Abstract
Bacterial microcompartments (BMCs) are proteinaceous organelles used by a broad range of bacteria to segregate and optimize metabolic reactions. Their functions are diverse, and can be divided into anabolic (carboxysome) and catabolic (metabolosomes) processes, depending on their cargo enzymes. The assembly pathway for the β-carboxysome has been characterized, revealing that biogenesis proceeds from the inside out. The enzymes coalesce into a procarboxysome, followed by encapsulation in a protein shell that is recruited to the procarboxysome by a short (∼17 amino acids) extension on the C-terminus of one of the encapsulated proteins. A similar extension is also found on the N- or C-termini of a subset of metabolosome core enzymes. These encapsulation peptides (EPs) are characterized by a primary structure predicted to form an amphipathic α-helix that interacts with shell proteins. Here, we review the features, function and widespread occurrence of EPs among metabolosomes, and propose an expanded role for EPs in the assembly of diverse BMCs.
Bacterial Microcompartments-Organelles of Bacteria
Until recently, subcellular compartmentalization was the hallmark feature of eukaryotic cells, in which metabolic segregation is achieved by a delimiting lipid membrane. However, over the past decade, an increasing number of studies (reviewed in refs.Citation1-5) have unveiled the sophisticated subcellular organization of bacteria. One striking example of this organization is the bacterial microcompartment (BMC), which can be considered the bacterial equivalent of the eukaryotic organelle.
In electron micrographs of thin-sectioned bacteria, BMCs appear as polyhedral bodies, typically ranging between 40 and 200 nm in diameter. Unlike their eukaryotic counterparts, they are composed entirely of protein.Citation3-5 BMCs consist of a semi-permeable protein shell that encloses multiple proteins, typically including several enzymes constituting a specific metabolic reaction sequence. Although only a few types have been experimentally characterized, bioinformatic analyses of the presumed contents of diverse BMCs suggest that the shell serves to protect the cell from toxic compounds generated by the encapsulated enzymes,Citation6 prevent the loss of volatile intermediates,Citation5,7 and/or exclude adverse competing substrates or enzymes. Furthermore, because they concentrate enzymes with their substrates, the encapsulated reactions are assumed to achieve higher efficiency, while confinement may enhance protein stability and the exclusion of oxygen makes the resident enzymes less prone to oxidative damage.Citation8,9
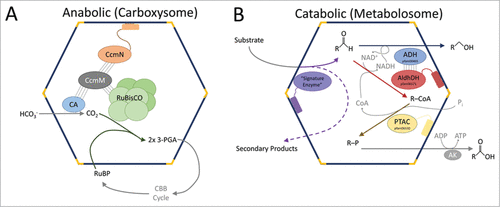
The functions of BMCs are defined by the enzymes they package. The most extensively characterized BMC is the carboxysome, which is found in cyanobacteria, chemoautotrophs and a few purple phototrophs. The carboxysome encapsulates ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) and carbonic anhydrase, and plays a key role in carbon fixation ().Citation10 In addition to carboxysomes, there are catabolic BMCs (metabolosomes) that degrade a variety of different carbon compounds (). Examples of these include the well-characterized propanediol utilizationCitation11 (PDU) and ethanolamine utilizationCitation12 (EUT) microcompartments and the recently characterized BMCs that degrade ethanolCitation13,14 and fucose/rhamnose.Citation15,16 BMCs are also phylogenetically diverse; a recent bioinformatics survey found 23 distinct loci types distributed across 23 bacterial phyla, illustrating their surprising ubiquity.Citation17
Figure 1. Schematic of carboxysome and metabolosome organization. (A) The RuBisCO small subunit-like domains of CcmM interact with RuBisCO (piggybacking); CcmM also associates with CcmN which interacts with shell proteins through its EP. (B) The signature and core enzymes of various metabolosomes contain EPs. Other proteins/subunits may interact with these proteins to be encapsulated by piggybacking. The α-helical portion of the EP is shown as a cylinder connected to the functional core of its cognate protein via an extended, poorly conserved linker. CA, carbonic anhydrase; 3-PGA, 3-phosphoglycerate; RuBP, ribulose 1,5-bisphosphate; CBB cycle, Calvin-Benson-Bassham cycle; AldhDH, aldehyde dehydrogenase; ADH, alcohol dehydrogenase; PTAC, phosphotransacylase; AK, acyl kinase.
