ABSTRACT
Mammalian Smaug1/Samd4a is an mRNA regulator involved in synapse plasticity and additional non-neuronal functions. Here we analyzed the expression of Smaug1/Samd4a variants and Smaug2/Samd4b in primary hippocampal neurons and non-neuronal cell lines. We found that multiple Smaug proteins are present in several mammalian cell lines, including a canonical full length Smaug1, a Smaug1 variant that lacks the third exon, termed ΔEIII, and Smaug2, the product of a highly homologous gene. These three major isoforms are expressed differentially along neuron development and form cytosolic bodies when transfected in cell lines. By using luciferase reporters, we found that the ΔEIII isoform, which lacks 10 amino acids in the sterile α motif involved in RNA binding, shows a RNA-binding capacity and repressor activity comparable to that of the full length Smaug1. These observations are an important groundwork for molecular studies of the Smaug post-transcriptional pathway, which is relevant to neuron development, mitochondrial function and muscle physiology in health and disease.
Introduction
In several cellular contexts, translational repression is linked to the formation of mRNA-silencing bodies, which are supramolecular aggregates that store mRNAs excluded from the translational pool together with repressor proteins. By definition, mRNA-silencing bodies can dissolve in response to specific stimuli, allowing the translation of specific transcripts. Stress granules (SGs) and Processing Bodies (PBs) are the best described mRNA-silencing bodies (reviewed in ref. Citation1). SGs form transiently upon cellular stress. PBs instead, are ubiquitous foci that contain molecules involved in mRNA repression and decay.
We have previously described a specific type of neuronal mRNA-silencing body called S-foci, as they contain the repressor protein Smaug1/Samd4a.Citation2 More recently, we reported that the 5′-3′ exoribonuclease XRN1 forms discrete aggregates, termed Synaptic XRN1-bodies (SX-bodies) apparently also involved in translational regulation at the post-synapse.Citation3 Both S-foci and SX-bodies are different from PBs or SGs, respond differently to a number of synaptic stimuli and belong to a growing family of neuronal bodies that contributes to the fine tuning of protein synthesis at the post-synapse, which we termed SyAS foci.Citation2-4 Both XRN1 and Smaug are expressed also in non-neuronal cell types and here we used several mammalian cell lines to study the expression and localization of XRN1 and Smaug molecules in cytoplasmic bodies. We found that at least 3 Smaug1/2 isoforms and splicing variants are present in mammalian cell lines. In addition, these 3 major Smaug molecules aggregate in cytosolic bodies. A Smaug1 splicing variant lacking the exon III binds RNA and displays a repressor activity comparable to that of the full length canonical Smaug1. These observations help to establish the molecular groundwork for future research on the Smaug regulatory pathway, which affect a wide range of key functions including insect early development, mammalian synapse plasticity and mitochondrial biogenesis.Citation2,5-8
Results and discussion
Two highly homologous genes encoding Smaug1/Samd4a and Smaug2/Samd4b are present in vertebrates,Citation9,10 opening the question on target specificity, redundancy and regulation diversity. In addition, Smaug1 has an alternative splicing isoform that lacks the third exon (amino acids 239 to 327, NM_001161576),Citation11,12 which we called ΔEIII. The excision of this exon removes the first 10 amino acids of a conserved sterile α motif (SAM), which is involved in the binding to a specific RNA motif termed Smaug Recognition Element (SRE)Citation9,10,13 ().
Figure 1. Smaug1 and Smaug2 variants in neurons and cell lines. (A) Schematic representation of full length (FL, NM_015589) and ΔEIII (NM_001161576) Smaug1. The ΔEIII lacks 10 amino acids in the SAM domain, which binds RNA. (B) RT-PCR of Smaug1 splicing variants. The presence of the full length and ΔEIII mRNAs was analyzed in neurons at 4, 8, 12 and 14 DIV by RT-PCR using isoform-specific primers (Material and Methods). Arrows indicate the position of each primer. C-, negative control (RNA sample with no reverse transcription). (C) Quantitative RT-PCR for total Smaug1 isoforms or Smaug2 in hippocampal neurons cultured during 8 or 14 d in vitro (DIV) was performed using the oligonucleotides indicated in Material and Methods. Results are expressed relative to β-actin mRNA levels. Both Smaug1 and Smaug2 transcripts accumulate during synaptogenesis in vitro. Error bars, s.e.m. *** p<0.001, Student's t-Test.
