.11158.
- Stavenhagen JB, Gorlatov S, Tuaillon N, Rankin CT, Li H, Burke S, Huang L, Johnson S, Bonvini E, Koenig S. Fc optimization of therapeutic antibodies enhances their ability to kill tumor cells in vitro and controls tumor expansion in vivo via low-affinity activating Fc gamma receptors. Cancer Res. 2007;67(18):8882–90. doi:10.1158/0008-5472.Can-07-0696.
- Nordstrom JL, Gorlatov S, Zhang W, Yang Y, Huang L, Burke S, Li H, Ciccarone V, Zhang T, Stavenhagen J. et al. Anti-tumor activity and toxicokinetics analysis of MGAH22, an anti-HER2 monoclonal antibody with enhanced Fcgamma receptor binding properties. Breast Cancer Res. 2011;13(6):R123. doi:10.1186/bcr3069.
- Zalevsky J, Chamberlain AK, Horton HM, Karki S, Leung IWL, Sproule TJ, Lazar GA, Roopenian DC, Desjarlais JR. Enhanced antibody half-life improves in vivo activity. Nat Biotechnol. 2010;28(2):157–59. doi:10.1038/nbt.1601.
- Dall’acqua WF, Woods RM, Ward ES, Palaszynski SR, Patel NK, Brewah YA, Wu H, Kiener PA, Langermann S. Increasing the affinity of a human IgG1, for the neonatal Fc receptor: biological consequences. J Immunol. 2002;169(9):5171–80. doi:10.4049/jimmunol.169.9.5171.
- Walker MR, Lund J, Thompson KM, Jefferis R. Aglycosylation of human IgG1 and IgG3 monoclonal-antibodies can eliminate recognition by human-cells expressing Fc-gamma-RI and or Fc-gamma-RII receptors. Biochem J. 1989;259(2):347–53. doi:10.1042/bj2590347.
- Atwell S, Ridgway JB, Wells JA, Carter P. Stable heterodimers from remodeling the domain interface of a homodimer using a phage display library. J Mol Biol. 1997;270(1):26–35. doi:10.1006/jmbi.1997.1116.
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. doi:10.1002/pro.3943.
- Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Design of therapeutic proteins with enhanced stability. Proc Natl Acad Sci USA. 2009;106(29):11937–42. doi:10.1073/pnas.0904191106.
- Lai PK, Fernando A, Cloutier TK, Kingsbury JS, Gokarn Y, Halloran KT, Calero-Rubio C, Trout BL. Machine learning feature selection for predicting high concentration therapeutic antibody aggregation. J Pharm Sci. 2021;110(4):1583–91. doi:10.1016/j.xphs.2020.12.014.
- Edelman GM, Cunningham BA, Gall WE, Gottlieb PD, Rutishauser U, Waxdal MJ. The covalent structure of an entire gammaG immunoglobulin molecule. Proc Natl Acad Sci USA. 1969;63(1):78–85. doi:10.1073/pnas.63.1.78.
- Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest. Bethesda (MD): NIH; 1991.
- Jensen RK, Plum M, Tjerrild L, Jakob T, Spillner E, Andersen GR. Structure of the omalizumab fab. Acta Crystallogr F Struct Biol Commun. 2015;71(Pt 4):419–26. doi:10.1107/S2053230X15004100.
- Bethea D, Wu SJ, Luo J, Hyun L, Lacy ER, Teplyakov A, Jacobs SA, Kt O, Gilliland GL, Feng Y. Mechanisms of self-association of a human monoclonal antibody CNTO607. Protein Eng Des Sel. 2012;25(10):531–37. doi:10.1093/protein/gzs047.
- Geoghegan JC, Fleming R, Damschroder M, Bishop SM, Sathish HA, Esfandiary R. Mitigation of reversible self-association and viscosity in a human IgG1 monoclonal antibody by rational, structure-guided fv engineering. MAbs. 2016;8(5):941–50. doi:10.1080/19420862.2016.1171444.
- Chow CK, Allan BW, Chai Q, Atwell S, Lu J. Therapeutic antibody engineering to improve viscosity and phase separation guided by crystal structure. Mol Pharm. 2016;13(3):915–23. doi:10.1021/acs.molpharmaceut.5b00817.
- Apgar JR, Tam ASP, Sorm R, Moesta S, King AC, Yang H, Kelleher K, Murphy D, D’Antona AM, Yan G. et al. Modeling and mitigation of high-concentration antibody viscosity through structure-based computer-aided protein design. PLoS One. 2020;15(5):e0232713. doi:10.1371/journal.pone.0232713.
- Lai PK, Ghag G, Yu Y, Juan V, Fayadat-Dilman L, Trout BL. Differences in human IgG1 and IgG4 S228P monoclonal antibodies viscosity and self-interactions: experimental assessment and computational predictions of domain interactions. MAbs. 2021;13(1):1991256. doi:10.1080/19420862.2021.1991256.
- Liu P, Pan Z, Gu C, Cao X, Liu X, Zhang J, Xiao Z, Wang X, Guo H, Ju D. et al. An omalizumab biobetter antibody with improved stability and efficacy for the treatment of allergic diseases. Front Immunol. 2020;11:596908. doi:10.3389/fimmu.2020.596908.