BMCs are proposed to have significant potential for the bioengineering of heterologous pathways.Citation18,19 For example, genetic loci coding for the BMC shell and the encapsulated pathway can be directly expressed in foreign hosts without any modification.Citation20,21 However, realizing the goal of engineering custom bacterial organelles containing multiple selected enzymes is predicated on understanding how BMCs assemble. Recently, the first elucidation of an assembly pathway for a BMC (the β-carboxysome) was reported, highlighting the key role of a short encapsulation peptide (EP). The carboxysome forms from the inside out ():Citation22 first, RuBisCO forms a procarboxysome by aggregating with CcmM, a protein composed of a γ-carbonic anhydrase domain and 3-5 additional domains that resemble the small subunit of RuBisCO.Citation22 CcmM, CcmN (a protein of unknown function) and a β-carbonic anhydrase form a complexCitation23,24 completing the procarboxysome. CcmN recruits the shell to the procarboxysome via its EP. The EP is composed of a short (∼17 amino acids) α-helix which interacts with the major shell protein CcmK2.Citation25 β-carboxysome assembly is abolished when the EP is deleted from CcmN, highlighting the importance of this peptide.Citation25
EPs have been predicted and characterized for both of the extensively studied metabolosomes, the EUT and PDU BMCs.Citation25-28 They were initially discovered in multiple sequence alignments comparing encapsulated and non-metabolosome homologs of PduP, the aldehyde dehydrogenase in the PDU BMC. The N-terminal extension found only on encapsulated homologs was sufficient to target foreign proteins to shells.Citation26 Kinney et al. experimentally identified the EP found on the C-terminus of CcmN, predicted it to be an amphipathic α-helix, and identified peptides with similar characteristics in different enzymes from diverse metabolosomes.Citation25 This led to the definition of the EP as a short sequence (typically 15–20 amino acids), predicted to form an amphipathic α-helix and separated from the functional domain of the protein by a poorly conserved linker. The α-helical structure of an EP was subsequently confirmed by NMR spectroscopy.Citation29
Here, we show that EPs are present in several of the key proteins encapsulated within BMCs where they presumably play crucial roles in assembly. Identifying and characterizing EPs is essential to both understanding the assembly of BMCs and for engineering BMCs for new functions. We focus on the diversity of EPs, their structural characteristics, as well as their widespread distribution.
Definitions
The majority of the BMCs characterized bioinformatically contain an enzyme that defines their substrate selectivity, which is referred to as the “signature enzyme.”Citation5,16,17 For example, in the carboxysome, the signature enzyme RuBisCO carboxylates the substrate ribulose-1,5-bisphosphate. In many types of metabolosomes, the signature enzyme acts on the initial and specific substrate of the BMC (e.g. propanediol dehydratase acting on propanediol in the PDU BMC, ethanolamine ammonia-lyase acting on ethanolamine in the EUT BMC); the reactions of these enzymes yield an aldehyde ( and ). Other BMC “core enzymes” complete the metabolic sequence of the metabolosome: the aldehyde is used by the core NAD+ and coenzyme A-dependent aldehyde dehydrogenase.Citation17 Recycling of NAD+ and coenzyme A inside the BMC is achieved by an alcohol dehydrogenase and a phosphotransacylase, respectively ( and ). This general model for metabolosome core function is thought to apply to the majority of diverse BMC types.Citation5,16,17
Table 1. List of the proteins analyzed in this study. The presence and the position of the EP are shown on its cognate protein as an amphipathic helix
Methods
To expand the available data on EPs, we surveyed the signature and core gene products of carboxysomes and functionally defined metabolosomes. These include CcmN orthologs, CcaA (the carboxysomal β-carbonic anhydrase), the propanediol dehydratase from the PDU BMC, the ethanolamine ammonia-lyase from the EUT BMC and a class II aldolase, as well as the core enzymes of diverse bioinformatically characterized metabolosomes:Citation17 the aldehyde and the alcohol dehydrogenases, and the phosphotransacylase (). To our knowledge, no other BMC encapsulated proteins contain EPs.
CcaA and CcmN orthologs were extracted from the Integrated Microbial Genomes (IMG) database (https://img.jgi.doe.gov/) by BLAST search. For the metabolosomes, sequences were extracted from the Dataset S1 from a recently constructed taxonomy of all BMCs.Citation17 Multiple sequence alignments were performed using MUSCLECitation30 and visualized with Jalview.Citation31 Secondary structure was predicted using the JPred server.Citation32
The EP in β-carboxysomes
From our analysis of carboxysome-encapsulated proteins, only CcmN contains a clearly identifiable EP. CcmN is an essential β-carboxysome assembly protein composed of a N-terminal domain constituted of multiple bacterial hexapeptide repeat domains (Pfam00132), separated from the C-terminal domain (containing the EP) by a linker of variable sequence and length.Citation25 In the alignment of the 151 CcmN orthologs, the length of the linkers ranged from 23 to 277 amino acids; there is no discernible sequence homology among linkers and even the amino acid composition is variable (Fig. S1; ). This wide range of linker length is unique to CcmN and we were unable to correlate linker length with any other sequence feature. In the majority of the CcmN sequences analyzed, a short (typically 5 to 15 residues) terminal peptide fragment follows the EP (Fig. S1). We examined this terminal fragment as well as the linker and the EP of CcmN to determine the proportion of amino acids with similar physicochemical characteristics. While the EP is enriched in hydrophobic residues, the linker and the terminal fragment are predominantly and consistently hydrophilic (). Notably, in the 3 cases lacking the terminal fragment (strains PCC 6301, PCC 7942 and PCC 7429), the EP is enriched in hydrophilic residues, compared to the other CcmN EP sequences (). Overall, this suggests that the hydrophilic character of both the linker and the terminal fragment may be important for the correct assembly of the β-carboxysome, whether by aiding in the solubility of the EP or providing a specific binding face for interaction with shell proteins.
