Abstract
The phytohormone jasmonic acid (JA) plays a central role in defense against necrotrophic pathogens and herbivores in Nicotiana attenuata. Recently Santhanam et al.Citation1 showed that JA does not have a major role in shaping the root- and shoot associated bacterial communities, though a few taxa differed among control (empty vector, EV) plants and plants impaired in their capacity to produce JA (irAOC). In this addendum, we provide additional data showing that the composition of the plant bacterial communities is mainly shaped by tissue type. The qualitative data analysis revealed that at the order level, 5 bacterial OTUs formed a core community found in all tissues irrespective of genotypes, while 9 OTUs were different among roots and shoots. The heterogeneity among individual plants was high masking the potential genotype effect on bacterial communities. Using a culture-dependent approach, 3 of 18 bacterial taxa retrieved either only from one of the genotypes or from both had a growth promoting effect on EV and irAOC seedlings. The data suggest that the local soil niche in which the roots grows is a major driver of the variability in root bacterial communities recruited by different individuals, and the plant growth-promoting effects of some taxa are independent of the genotype.
Plants harbor a diverse range of bacterial communitiesCitation2,3 which are influenced by many biotic and abiotic factors.Citation2 Several studies showed that tissue types such as leaves and roots influence the bacterial community composition, and harbor distinct communities.Citation1,4 It is often assumed that root bacterial communities are shaped by soil microbiota,Citation5-7 and leaf bacterial communities by air, sunlight irradiation, stomata and mineral content of the leaves.Citation8-10 However, 4 independent studies using Arabidopsis as a model system indicated that at the phylum level, core communities such as Actinobacteria, Bacteriodetes and Proteobacteria can be found in all roots independent of soil type and genotypes,Citation2,4,5-7 strongly indicating that bacteria do not randomly colonize roots, but certain phyla preferentially colonize plant roots.
In a previous study,Citation1 we analyzed leaf and root bacterial community of isogenic field grown plants impaired in JA-production (irAOC) and control plants (empty vector, EV) by culture dependent and independent (pyrosequencing) approaches. Based on the quantitative data, we showed that leaf bacterial communities are different from those of roots.Citation1 Here, we demonstrate that based on qualitative data (presence and absence of OTUs at 97% similarity) leaf bacterial communities are clearly distinct from roots (), and within each tissue type (root vs leaf) plants impaired in JA production and EV plants do not show a genotype-specific pattern. These data are consistent with our previous findings, and similar results were obtained by Bodenhausen et al,Citation4 who showed that leaf bacterial communities of Arabidopsis thaliana are different from those of roots and concluded that organ type (root vs leaf) type influences the composition of the bacterial communities.
Figure 1. Bacterial communities are clearly different among roots and shoots (A), but not by the plant's capacity to produce jasmonic acid (JA). Venn diagram represents the core and root specific OTUs among EV and irAOC genotype tissues (B). Here we assigned core communities as OTUs recovered from all tissue samples (n = 20) and root specific OTUs (n = 10) irrespective of genotypes. The group-average dendogram was constructed by unweighted UniFrac distance metric and the Venn diagram based on presence and absence data of OTUs at the order level in roots and shoots of EV and irAOC plants. Only OTUs which were retrieved at least from all 5 replicates per tissue/genotype combination were considered, and not the total number of orders found. Abbreviations: R, roots; L, leaves
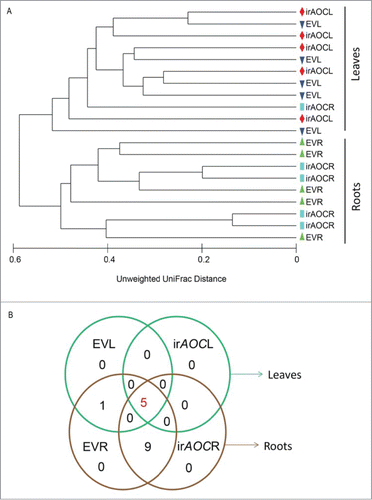
Though root and leaf bacterial communities were clearly distinct,Citation1 a bacterial core community was present in all samples of field-grown N. attenuata plants irrespective of the genotype (). At the order level, this core community consisted of 5 OTUs retrieved from all roots and shoots from 27 OTUs found in total. These 5 OTUs belonged to the bacterial phyla Bacteriodetes, Proteobacteria and Firmicutes. Additionally, 9 OTUs were present in all root samples, representing also 2 additional bacterial phyla, Actinobacteria and Deinococcus-thermus, indicating a root-specific enrichment. In accordance with our study, Actinobacteria were shown in a previous studies to be enriched in roots irrespective of soil types.Citation5,6 Deinococcus-thermus taxa are described as highly resistant to environmental hazards and can survive high doses of gamma and UV radiation.Citation11,12 Interestingly, N. attenuata's native habitat, the Great Basin Desert, Utah, USACitation13 is characterized by high light intensities and high UV-B fluence rates.