- Arora J, Hu Y, Esfandiary R, Sathish HA, Bishop SM, Joshi SB, Middaugh CR, Volkin DB, Weis DD. Charge-mediated Fab-Fc interactions in an IgG1 antibody induce reversible self-association, cluster formation, and elevated viscosity. MAbs. 2016;8(8):1561–74. doi:10.1080/19420862.2016.1222342.
- Chowdhury AA, Manohar N, Witek MA, Woldeyes MA, Majumdar R, Qian KK, Kimball WD, Xu S, Lanzaro A, Truskett TM. et al. Subclass effects on self-association and viscosity of monoclonal antibodies at high concentrations. Mol Pharm. 2023;20(6):2991–3008. doi:10.1021/acs.molpharmaceut.3c00023.
- Edgcomb SP, Murphy KP. Variability in the pKa of histidine side-chains correlates with burial within proteins. Proteins. 2002;49(1):1–6. doi:10.1002/prot.10177.
- Yadav S, Liu J, Shire SJ, Kalonia DS. Specific interactions in high concentration antibody solutions resulting in high viscosity. J Pharm Sci. 2010;99(3):1152–68. doi:10.1002/jps.21898.
- Tomar DS, Kumar S, Singh SK, Goswami S, Li L. Molecular basis of high viscosity in concentrated antibody solutions: strategies for high concentration drug product development. MAbs. 2016;8(2):216–28. doi:10.1080/19420862.2015.1128606.
- Inoue N, Takai E, Arakawa T, Shiraki K. Specific decrease in solution viscosity of antibodies by arginine for therapeutic formulations. Mol Pharm. 2014;11(6):1889–96. doi:10.1021/mp5000218.
- Oganesyan V, Damschroder MM, Woods RM, Cook KE, Wu H, Dall’acqua WF. Structural characterization of a human Fc fragment engineered for extended serum half-life. Mol Immunol. 2009;46(8–9):1750–55. doi:10.1016/j.molimm.2009.01.026.
- Matsumiya S, Yamaguchi Y, Saito J, Nagano M, Sasakawa H, Otaki S, Satoh M, Shitara K, Kato K. Structural comparison of fucosylated and nonfucosylated Fc fragments of human immunoglobulin G1. J Mol Biol. 2007;368(3):767–79. doi:10.1016/j.jmb.2007.02.034.
- Majumdar R, Esfandiary R, Bishop SM, Samra HS, Middaugh CR, Volkin DB, Weis DD. Correlations between changes in conformational dynamics and physical stability in a mutant IgG1 mAb engineered for extended serum half-life. MAbs. 2015;7(1):84–95. doi:10.4161/19420862.2014.985494.
- Tavakoli-Keshe R, Phillips JJ, Turner R, Bracewell DG. Understanding the relationship between biotherapeutic protein stability and solid-liquid interfacial shear in constant region mutants of IgG1 and IgG4. J Pharm Sci. 2014;103(2):437–44. doi:10.1002/jps.23822.
- Jacobsen FW, Stevenson R, Li C, Salimi-Moosavi H, Liu L, Wen J, Luo Q, Daris K, Buck L, Miller S. et al. Engineering an IgG scaffold lacking effector function with optimized developability. J Biol Chem. 2017;292(5):1865–75. doi:10.1074/jbc.M116.748525.
- Park E, Izadi S. Molecular surface descriptors to predict antibody developability: sensitivity to parameters, structure models, and conformational sampling. MAbs. 2024;16(1). doi:10.1080/19420862.2024.2362788.
- Sharma VK, Patapoff TW, Kabakoff B, Pai S, Hilario E, Zhang B, Li C, Borisov O, Kelley RF, Chorny I. et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci U S A. 2014;111(52):18601–06. doi:10.1073/pnas.1421779112.
- Bos AB, Luan P, Duque JN, Reilly D, Harms PD, Wong AW. Optimization and automation of an end-to-end high throughput microscale transient protein production process. Biotechnol Bioeng. 2015;112(9):1832–42. doi:10.1002/bit.25601.
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the expasy server. In: Walker J. editor. The proteomics protocols handbook. Totowa, New Jersey: Humana Press Inc; 2005. p. 571–607.
- Molecular Operating Environment (MOE). 2022.02 chemical computing group ULC, 910-1010 sherbrooke st. W. Montreal (QC) H3A 2R7, 2024; 2022.
- Roe DR, Cheatham TE. 3rd. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9(7):3084–95. doi:10.1021/ct400341p.
- Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–713. doi:10.1021/acs.jctc.5b00255.
- Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ. GLYCAM06: a generalizable biomolecular force field. Carbohydrates J Comput Chem. 2008;29(4):622–55. doi:10.1002/jcc.20820.
- Jorgensen WL, Chandreasekhar J, Madura JD, I RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–35. doi:10.1063/1.445869.
- Joung IS, Cheatham TE. 3rd. Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J Phys Chem B. 2008;112(30):9020–41. doi:10.1021/jp8001614.
- Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham ITE, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Giambasu G. et al. Amber 2019. San Francisco: University of California; 2019.
- Le Grand S, Götz AW, Walker RC. SPFP: speed without compromise—A mixed precision model for GPU accelerated molecular dynamics simulations. Comp Phys Comm. 2013;184(2):374–80. doi:10.1016/j.cpc.2012.09.022.
- Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. J Chem Theory Comput. 2015;11(4):1864–74. doi:10.1021/ct5010406.
- Pastor RW, Brooks BR, Szabo A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol Phys. 1988;65(6):1409–19. doi:10.1080/00268978800101881.