Table 2. Amino acid composition and lengths of the EPs and linkers of the various proteins analyzed. Amino acids were grouped by sidechain characteristics. Standard deviations are indicated as +/−
The EPs of Metabolosome Signature Enzymes
Ethanolamine ammonia-lyase, the signature enzyme of the EUT BMC, is composed of 2 different subunits - EutB and EutC - which assemble into a trimer of (αβ) dimers.Citation33 A previously reported sequence alignment identified a N-terminal extension on the BMC-encapsulated EutC from Escherichia coli K-12.Citation26 This was later experimentally confirmed to be an EP when the first 19 residues of EutC targeted GFP to the EUT shells.Citation28
We extracted 125 EutC sequences from the LoClass Data set S1Citation17 and aligned them with 22 EutC sequences from organisms that do not encode any BMC shell protein genes (Fig. S2). All but 2 (from strains of Shigella boydii, perhaps indicating that this BMC is no longer functional, or that the encapsulation is achieved by another process) of the EutC orthologs encoded in BMC loci contain an N-terminal extension. Secondary structure prediction and amino acid composition indicate that this N-terminal extension may form an amphipathic α-helix, meeting the criteria for an EP defined by Kinney et al.Citation25 The EP is joined to the enzymatic domain of EutC by a linker (36–91 amino acids) of poorly conserved primary structure and composition (; Fig. S2). We similarly analyzed EutB, but no peptide extension was detected in any part of the protein, suggesting that EutC is the only subunit of ethanolamine ammonia-lyase to have an EP. Because the native form of this signature enzyme is a complex formed by the 2 subunits, one EP is presumably sufficient to encapsulate the holoenzyme within the BMC, with EutB internalized through its interaction with EutC (“piggybacking”).
The other well-characterized metabolosome, the PDU BMC, contains B12-dependent propanediol dehydratase as its signature enzyme. This is formed by PduC, PduD and PduE in an α2β2γ2 complex.Citation34,35 We did not detect an EP on PduC, consistent with the experimental observation that it cannot be encapsulated in the absence of other PDU core proteins.Citation27 However, in another study,Citation20 GFP-fused to full-length PduC was successfully targeted to empty, recombinant PDU shells, suggesting that for some encapsulated proteins other determinants besides the EP play a role in encapsulation.
An EP was previously predicted for the N-terminus of PduD.Citation25,26 Experimental data showed that the PduD EP is sufficient to package the PduCDE holoenzyme into the BMC.Citation27 Fusion of PduD EP to GFP associates it with empty PDU shells.Citation20 We aligned 84 PduD sequences with 6 control (non BMC-associated) PduD sequences: 82 of the orthologs contain the characteristic N-terminal extension (the 2 exceptions are Lactobacillus brevis sequences) (Fig. S4). The PduD EP is followed by a linker ranging from 22 to 58 amino acids of variable composition ().
The alignment of 103 PduE sequences with 6 control PduE sequences from the genomes of organisms that do not encode BMCs shows the presence of a N-terminal extension in the BMC-associated PduE orthologs (except for 2 Thermoanaerobacterium and 4 Mycobacterium species; Fig. S3). These extensions have the typical characteristics of the EPs: about 18 residues predicted to form an amphipathic α-helix followed by a linker region that connects the EP to the core of the protein.Citation25 Again, the composition of the linker region is poorly conserved, however, its length is relatively more consistent, ranging from 26 to 39 amino acids (). Despite the predicted EP on PduE, the protein cannot be encapsulated in empty shells.Citation27 Presumably it is packaged through its interaction with PduC/PduD.
Collectively, these data for assembly of metabolosome signature enzymes suggest that inter-protein interactions are not only essential for formation of the holoenzyme but likewise provide a means to pack multiple proteins in the core of the metabolosome. The signature enzyme subunits EutB and PduC lack EPs and are probably encapsulated through association with the other enzyme(s) of the complex. A major difference between PduCDE and EutBC, however, is that EutBC contains only one N-terminal EP, while 2 N-terminal EPs are present in PduCDE, even though, presumably, only one would be enough for encapsulation. Notably, the amino acid sequences of the EPs of PduD and PduE are quite different which may reflect interactions with different shell protein binding sites, or suggest that the EP on PduE has a distinct function, such as aggregating lumenal enzymes ().
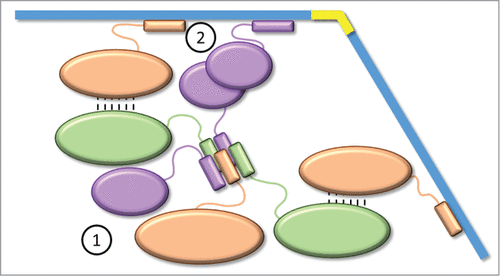
Figure 4. Aggregation model for the metabolosome assembly. For clarity, the model is represented by only 3 proteins. EPs interact together to form a cluster (prometabolosome) as the first step of the metabolosome assembly. Through interaction network (piggybacking) and oligomerization, some EPs are free to recruit the shell proteins to enclose the cluster as the second step.