In earlier studies, independent of the soil type, the bacterial community composition was found to be similar at the phylum level in different genotypes of the same and related species. Only few bacterial taxa were quantitatively different among plant genotypesCitation1,5-7 suggesting that genotypes have a minor role in structuring bacterial communities. Based on 16S rDNA gene pyrosequencing of 8 Arabidopsis ecotypes roots Lundberg et alCitation5 showed that of 778 OTUs, only 12 OTUs exhibited host genotype specific quantitative enrichment. In another study, using the same technique, Bulgarelli et al Citation6 found only one OTU was significantly different among 2 Arabidopsis ecotypes. In our study, based on ANOSIM β diversity, the overall bacterial diversity of leaves and roots of EV and irAOC genotypes was not significantly different,Citation1 and we did not find a consistent clustering based on genotype () by qualitative data, and the bacterial community composition was highly heterogeneous among replicate plants based on qualitative and quantitative data. At the genera level, 21 OTUs significantly differed among EV and irAOC roots,Citation1 and at the order level, 1 OTU was distinct among the genotypes (). We hypothesize that sample-to-sample differences are due to differences in the local soil bacterial community in which the plant grows (), leading to the recruitment of soil-specific taxa. This hypothesis is in line with a large-scale study using 27 maize genotypes growing at 5 different locations in the US. Bacterial communities clearly clustered by soil, but not by genotype.Citation14
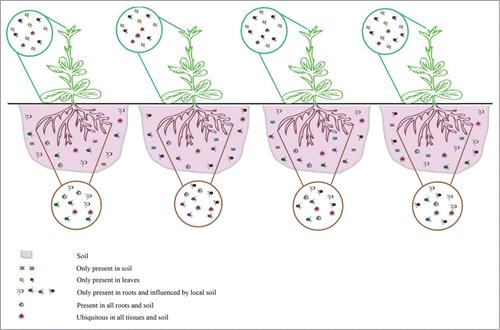
Figure 2. Summarizing scheme illustrating the intraspecific variation of root bacterial communities among individual plants of N. attenuata. Shoot bacterial endophytes are similar among individual plants and independent of the plant's capacity to produce JA, while root-associated bacterial communities are highly variable among different individuals irrespective of genotypes. We hypothesize that the intraspecific differences are due to local soil niches leading to the recruitment of plant-specific taxa that could be recruited for “opportunistic mutualisms.”
Bacterial communities, which reside in plants can either have beneficial or detrimental effects on their hosts.Citation15,16 Detrimental effects are caused by necrotrophic and biotrophic pathogens Citation17,18 Beneficial effects can be direct or indirect, resulting in plant growth promotion. Direct plant growth promotion (PGP) can result from improved nutrient acquisition (e.g. nitrogen, phosphorous), the production of phytohormones (IAA, gibberellins) or the synthesis of stress modulators such as 1-amino cyclopropane-1- carboxylate (ACC) deaminase which lowers the plant's endogenous ethylene levels.Citation3,19 Indirect plant growth promotion can be due to the prevention or reduction of pathogen infection either by direct suppression, e.g., outcompeting pathogens for nutrients or by priming tissues for enhanced defense against pathogen or herbivore attack.Citation20,21 The enrichment of certain beneficial bacterial taxa can lead to opportunistic mutualisms between plants and microbes, as exemplified by a recent study of Meldau et alCitation22,23 showing that an ethylene insensitive N. attenuata genotype can recruit beneficial microbes to compensate its growth deficiency. PGP traits of certain bacterial isolates can be host dependent; e.g. Long et alCitation24 showed that bacterial strains isolated from Solanum nigrum roots were unable to promote growth of N. attenuata. However, in general, large numbers of bacterial isolates enhance growth of plants independent of their hosts.Citation3,19,20 The exact mechanism responsible for the recruitment and fine tuning of bacterial taxa from the local soil community remains to be elucidated. Some studies indicate that the lignin content and cell wall composition may play a role,Citation2,25 in addition to ethylene signaling.Citation26
