ABSTRACT
The post-synaptic spines of neuronal dendrites are highly elaborate membrane protrusions. Their anatomy, stability and density are intimately linked to cognitive performance. The morphological transitions of spines are powered by coordinated polymerization of actin filaments against the plasma membrane, but how the membrane-associated polymerization is spatially and temporally regulated has remained ill defined. Here, we discuss our recent findings showing that dendritic spines can be initiated by direct membrane bending by the I-BAR protein MIM/Mtss1. This lipid phosphatidylinositol (PI(4,5)P2) signaling-activated membrane bending coordinated spatial actin assembly and promoted spine formation. From recent advances, we formulate a general model to discuss how spatially concentrated protein-lipid microdomains formed by multivalent interactions between lipids and actin/membrane regulatory proteins might launch cell protrusions.
The initiation of dendritic filopodia is promoted by membrane bending while their elongation requires actin polymerization
It is widely acknowledged that the actin cytoskeleton drives the morphological changes underlying dendritic spine morphogenesis.Citation1,2 However, actin polymerization is not the only mechanism shaping spines. We recently discovered that during the initiation of dendritic protrusions, actin polymerization is preceded by a separate membrane-bending step.Citation3 This direct membrane bending is likely to involve different I-BAR/IF-BAR domain proteins, which mediate the bending of the plasma membrane outwards.Citation3-7 I-BAR and IF-BAR proteins are multidomain scaffolding proteins that bind phosphoino-sitides (in particular PI(4,5)P2 and PI(3,4,5)P3) via a slightly curved lipid-binding interface.Citation4,5,8 Above a critical concentration, these proteins oligomerize, which results in the directional bending of the membrane into a tubular protrusion.Citation5,8,9 Remarkably, this outward bending of the dendritic membrane might be counteracted by canonical BAR-proteins such as ArhGAP44 that induce opposite membrane curvature.Citation10 In neurons, we found that MIM was recruited to the dendritic membrane by phosphoinositides, resulting in its oligomerization and the induction of small membrane buds that gave rise to filopodia ().Citation3 The growth of these buds into filopodia critically required actin filament assembly by the Arp2/3 complex.Citation3
Figure 1. The role of MIM during dendritic filopodia initiation. (A) MIM domain structure. (B) A schematic representation of an MIM monomer, which dimerizes via its I-BAR domain. The central region contains polyproline stretches and sites that are modified post-translationally. The C-terminus contains an ATP-actin binding WH2 domain. (C-E) The upper panel displays time frames of initiating dendritic filopodia and below is a schematic summary of each stage according to our findings.Citation3 (C) phosphoinositide-dependent recruitment of MIM results in its site-specific oligomerization and (D) outward membrane bending. (E) The subsequent elongation of the protrusion requires Arp2/3-mediated actin assembly. Scale bar 0.5 μm.
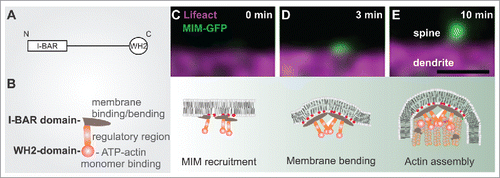
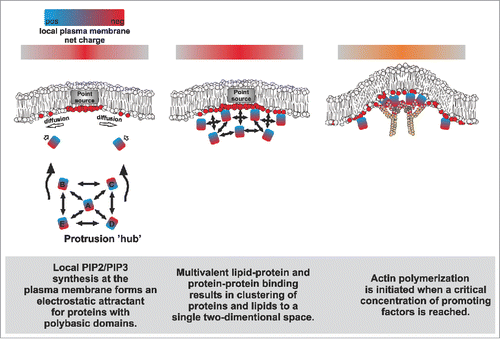
Figure 2. A simplified hypothetical model on how proteins that promote protrusion formation might be sustainably concentrated on a specific membrane domain via multivalent protein-protein and protein-membrane interactions.
In cultured neurons, the expression levels of both MIM and srGAP3 positively correlate with the density of membrane protrusions.Citation3,4 Because filopodia are pre-cursors for the dendritic spines that host excitatory synapses,Citation11-13 dysfunctional filopodia initiation is likely to cause aberrant circuits and neurological disturbances. Indeed, MIM knockout mice displayed dysfunctional synaptic transmission and both MIM and SrGAP3 knockout mice displayed abnormal behavior.Citation3,4 Taken together, a molecular assembly that couples specific lipids, membrane bending molecules and actin assembly factors cooperate during the launching of spines.