A class II aldolase (Pfam00596) is predicted to serve as a signature enzyme for 2 distinct types of BMCs; it cleaves fucose and rhamnose to dihydroxyacetone phosphate or pyruvate (depending on whether the sugar is phosphorylated), and lactaldehyde, which is then processed by the BMC-encapsulated enzymes. In the Planctomycetes and Verrucomicrobia-Type (PVM)Citation16 and likely the PVM-likeCitation17 BMCs, lactaldehyde is directly processed by the BMC core enzymes.Citation16 For a bioinformatically described BMC (GRM5), which presumably encapsulates a Glycyl Radical Enzyme,Citation15,17 lactaldehyde is reduced to propanediol.Citation15 Twentythree BMC-associated and 12 non-BMC associated class II aldolase sequences were aligned. An ∼17 amino acid peptide extension that contains the EP characteristics defined by Kinney et al.Citation25 is found at the C-terminus of 21 sequences (Anaerobaculum mobile and Ruminococcus torques do not have the extension) (Fig. S5) separated from the functional domain of the protein by a linker ranging from 23 to 81 residues without amino acid composition or sequence conservation.
Collectively, these data show that all of the signature enzymes of experimentally characterized metabolosomes contain at least one EP, either N-terminal (PduD, PduE and Eut C) of C-terminal (class II aldolase) (). These EPs from different signature enzymes do not share a conserved sequence, but they do share sequence properties ().
The EPs of the Metabolosome Core Enzymes, Aldehyde and Alcohol Dehydrogenases and Phosphotransacylase
Aldehyde dehydrogenase is the most ubiquitous core enzyme found among metabolosomes.Citation17 Several studies have focused on the EP on the N-terminus of this enzyme,Citation26,29,36,37 and, in one case, a solution structure was solved, confirming the predictionCitation25 that the EP forms an amphipathic helix.Citation29
A multiple sequence alignment of 362 aldehyde dehydrogenase sequences shows that the encapsulation extension (EP and poorly conserved linker region) can be on either the N- or the C-terminus of the enzyme (Fig. S6), in contrast to the signature enzymes of metabolosomes (PduD, PduE, EutC and class II aldolase) and CcmN, where the extension is consistently observed only on one terminus of the protein. The location of the EP correlates with the BMC subfamily (for a review of the different families and subfamilies, see reference Citation17). For example, the EP is found on the N-terminus of the EUT1-associated aldehyde dehydrogenase, and on the C-terminus of the other EUT BMCs (EUT2A, B, C, D, EUT3 and EUT-other). In contrast, all aldehyde dehydrogenase orthologs in the PDU subfamilies contain a N-terminal EP. In our alignment, only a few aldehyde dehydrogenase sequences do not appear to have an EP. Upon closer inspection, these were identified as paralogs, present in the loci in addition to an aldehyde dehydrogenase gene that contains an EP. For example, the GRM1 locus found in 5 Streptococcus strains encodes 2 copies of an aldehyde dehydrogenase: one is predicted to be inactive due to a mutation of a catalytic residue;Citation17 this paralog lacks an EP while the other one is presumably active (based on residues conserved in the active site) and contains an EP on its C-terminus. However, the GRM1 locus of other organisms (Desulfovibrio, Desulfitobacterium, Clostridium) also possesses 2 copies of the aldehyde dehydrogenase, one active and the other one inactive, both with an EP on the C-terminus. It is tempting to posit that the presumably inactive forms serve as a scaffold for protein interaction and aggregation in the lumen of the BMC (see below).
We also examined the next enzyme in the metabolosome core metabolic sequence (), the alcohol dehydrogenase. Despite the prediction of an N-terminal EP for EutG (EUT-associated alcohol dehydrogenase) from Escherichia coli based on a sequence alignment,Citation26 experimental data showed that the first 19 amino acids of EutG from Salmonella enterica do not target fused GFP to the shells.Citation28 Our analysis did not identify an obvious EP (Fig. S7). However, all but one of the alcohol dehydrogenase orthologs from a specific EUT BMC subtype (EUT1), have a short additional peptide (about 20 amino acids) directly preceding the conserved N-terminus; it is predicted to fold as a α-helix, but does not otherwise fit the definition of an EP (no apparent amphipathicity of the helix, and no linker). It has been shown that alcohol and aldehyde dehydrogenase form a complex,Citation38 thus protein-protein interactions (piggy-backing), as proposed for PduCDE and EutBC (above), may be the means for encapsulation of the alcohol dehydrogenase ().
Finally, we examined the last enzyme of the metabolosome core metabolic reaction, the phosphotransacylase. The alignment of 227 sequences of BMC-associated phosphotransacylases with 20 control (non-BMC associated) sequences shows the presence of a characteristic EP in the N-terminus of the enzyme (Fig. S8). The linker connecting the EP to the core of the enzyme is shorter than what is typically observed. In most cases, the linker is approximately 5 amino acids (), suggesting reduced flexibility relative to other linkers.