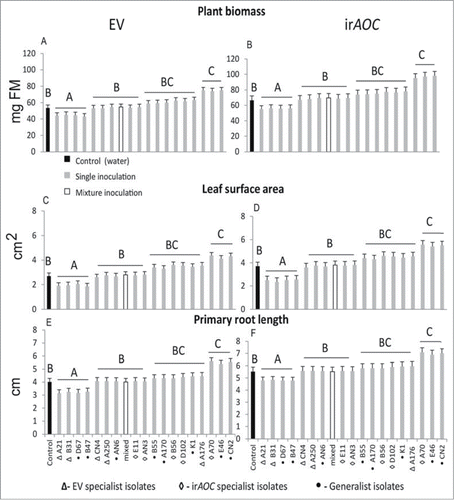
In our study, we isolated 414 bacterial strains from surface sterilized roots and leaves of both genotypes using a culture-dependent approach.Citation1 18 strains were classified as putative specialists and generalists based on the isolation of a particular strain from either of the 2 plant genotypes (EV or irAOC). PGP effects of these putative generalist and specialist bacterial strains were investigated (for experimental details see “In-vitro re-isolation” in Santhanam et alCitation1). Three of the 18 strains (B. cereusi CN2, P. azotoformans A70 A. nitroguajacolicus E46, n = 6, P < 0.05, Fisher's PLSD) used in this assay significantly promoted plant growth with respect to plant biomass (ANOVA; F19,90 = 6.81, P < 0.001), leaf surface area (ANOVA; F19,90 = 3.58, P < 0.001) and primary root length (ANOVA; F19,90 = 19.84, P < 0.001, ). Four isolates significantly reduced plant growth (n = 6, P < 0.05, Fisher's PLSD, Fig. 3), while 11 isolates had no significant effect (). The PGP effects did not depend on the plant's genotype from which they were isolated (2-way ANOVA plant biomass- bacterial type*genotype: p = 0 .98, leaf surface area- bacterial type*genotype: p = 0 .99, root length- bacterial type*genotype: p = 0 .97). Interestingly, inoculation with a mixture of all 18 bacterial isolates also did not promote growth, though - based on the single inoculations - plant growth promoting bacteria were present. We assume that under in-vitro conditions growth promotion by these bacterial isolates might be inhibited by competitive interactions among different isolates in mixed inoculations. In order to test whether colonization of bacterial isolates correlate with the PGP effects, we performed a linear regression model with both genotypes; however, no correlation was observed between PGP effects and the colonization pattern (EV- plant biomass: R2 = 0 .07, p = 0 .274, leaf surface area:R2 = 0.05, p = 0 .337, root length:R2 = 0.05,p = 0 .336; irAOC- plant biomass: R2 = 0 .074, p = 0 .258, leaf surface area:R2 = 0.04, p = 0 .39, root length:R2 = 0.05,p = 0 .319). This analysis corroborates that PGP effects were independent of quantitative root colonization and genotype and might be influenced by bacterial PGP traits such as production of IAA and ACC deaminase activity as exemplified by Long et al.Citation24
Figure 3. Putative genotype specialist and generalist bacterial taxa were isolated by culture-dependent approach and plant growth promoting (PGP) effects were performed under in-vitro conditions.Citation1 Plant biomass (A and B), leaf surface area (C and D) and primary root length (E and F) of EV and irAOC plants were measured 24 d after 7-day old seedlings were inoculated. PGP effects of 3 bacterial taxa are independent of genotypes. The experimental setup is the same as described in Santhanam et al.Citation1 Mean (±SE, n = 6, different letters indicate significant differences among mock-and bacterial inoculations, one-way ANOVA with Fisher's PLSD test; P < 0.05).
Based on these results, we conclude that leaves and roots of field grown N. attenuata plants harbor distinct bacterial communities. Though all field-grown plants show a core community of the same bacterial orders, we assume the local soil niche determines the overall variability in the composition of the root-associated bacterial communities. Some of the recruited bacteria have a beneficial effect on plant growth independent of the genotype and may increase the plant's fitness depending on the environmental conditions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the Brigham Young University, Utah, USA, for use of their field station, the Lytle Ranch Preserve, and D. Kessler and C. Diezel for field sample collections.
Funding
We thank the Max Planck Society and the European Research Council (ERC Advanced Grant 293926) for funding.