The upstream signaling of dendritic filopodia initiation
An important, unresolved aspect about dendritic filopodia is whether their initiation involves specific neuronal activity, signaling cues and/or some pre-existing structures. Filopodia initiation in axons of chick DRG neurons has been associated with preassembled actin patches.Citation14 In our experiments using the actin filament-binding peptide Lifeact, we did not observe clear preassembled actin patches at sites of future filopodia initiation (see e.g. ), which might reflect the structural differences of filamentous actin assemblies involved in filopodia formation between axons and dendrites. The axonal filopodia comprise of straight, fascin-bundled actin filaments,Citation15 whereas the dendritic filopodia display a mixture of branched and straight actin filaments that are not tightly bundled.Citation16 Moreover, since MIM was not found in axons,Citation3 it seems likely that the filopodia initiation mechanisms differ between axons and dendrites. It has also been shown that dendritic filopodia often emerge close to pre-assembled PSD95 clusters found along the dendritic shafts, and that filopodia start to accumulate PSD95 after 30-120 min of their appearance.Citation12 In agreement, we observed cases where new filopodia emerged proximal to a pre-existing shaft synapse (marked by SAP97), however, we were unable to observe clear accumulation of SAP97 to filopodia within our 40 minute movies ((3) and data not shown). Thus, our data suggests that shaft synapses might be somehow involved in stimulating the initiation of new filopodia, but they are not directly converted to spine synapses. Altogether, our data implies that MIM is among the first molecules accumulating to future spine initiation sites.
Little is known about the signaling events that promote spine formation. Glutamatergic activity positively correlates with dendritic filopodia density,Citation17 but whether this effect is more attributable to increased initiation frequency or lifetime of filopodia, is not known. Similarly, the signaling molecule brain-derived neurotrophic factor (BDNF) increases dendritic filopodia density.Citation18 Interestingly, PI(3,4,5,)P3 seems to be the down-stream signal for both glutamate and BDNF.Citation19,20 PI(3,4,5,)P3 was shown to accumulate to dendritic filopodia and to spinules (filopodia-like protrusions that arise from spine heads), and this accumulation was promoted by BDNF applicationCitation20 and neuronal activation,Citation19 respectively. While the generation of plasma membrane PI(4,5)P2 and PI(3,4,5)P3 is mediated by PI(4)P5 and PI3-kinases; fairly little is known of how the activities of these kinases are regulated.Citation21 Moreover, it remains to be determined whether the I-BAR/IF-BAR proteins respond to PI(4,5)P2 and PI(3,4,5)P3 in a similar manner. At least in vitro, these proteins do not seem to display a high degree of specificity to any particular phosphoinositide,Citation4,5,8 and since they all bind to the most abundant phosphoinositide at the plasma membrane PI(4,5)P2,Citation22 most studies have focused on this interaction.
Based recent advances, we articulate in the next chapter a hypothetical model to explain how transient phosphoinositide lipid signals can be sustained spatially to initiate protrusion assembly ().
Self-organization of the biochemical constituents of protrusion initiation into a single membrane domain
The phosphorylated phosphoinositides PI(4,5)P2/PI(3,4,5)P3 are intimately linked to both synaptic functionCitation23 and the formation of cell protrusionsCitation24 by promoting actin polymerization through selective binding and activation/inactivation of molecules associated with actin polymerization.Citation21,25 For example, the actin filament nucleation-promoting factor (NPF) N-WASP is sharply activated in response to a small (0.5–1.5 fold) increase in PI(4,5)P2 density,Citation26 whereas the actin filament disassembly factor ADF/cofilin is inactivated in a PI(4,5)P2-density dependent manner.Citation27 Importantly however, due to fast diffusion of lipids on membranes, it has remained unclear how spatially confined density differences of phosphoinositides are generated and sustained in cells in order to activate productive actin assembly for protrusion formation.