General Features of EPs
From our comparison of 1065 EPs from different encapsulated proteins in diverse BMCs, we can identify a characteristic pattern of alternation of hydrophobic and hydrophilic residues despite a lack of sequence conservation (). Secondary structure prediction indicates that they most probably form a short amphipathic α-helix (about 18 amino acids) present on either the N-terminus (EutC, PduD, PduE, PTAC) or the C-terminus (CcmN, class II aldolase) of the protein (). Interestingly, in the case of the aldehyde dehydrogenase, the EP can be on either side of the functional domain. For all of the proteins, the EP is separated from the core of the enzyme by a poorly conserved linker typically enriched in hydrophilic and small amino acids (). Finally, our data suggest that piggybacking by proteins lacking an EP (e.g., alcohol dehydrogenase, some subunits of signature enzymes), as shown for core components of the β-carboxysome, likely occurs in the assembly of diverse BMCs. Some BMC-specific differences emerge upon close inspection. For example, the CcmN (β-carboxysome) EP contains aromatic amino acids, whereas they are rarely found in metabolosome EPs. Furthermore, using a script based on a published formulae,Citation39,40 (available at Citation41) we calculated the hydrophobic moment of all the EPs surveyed here; metabolosome EPs have a consistently higher amphipathicity score (Fig. S9).
Future Directions and Questions
Two studies conducted on the PduP (aldehyde dehydrogenase of the PDU microcompartment) EP from Salmonella enterica have probed the residues important for encapsulation. Mutagenesis of the PduP EP with encapsulation assayed by fluorescence microscopy, demonstrated the importance of the hydrophobic residues L6, L9, I10, I13 and L14.Citation42 Likewise alanine scanning mutagenesis followed by BMC purification and enzyme assays identified E7, I10 and L14 (corresponding to one face of the helix) as key residues for encapsulation.Citation36 These residues were proposed to interact with the hydrophobic residues on the short C-terminal helix of the hexameric shell proteins PduA and PduJ (). Collectively, these results suggest that (at least for the PDU BMC) the hydrophobic surface of the EP is most likely responsible for the binding to the shell proteins (and so, for the encapsulation), while its solvent-exposed hydrophilic side provide either solubility or a surface to interact with other encapsulated enzymes.
Figure 3. Model of EPs interacting with shell proteins. (A) Proposed 1:1 modelCitation36 in which EPs (orange) interact with the C-terminal α-helices of a hexameric shell protein (blue). (B) Alternative model in which EPs interact with the epitope formed by the hexamer-hexamer interfaces. Distinct epitopes (denoted as 1 and 2) are formed by the interactions of different combinations of shell protein paralogs (denoted by different shades of blue).
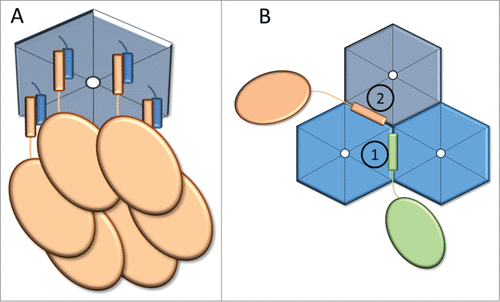
This model for EP association with the shell results in a 1:1 interaction between EP and each monomer of a hexamer, which seems difficult to accommodate sterically () Alternatively, considering that the basic building blocks of the shells (hexamers, pseudo-hexamers and pentamers) associate with each other into layers or strips in the absence of core BMC proteins,Citation4,20,37,43-47 binding sites for EPs may form at the interface between 2 or more hexamers (). This model also suggests how specific lumen proteins can be associated with specific shell proteins. Currently there are no data to explain how the stoichiometry and organization of the multiple shell paralogs of any BMC shell is achieved. Thus, having several EPs (each specific to different hexamer-hexamer interfaces) would be a way to organize the composition of the shell by selectively recruiting the different shell proteins. If this model is correct, the EP interaction with the shell would both encapsulate enzymes and spatially organize the different shell proteins in the facets.
Combining this model with the proposed piggybacking of core metabolosome proteins leads to a model for a prometabolosome-first assembly of BMCs, in a manner analogous to β-carboxysome assembly.Citation22 For example, given the number of cargo proteins (up to 9 in the case of the PDU BMC, some of them forming a complex, e.g. PduCDE) and the interaction network established among them, it is likely that a pro-PDU metabolosome forms followed by encapsulation by the shell. Consistent with this model is a role for at least a subset of EPs in nucleating a prometabolosome core (e.g., the EP on PduE); heterologous expression of proteins containing an EP from metabolosomes leads to their aggregation, whereas the removal of the EP renders them soluble.Citation33,48,49 This aggregation could be due to the amphipathicity of the helix (hydrophobic interactions between several EPs strongly favored in aqueous environment). Accordingly, EPs may have 2 roles in the metabolosomes: clustering of the different enzymes (analogous to the role played by CcmM in the carboxysome), and the recruitment of the shell proteins to enclose the resulting prometabolosome in a shell ().