References
- Santhanam R, Groten K, Meldau DG, Baldwin IT. Analysis of plant-bacteria interactions in their native habitat: bacterial communities associated with wild tobacco are independent of endogenous jasmonic acid levels and developmental stages. PLoS One [Internet] 2014; 9:e94710; http://dx.doi.org/10.1371/journal.pone.0094710
- Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol [Internet] 2012; 64:9.1-9.32; PMID:23157644
- Doornbos RF, Loon LC, Bakker PAHM. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. Agron Sustain Dev [Internet] 2011 [cited 2012 Jul 22];; 32:227-43; http://dx.doi.org/10.1007/s13593-011-0028-y
- Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS One [Internet] 2013; 8:e56329; PMID:23457551; http://dx.doi.org/10.1371/journal.pone.0056329
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, et al. Defining the core Arabidopsis thaliana root microbiome. Nature [Internet] 2012 [cited 2013 Feb 1]; 488:86-90; PMID:22859206; http://dx.doi.org/10.1038/nature11237
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012; 488:91-5; PMID:22859207; http://dx.doi.org/10.1038/nature11336
- Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci [Internet] 2013; PMID:24379374
- Kadivar H, Stapleton AE. Ultraviolet radiation alters maize phyllosphere bacterial diversity. Microb Ecol [Internet] 2003; 45:353-61; PMID:12704563; http://dx.doi.org/10.1007/s00248-002-1065-5
- Vorholt JA. Microbial life in the phyllosphere. Nat Rev Micro [Internet] 2012; 10:828-40; http://dx.doi.org/10.1038/nrmicro2910
- Reisberg EE, Hildebrandt U, Riederer M, Hentschel U. Distinct phyllosphere bacterial communities on Arabidopsis wax mutant leaves. PLoS One [Internet] 2013; 8:e78613; PMID:24223831; http://dx.doi.org/10.1371/journal.pone.0078613
- Daly MJ. A new perspective on radiation resistance based on Deinococcus radiodurans. Nat Rev Micro [Internet] 2009; 7:237-45; http://dx.doi.org/10.1038/nrmicro2073
- Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol Mol Biol Rev [Internet] 2001; 65:44-79; http://dx.doi.org/10.1128/MMBR.65.1.44-79.2001
- Baldwin IT. An ecologically motivated analysis of plant-herbivore interactions in native tobacco. Plant Physiol [Internet] 2001; 127:1449-58; PMID:11743088; http://dx.doi.org/10.1104/pp.010762
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL, Buckler ES, Ley RE. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci [Internet] 2013; 110:6548-53; PMID:23576752; http://dx.doi.org/10.1073/pnas.1302837110
- Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol [Internet] 1997; 43:895-914; http://dx.doi.org/10.1139/m97-131
- Reinhold-Hurek B, Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol [Internet] 2011 [cited 2013 Jan 29]; 14:435-43; http://dx.doi.org/10.1016/j.pbi.2011.04.004
- Alfano JR, Collmer A. Bacterial pathogens in plants: Life up against the wall. Plant Cell [Internet] 1996; 8:1683-98; http://dx.doi.org/10.1105/tpc.8.10.1683
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol [Internet] 2005; 43:205-27; PMID:16078883; http://dx.doi.org/10.1146/annurev.phyto.43.040204.135923
- Glick BR. Plant growth-promoting bacteria: mechanisms and applications. Scientifica (Cairo) [Internet] 2012; 2012:1-15; http://dx.doi.org/10.6064/2012/963401
- Compant S, Clément C, Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol Biochem [Internet] 2010 [cited 2013 Jan 30]; 42:669-78; http://dx.doi.org/10.1016/j.soilbio.2009.11.024.
- Lugtenberg B, Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol [Internet] 2009 [cited 2012 Jul 13]; 63:541-56; PMID:19575558; http://dx.doi.org/10.1146/annurev.micro.62.081307.162918
- Meldau DG, Long HH, Baldwin IT. A native plant growth promoting bacterium, Bacillus sp. B55, rescues growth performance of an ethylene-insensitive plant genotype in nature. Front Plant Sci [Internet] 2012 [cited 2012 Nov 8]; 3:112; PMID:22701461; http://dx.doi.org/10.3389/fpls.2012.00112
- Meldau DG, Meldau S, Hoang LH, Underberg S, Wünsche H, Baldwin IT. Dimethyl disulfide produced by the naturally associated bacterium Bacillus sp B55 promotes Nicotiana attenuata growth by enhancing sulfur nutrition. Plant Cell Online [Internet] 2013; 25:2731-47; http://dx.doi.org/10.1105/tpc.113.114744
- Long HH, Schmidt DD, Baldwin IT. Native bacterial endophytes promote host growth in a species-specific manner; phytohormone manipulations do not result in common growth responses. PLoS One [Internet] 2008 [cited 2012 Nov 2]; 3:e2702; PMID:18628963; http://dx.doi.org/10.1371/journal.pone.0002702
- Bennett AE, Grussu D, Kam J, Caul S, Halpin C. Plant lignin content altered by soil microbial community. New Phytol [Internet] 2014; 206:166-74.
- Long HH, Sonntag DG, Schmidt DD, Baldwin IT. The structure of the culturable root bacterial endophyte community of Nicotiana attenuata is organized by soil composition and host plant ethylene production and perception. New Phytol [Internet] 2010 [cited 2012 Nov 8]; 185:554-67; PMID:19906091; http://dx.doi.org/10.1111/j.1469-8137.2009.03079.x