Simulations and experimental evidence suggest that signaling events that lead to a point source generation of negatively charged lipids (e.g., via the local activation of PIP-modifying enzymes) can promote the spatial recruitment of proteins with polybasic domains ().Citation28,29 Importantly, both I-BAR proteins and many actin-associated proteins harbor polybasic domains that bind PI(4,5)P2 in a multivalent manner, promoting the formation of PI(4,5)P2-nanoclusters.Citation3,9,26,27,30,31 Due to their self-association, I-BAR proteins oligomerize upon a critical concentration, leading to the formation of PI(4,5)P2-enriched lipid phases underneath the dynamic protein coat, and the simultaneous bending of the membrane outwards.Citation9,32 These negatively charged microdomains are likely to serve as powerful attractants for phosphoinositide-binding moieties, such as polybasic domains.Citation33 Such domains are found in many actin regulatory proteins, including signaling proteins such as GTPases,Citation31 forminsCitation30,34 and nucleation promoting factors.Citation26 These actin assembly factors interact with each other via additional domains such as SH3 and polyproline stretches, and also bring non-lipid-binding molecules to the site. As an example, WH2 domain-containing proteins (e.g., I-BAR proteins) might increase the local concentration of ‘polymerization competent’ ATP-actin monomers at sites of protrusions to fuel for actin polymerization.
Altogether, the lipid-protein and protein-protein binding events are expected to sustain the state of lipid demixing and increase the likelihood of other interactions to occur in the same area, promoting the assembly of a complex, high-avidity meshwork at a focused area of the membrane (). The organizing aspects of such an assembly might bear some resemblance to the nephrin-Nck-N-Wasp membrane-signaling system, which undergoes a phase separation above a critical concentration to promote local actin assembly.Citation35,36 Importantly, we were able to partially recapitulate this self-organization phenomenon with our in vitro reductionist approach combining PI(4,5)P2-containing giant vesicles, actin, Arp2/3, N-WASP and MIM I-BAR domain. This combination was sufficient to induce the clustering of these molecules together at a single focus on the membrane to activate actin assembly.Citation3
The duration of the initial lipid synthesis is likely to play a defining role in the final output (actin polymerization), as a critical concentration of phosphoinositides and molecules responding to them needs to be reached in order to initiate the protrusion. Here, molecules with a sharp activation threshold to phosphoinositide density (such as N-WASP) play a key role by keeping the system inactive unless a threshold stimulus is reached. Moreover, specificity could be enforced through selective recruitment of molecules that activate key components. For example, many guanine nucleotide exchange factors of GTPases contain PH domains that are highly sensitive to particular phosphoinositide-species, and thus they regulate the GTPase activity spatially in a lipid-specific manner.Citation37,38 In addition, local membrane curvature, in conjugation with phosphoinositides, has an important role in modulating the efficiency of actin polymerization,Citation39,40 altogether suggesting that actin assembly in initiating protrusion is fine-tuned according to the geometrical and chemical nature of the environment.
Collectively, we envision that the initiation of (dendritic) filopodia involves the formation of a complex multicomponent lipid-protein assembly that supports lipid demixing to ensure the polymerization of actin filaments in a spatially confined area.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Hotulainen P, Hoogenraad CC. Actin in dendritic spines: connecting dynamics to function. J Cell Biol 2010; 189:619-29; PMID:20457765; http://dx.doi.org/10.1083/jcb.201003008