However there are anomalies yet to be reconciled with this model. For example, the first 42 amino acids of PduV (a protein of unknown function with weak GTPase activity and lacking an EP) target GFP to the PDU BMC, and thus presumably interacts with the shell.Citation47 PduV is associated to the exterior of the shell, potentially playing a role in the spatial localization of the BMC,Citation47 suggesting that a mechanism independent from EPs exists to associate proteins with the shell. Also, as noted above, full-length PduC (lacking an EP) can target GFP to empty, recombinant shells.Citation47 Thus, this subunit likely contains a yet-unrecognized determinant for binding to the shell.
Our results demonstrate that EPs can be located on either the N- or C-terminus of cargo proteins ( and ). An interesting question is: Does location matter? In other words, what would happen if a N-terminal EP for example is transferred to the C-terminus of the same protein? The presence of the poorly conserved linker, which “isolates” the EP from the core of the protein, suggests that an EP is an independent module that would be expected to successfully encapsulate the protein irrespective of its location. Likewise, it is possible that the strict positioning of the EP on one end of a metabolosome enzyme is required so that the remainder of the protein is unobstructed in forming an interaction with a second core protein, such as the case of the interaction between the N-terminal domains of CcmM and CcmN in carboxysome formation.Citation22,25 Such a hypothesis about the importance of the EP location would be interesting to investigate experimentally.
Understanding the interaction between EPs with shell proteins is important for the bioengineering of BMCs. The simplest strategy currently undertaken consists in fusing different EPs to enzymes of interest to create new encapsulated pathways. For example, a PDU BMC was engineered into an ethanol reactor when pyruvate decarboxylase and alcohol dehydrogenase where fused to the EPs of PduD and PduP.Citation29 Promising results for improving plant carbon fixation were obtained when Cyan Fluorescent Protein fused to the EP of CcmN compartmentalized into engineered protein shells resembling β-carboxysomes in tobacco chloroplasts.Citation50 This study shows that EP-shell interactions can be maintained across kingdoms, thus paving the way for BMC biotechnology applications in higher organisms.
However many enzymatic pathways of biotechnological interest require a large number of enzymes and supporting proteins. The limitation on the number of EPs per BMC can be overcome by considering protein domains as modular building blocks that could be engineered to associate via piggybacking.Citation5 For example, by designing fusion proteins composed of one EP and several protein domains that interact to form a metabolosome core.Citation51 These different modules may require separation by flexible linkers based on the ones that connect the EP to the functional domain in the native proteins. Likewise, if the EP indeed plays a role in organizing the composition of the shell, regulating the relative abundance of the different EPs in a system could permit control of the stoichiometry of the shell proteins, and therefore fine-tuning of the selective permeability of the shell. Finally, using the knowledge of EP characteristics, synthetic EPs with different features (e.g., more robust interaction with shell proteins or with other encapsulated enzymes) can be designed. By combining these different strategies, it will be possible to assemble and control new pathways in BMCs that are of direct relevance for biotechnologies, in both bacteria and plants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Download Zip (1.2 MB)Acknowledgments
The authors would like to acknowledge Seth Axen for his help in the bioinformatics analysis, Dr. Markus Sutter for his help with amino acid composition analysis, and the members of the Kerfeld Lab for collegial discussions.
Funding
Support for this work was provided by the US Department of Energy, Basic Energy Sciences, DE-FG02–91ER20021.
References
- Shively JM, Cannon GC, Heinhorst S, Fuerst JA, Bryant DA, Gantt E, et al. Intracellular Structures of Prokaryotes: Inclusions, Compartments and Assemblages. In: Encyclopedia of Microbiology. Schaechter M, ed. (Third Edition). Oxford: Academic Press; 2009:404-24.
- Cornejo E, Abreu N, Komeili A. Compartmentalization and organelle formation in bacteria. Curr Opin Cell Biol 2014; 26:132-8; PMID:24440431; http://dx.doi.org/10.1016/j.ceb.2013.12.007
- Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol 2010; 64:391-408; PMID:20825353; http://dx.doi.org/10.1146/annurev.micro.112408.134211
- Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev 2014; 78:438-68; PMID:25184561; http://dx.doi.org/10.1128/MMBR.00009-14
- Kerfeld CA, Erbilgin O. Bacterial microcompartments and the modular construction of microbial metabolism. Trends Microbiol 2015; 23:22-34; PMID:25455419; http://dx.doi.org/10.1016/j.tim.2014.10.003
- Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol 2006; 188:2865-74; PMID:16585748; http://dx.doi.org/10.1128/JB.188.8.2865-2874.2006
- Sampson EM, Bobik TA. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 2008; 190:2966-71; PMID:18296526; http://dx.doi.org/10.1128/JB.01925-07
- Huseby DL, Roth JR. Evidence that a metabolic microcompartment contains and recycles private cofactor pools. J Bacteriol 2013; 195:2864-79; PMID:23585538; http://dx.doi.org/10.1128/JB.02179-12
- Brinsmade SR, Paldon T, Escalante-Semerena JC. Minimal functions and physiological conditions required for growth of salmonella enterica on ethanolamine in the absence of the metabolosome. J Bacteriol 2005; 187:8039-46; PMID:16291677; http://dx.doi.org/10.1128/JB.187.23.8039-8046.2005
- Yeates TO, Tsai Y, Tanaka S, Sawaya MR, Kerfeld CA. Self-assembly in the carboxysome: a viral capsid-like protein shell in bacterial cells. Biochem Soc Trans 2007; 35:508-11; PMID:17511640; http://dx.doi.org/10.1042/BST0350508
- Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B(12)-dependent 1, 2-propanediol degradation. J Bacteriol 1999; 181:5967-75; PMID:10498708
- Kofoid E, Rappleye C, Stojiljkovic I, Roth J. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J Bacteriol 1999; 181:5317-29; PMID:10464203
- Lurz R, Mayer F, Gottschalk G. Electron microscopic study on the quaternary structure of the isolated particulate alcohol-acetaldehyde dehydrogenase complex and on its identity with the polygonal bodies of Clostridium kluyveri. Arch Microbiol 1979; 120:255-62; http://dx.doi.org/10.1007/BF00423073
- Seedorf H, Fricke WF, Veith B, Bruggemann H, Liesegang H, Strittmatter A, et al. The genome of Clostridium kluyveri, a strict anaerobe with unique metabolic features. Proc Natl Acad Sci U S A 2008; 105:2128-33; PMID:18218779; http://dx.doi.org/10.1073/pnas.0711093105
- Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, et al. Involvement of a bacterial microcompartment in the metabolism of fucose and rhamnose by Clostridium phytofermentans. PLoS One 2013; 8:e54337; PMID:23382892; http://dx.doi.org/10.1371/journal.pone.0054337
- Erbilgin O, McDonald KL, Kerfeld CA. Characterization of a planctomycetal organelle: a novel bacterial microcompartment for the aerobic degradation of plant saccharides. Appl Environ Microbiol 2014; 80:2193-205; PMID:24487526; http://dx.doi.org/10.1128/AEM.03887-13
- Axen SD, Erbilgin O, Kerfeld CA. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol 2014; 10:e1003898; PMID:25340524; http://dx.doi.org/10.1371/journal.pcbi.1003898
- Frank S, Lawrence AD, Prentice MB, Warren MJ. Bacterial microcompartments moving into a synthetic biological world. J Biotechnol 2013; 163:273-9; PMID:22982517; http://dx.doi.org/10.1016/j.jbiotec.2012.09.002
- Fischbach M, Voigt CA. Prokaryotic gene clusters: a rich toolbox for synthetic biology. Biotechnol J 2010; 5:1277-96; PMID:21154668; http://dx.doi.org/10.1002/biot.201000181
- Parsons JB, Dinesh SD, Deery E, Leech HK, Brindley AA, Heldt D, et al. Biochemical and structural insights into bacterial organelle form and biogenesis. J Biol Chem 2008; 283:14366-75; PMID:18332146; http://dx.doi.org/10.1074/jbc.M709214200
- Bonacci W, Teng PK, Afonso B, Niederholtmeyer H, Grob P, Silver PA, et al. Modularity of a carbon-fixing protein organelle. Proc Natl Acad Sci U S A 2012; 109:478-83; PMID:22184212; http://dx.doi.org/10.1073/pnas.1108557109
- Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: the carboxysome assembly pathway. Cell 2013; 155:1131-40; PMID:24267892; http://dx.doi.org/10.1016/j.cell.2013.10.044
- Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem 2007; 282:29323-35; PMID:17675289; http://dx.doi.org/10.1074/jbc.M703896200
- Cot SS, So AK, Espie GS. A multiprotein bicarbonate dehydration complex essential to carboxysome function in cyanobacteria. J Bacteriol 2008; 190:936-45; PMID:17993516; http://dx.doi.org/10.1128/JB.01283-07
- Kinney JN, Salmeen A, Cai F, Kerfeld CA. Elucidating essential role of conserved carboxysomal protein CcmN reveals common feature of bacterial microcompartment assembly. J Biol Chem 2012; 287:17729-36; PMID:22461622; http://dx.doi.org/10.1074/jbc.M112.355305
- Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, et al. Short N-terminal sequences package proteins into bacterial microcompartments. Proc Natl Acad Sci U S A 2010; 107:7509-14; PMID:20308536; http://dx.doi.org/10.1073/pnas.0913199107