- Svitkina T, Lin WH, Webb DJ, Yasuda R, Wayman GA, Van Aelst L, Soderling SH. Regulation of the postsynaptic cytoskeleton: roles in development, plasticity, and disorders. J Neurosci 2010; 30:14937-42; PMID:21068295; http://dx.doi.org/10.1523/JNEUROSCI.4276-10.2010
- Saarikangas J, Kourdougli N, Senju Y, Chazal G, Segerstråle M, Minkeviciene R, Kuurne J, Mattila PK, Garrett L, Hölter SM, et al. MIM-Induced Membrane Bending Promotes Dendritic Spine Initiation. Dev Cell 2015; 33:644-59; PMID:26051541; http://dx.doi.org/10.1016/j.devcel.2015.04.014
- Carlson BR, Lloyd KE, Kruszewski A, Kim IH, Rodriguiz RM, Heindel C, Faytell M, Dudek SM, Wetsel WC, Soderling SH. WRP/srGAP3 facilitates the initiation of spine development by an inverse F-BAR domain, and its loss impairs long-term memory. J Neurosci 2011; 31:2447-60; PMID:21325512; http://dx.doi.org/10.1523/JNEUROSCI.4433-10.2011
- Coutinho-Budd J, Ghukasyan V, Zylka MJ, Polleux F. The F-BAR domains from srGAP1, srGAP2 and srGAP3 regulate membrane deformation differently. J Cell Sci 2012; 125:3390-401; PMID:22467852; http://dx.doi.org/10.1242/jcs.098962
- Kessels MM, Qualmann B. Different functional modes of BAR domain proteins in formation and plasticity of mammalian postsynapses. J Cell Sci 2015; 128:3177-85; PMID:26285709; http://dx.doi.org/10.1242/jcs.174193
- Carlson B, Soderling SH. Mechanisms of cellular protrusions branch out. Dev Cell 2009; 17:307-9; PMID:19758555; http://dx.doi.org/10.1016/j.devcel.2009.08.015
- Mattila PK, Pykäläinen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol 2007; 176:953-64; PMID:17371834; http://dx.doi.org/10.1083/jcb.200609176
- Saarikangas J, Zhao H, Pykäläinen A, Laurinmäki P, Mattila PK, Kinnunen PK, Butcher SJ, Lappalainen P. Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol 2009; 19:95-107; PMID:19150238; http://dx.doi.org/10.1016/j.cub.2008.12.029
- Galic M, Tsai FC, Collins SR, Matis M, Bandara S, Meyer T. Dynamic recruitment of the curvature-sensitive protein ArhGAP44 to nanoscale membrane deformations limits exploratory filopodia initiation in neurons. eLife 2014; 3:e03116; PMID:25498153; http://dx.doi.org/10.7554/eLife.03116
- Lohmann C, Bonhoeffer T. A role for local calcium signaling in rapid synaptic partner selection by dendritic filopodia. Neuron 2008; 59:253-60; PMID:18667153; http://dx.doi.org/10.1016/j.neuron.2008.05.025
- Marrs GS, Green SH, Dailey ME. Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat Neurosci 2001; 4:1006-13; PMID:11574832; http://dx.doi.org/10.1038/nn717
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 1996; 17:91-102; PMID:8755481; http://dx.doi.org/10.1016/S0896-6273(00)80283-4
- Ketschek A, Gallo G. Nerve growth factor induces axonal filopodia through localized microdomains of phosphoinositide 3-kinase activity that drive the formation of cytoskeletal precursors to filopodia. J Neurosci 2010; 30:12185-97; PMID:20826681; http://dx.doi.org/10.1523/JNEUROSCI.1740-10.2010
- Menna E, Disanza A, Cagnoli C, Schenk U, Gelsomino G, Frittoli E, Hertzog M, Offenhauser N, Sawallisch C, Kreienkamp HJ, et al. Eps8 regulates axonal filopodia in hippocampal neurons in response to brain-derived neurotrophic factor (BDNF). PLoS Biol 2009; 7:e1000138; PMID:19564905; http://dx.doi.org/10.1371/journal.pbio.1000138
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell 2010; 21:165-76; PMID:19889835; http://dx.doi.org/10.1091/mbc.E09-07-0596
- Portera-Cailliau C, Pan DT, Yuste R. Activity-regulated dynamic behavior of early dendritic protrusions: evidence for different types of dendritic filopodia. J Neurosci 2003; 23:7129-42; PMID:12904473.