- Fan C, Bobik TA. The N-terminal region of the medium subunit (PduD) packages adenosylcobalamin-dependent diol dehydratase (PduCDE) into the Pdu microcompartment. J Bacteriol 2011; 193:5623-8; PMID:21821773; http://dx.doi.org/10.1128/JB.05661-11
- Choudhary S, Quin MB, Sanders MA, Johnson ET, Schmidt-Dannert C. Engineered protein nano-compartments for targeted enzyme localization. PLoS One 2012; 7:e33342; PMID:22428024; http://dx.doi.org/10.1371/journal.pone.0033342
- Lawrence AD, Frank S, Newnham S, Lee MJ, Brown IR, Xue WF, et al. Solution structure of a bacterial microcompartment targeting peptide and its application in the construction of an ethanol bioreactor. ACS Synth Biol 2014; 3:454-65; PMID:24933391; http://dx.doi.org/10.1021/sb4001118
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792-7; PMID:15034147; http://dx.doi.org/10.1093/nar/gkh340
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009; 25:1189-91; PMID:19151095; http://dx.doi.org/10.1093/bioinformatics/btp033
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res 2008; 36:W197-201; PMID:18463136; http://dx.doi.org/10.1093/nar/gkn238
- Shibata N, Tamagaki H, Hieda N, Akita K, Komori H, Shomura Y, et al. Crystal structures of ethanolamine ammonia-lyase complexed with coenzyme B12 analogs and substrates. J Biol Chem 2010; 285:26484-93; PMID:20519496; http://dx.doi.org/10.1074/jbc.M110.125112
- Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol 1997; 179:6633-9; PMID:9352910
- Sauvageot N, Pichereau V, Louarme L, Hartke A, Auffray Y, Laplace JM. Purification, characterization and subunits identification of the diol dehydratase of Lactobacillus collinoides. Eur J Biochem 2002; 269:5731-7; PMID:12423373; http://dx.doi.org/10.1046/j.1432-1033.2002.03288.x
- Fan C, Cheng S, Sinha S, Bobik TA. Interactions between the termini of lumen enzymes and shell proteins mediate enzyme encapsulation into bacterial microcompartments. Proc Natl Acad Sci U S A 2012; 109:14995-5000; PMID:22927404; http://dx.doi.org/10.1073/pnas.1207516109
- Lassila JK, Bernstein SL, Kinney JN, Axen SD, Kerfeld CA. Assembly of robust bacterial microcompartment shells using building blocks from an organelle of unknown function. J Mol Biol 2014; 426:2217-28; PMID:24631000; http://dx.doi.org/10.1016/j.jmb.2014.02.025
- Cheng S, Fan C, Sinha S, Bobik TA. The PduQ enzyme is an alcohol dehydrogenase used to recycle NAD+ internally within the Pdu microcompartment of Salmonella enterica. PLoS One 2012; 7:e47144; PMID:23077559; http://dx.doi.org/10.1371/journal.pone.0047144
- Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature 1982; 299:371-4; PMID:7110359; http://dx.doi.org/10.1038/299371a0
- Fauchère J, Pliska V. Hydrophobic parameters pi of amino-acid side chains from the partitioning of N-acetyl-amino-acid amides. Eur J Med Chem 1983; 8:369-75
- http://www.kerfeldlab.org/amphihelix-scripts-and-files.html.
- Kim EY, Tullman-Ercek D. A rapid flow cytometry assay for the relative quantification of protein encapsulation into bacterial microcompartments. Biotechnol J 2014; 9:348-54; PMID:24323373; http://dx.doi.org/10.1002/biot.201300391
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, et al. Protein structures forming the shell of primitive bacterial organelles. Science 2005; 309:936-8; PMID:16081736; http://dx.doi.org/10.1126/science.1113397
- Kinney JN, Axen SD, Kerfeld CA. Comparative analysis of carboxysome shell proteins. Photosynth Res 2011; 109:21-32; PMID:21279737; http://dx.doi.org/10.1007/s11120-011-9624-6
- Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA. The structure of CcmP, a tandem bacterial microcompartment domain protein from the beta-carboxysome, forms a subcompartment within a microcompartment. J Biol Chem 2013; 288:16055-63; PMID:23572529; http://dx.doi.org/10.1074/jbc.M113.456897
- Pang A, Frank S, Brown I, Warren MJ, Pickersgill RW. Structural insights into higher order assembly and function of the bacterial microcompartment protein PduA. J Biol Chem 2014; 289:22377-84; PMID:24873823; http://dx.doi.org/10.1074/jbc.M114.569285
- Parsons JB, Frank F, Bhella D, Liang M, Prentice MB, Mulvihill DP, et al. Synthesis of empty bacterial microcompartments, directed organelle protein incorporation, and evidence of filament-associated organelle movement. Mol Cell 2010; 38:305-15; PMID:20417607; http://dx.doi.org/10.1016/j.molcel.2010.04.008
- Akita K, Hieda N, Baba N, Kawaguchi S, Sakamoto H, Nakanishi Y, et al. Purification and some properties of wild-type and N-terminal-truncated ethanolamine ammonia-lyase of Escherichia coli. J Biochem 2010; 147:83-93; PMID:19762342; http://dx.doi.org/10.1093/jb/mvp145
- Tobimatsu T, Kawata M, Toraya T. The N-terminal regions of beta and gamma subunits lower the solubility of adenosylcobalamin-dependent diol dehydratase. Biosci Biotechnol Biochem 2005; 69:455-62; PMID:15784971; http://dx.doi.org/10.1271/bbb.69.455
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MA, et al. β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 2014; 79:1-12; PMID:24810513; http://dx.doi.org/10.1111/tpj.12536
- Gonzalez-Esquer CR, Shubitowski T, Kerfeld CA. Streamlined construction of a cyanobacterial organelle via protein domains fusions. In revision.