- Ji Y, Pang PT, Feng L, Lu B. Cyclic AMP controls BDNF-induced TrkB phosphorylation and dendritic spine formation in mature hippocampal neurons. Nat Neurosci 2005; 8:164-72; PMID:15665879; http://dx.doi.org/10.1038/nn1381
- Ueda Y, Hayashi Y. PIP(3) regulates spinule formation in dendritic spines during structural long-term potentiation. J Neurosci 2013; 33:11040-7; PMID:23825409; http://dx.doi.org/10.1523/JNEUROSCI.3122-12.2013
- Luikart BW, Zhang W, Wayman GA, Kwon CH, Westbrook GL, Parada LF. Neurotrophin-dependent dendritic filopodial motility: a convergence on PI3K signaling. J Neurosci 2008; 28:7006-12; PMID:18596174; http://dx.doi.org/10.1523/JNEUROSCI.0195-08.2008
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 2013; 93:1019-137; PMID:23899561; http://dx.doi.org/10.1152/physrev.00-028.2012
- McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 2005; 438:605-11; PMID:16319880; http://dx.doi.org/10.1038/nature04398
- Dotti CG, Esteban JA, Ledesma MD. Lipid dynamics at dendritic spines. Front Neuroanatomy 2014; 8:76; PMID:25152717; http://dx.doi.org/10.3389/fnana.2014.00076
- Lee K, Gallop JL, Rambani K, Kirschner MW. Self-assembly of filopodia-like structures on supported lipid bilayers. Science 2010; 329:1341-5; PMID:20829485; http://dx.doi.org/10.1126/science.1191710
- Saarikangas J, Zhao H, Lappalainen P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol Rev 2010; 90:259-89; PMID:20086078; http://dx.doi.org/10.1152/physrev.000-36.2009
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA. A polybasic motif allows N-WASP to act as a sensor of PIP(2) density. Mol Cell 2005; 17:181-91; PMID:15664188; http://dx.doi.org/10.1016/j.molcel.2004.11.054
- Zhao H, Hakala M, Lappalainen P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys J 2010; 98:2327-36; PMID:20483342; http://dx.doi.org/10.1016/j.bpj.2010.01.046
- Kiselev VY, Marenduzzo D, Goryachev AB. Lateral dynamics of proteins with polybasic domain on anionic membranes: a dynamic Monte-Carlo study. Biophys J 2011; 100:1261-70; PMID:21354399; http://dx.doi.org/10.1016/j.bpj.2011.01.025
- Brown DA. PIP2Clustering: From model membranes to cells. Chem Phys Lipids 2015; 192:33-40.
- Gorelik R, Yang C, Kameswaran V, Dominguez R, Svitkina T. Mechanisms of plasma membrane targeting of formin mDia2 through its amino terminal domains. Mol Biol Cell 2011; 22:189-201; PMID:21119010; http://dx.doi.org/10.1091/mbc.E10-03-0256
- Heo WD, Inoue T, Park WS, Kim ML, Park BO, Wandless TJ, Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science 2006; 314:1458-61; PMID:17095657; http://dx.doi.org/10.1126/science.1134389
- Zhao H, Michelot A, Koskela EV, Tkach V, Stamou D, Drubin DG, Lappalainen P. Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep 2013; 4:1213-23; PMID:24055060; http://dx.doi.org/10.1016/j.celrep.2013.08.024
- Picas L, Viaud J, Schauer K, Vanni S, Hnia K, Fraisier V, Roux A, Bassereau P, Gaits-Iacovoni F, Payrastre B, et al. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat Commun 2014; 5:5647; PMID:25487648; http://dx.doi.org/10.1038/ncomms6647
- Ramalingam N, Franke C, Jaschinski E, Winterhoff M, Lu Y, Brühmann S, Junemann A, Meier H, Noegel AA, Weber I, et al. A resilient formin-derived cortical actin meshwork in the rear drives actomyosin-based motility in 2D confinement. Nat Commun 2015; 6:8496; PMID:26415699; http://dx.doi.org/10.1038/ncomms9496
- Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife 2014; 3:e04123; PMID:25321392; http://dx.doi.org/10.7554/eLife.04123
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012; 483:336-40; PMID:22398450; http://dx.doi.org/10.1038/nature10879
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nature reviews. Mol Cell Biol 2008; 9:99-111; PMID:18216767.
- Welch HC. Regulation and function of P-Rex family Rac-GEFs. Small GTPases 2015; 6:49-70; PMID:25961466; http://dx.doi.org/10.4161/21541248.2014.973770
- Takano K, Toyooka K, Suetsugu S. EFC/F-BAR proteins and the N-WASP-WIP complex induce membrane curvature-dependent actin polymerization. EMBO J 2008; 27:2817-28; PMID:18923421; http://dx.doi.org/10.1038/emboj.2008.216
- Gallop JL, Walrant A, Cantley LC, Kirschner MW. Phosphoinositides and membrane curvature switch the mode of actin polymerization via selective recruitment of toca-1 and Snx9. Proc Natl Acad Sci U S A 2013; 110:7193-8; PMID:23589871; http://dx.doi.org/10.1073/pnas.1305-286110