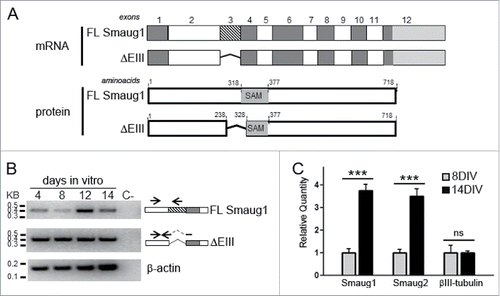
First, we investigate by RT-PCR the presence of transcripts encoding the full length Smaug1 (FL Smaug1) and the ΔEIII variant during hippocampal neuron maturation in vitro at day 4, 8, 12 and 14 after plating (). We found that the transcript encoding the ΔEIII variant was present all along neuron development in vitro at about constant levels, whereas the transcript encoding the full length molecule greatly increased at 12 DIV, when synaptogenesis is massively triggered. Relevantly, we have shown before that Smaug1 proteins are not detected in hippocampal neurons before synaptogenesis and Smaug1 protein levels increase at day 10 along with synapse markers.Citation2 The expression of Drosophila Smaug is regulated at the translational level, with a strong repression of dSmaug mRNA during oogenesis,Citation14 and our observations suggest a similar repression of Smaug1 ΔEIII transcripts before synaptogenesis that remains to be investigated. A minor expression of Smaug1 protein variants at early times during hippocampal development cannot be ruled out and its putative relevance remains unknown.
Next, we compared the expression of Smaug1 (full length and ΔEIII together) and Smaug2 by quantitative PCR in cultured neurons before (8 days) and after synaptogenesis (14 days), and we found that the expression of both Smaug1 and Smaug2 transcripts increased 4 times at this developmental time (). This is in accordance with previous work reporting that the presence of Smaug proteins increases dramatically during synaptogenesis.Citation2 Smaug1 knockdown leads to the accumulation of immature synapsisCitation2 and the presence of Smaug2 transcripts in developing hippocampal neurons suggests an additional role for Smaug2 in synapse formation or neuron maturation, which remains to be investigated. It was shown recently that Smaug2 protein, but not Smaug1, is expressed during embryonic cortical neurogenesis. Smaug2 forms a translational repression complex that helps precursor maintenance.Citation15 As in hippocampal neurons, Smaug1 is expressed later during the development of cortical neurons, and altogether these observations suggest that Smaug1 and 2 are important at different times during neuron differentiation and maturation.
In addition to its role in neuronal precursors and in mRNA regulation at the post-synapse, recent work indicates that Smaug functions beyond the CNS. Smaug molecules are involved in translation homeostasis in Myotonic Dystrophy Type 1 (DM1) models and affect several transcripts involved in mitochondrial function in both Drosophila and mammals.Citation5-7 We analyzed the expression of Smaug1/2 isoforms and variants in several mammalian cell lines by RT-PCR and found that the 3 major Smaug1/2 transcripts expressed in primary hippocampal neurons are also present in cell lines derived from bone (U2OS), embryonic renal (HEK293T) and nervous tissue (SH-SY5Y) (). Quantitative analysis indicates that both the full length Smaug1 isoform and the ΔEIII mRNAs are expressed at comparable levels in HEK293T and U2OS cells exponentially growing. In turn, Smaug2 mRNA is expressed at higher levels than Smaug1 mRNA, a feature more pronounced in HEK293T than in U2OS ().
Figure 2. (A) The expression of Smaug1 variants and of Smaug2 was analyzed in the following cell lines: U2OS (U2), COS7 (C7), HEK293T (HK), SH-SY5Y (SH). A plasmid containing the complete Smaug1 or Smaug2 cDNA sequence was used in each case as a positive control (C+); C-, negative control (RNA sample with no reverse transcription). β-actin was analyzed for comparison. All cell lines express both Smaug1 isoforms and Smaug2. (B) Quantification of full length, ΔEIII, total Smaug1 or Smaug2 mRNAs in U2OS and HEK293T cells. Results are expressed as absolute cDNA in pg/µl, calculated using a standard curve obtained with plasmids containing the respective cDNAs as templates. Error bars, s.e.m. Statistical significance from Smaug2 relative to full length, ΔEIII and Smaug1 according to one-way ANOVA, Bonferroni post-test was *** p<0.0001. (C, D) Full length and ΔEIII Smaug1 bind and repress SRE-luciferase containing reporters. (C) The indicated Smaug1 constructs fused to a SBP-tag were co-transfected with firefly luciferase reporters carrying or not SREs (Materials and Methods). A plasmid encoding SBP-MBP was used as a negative control. The ratio in arbitrary units of luciferase mRNA in the pull down relative to that in total extracts is plotted. Western blot of pull down (PD) fractions or inputs (IN) and PD fractions are shown. The two Smaug1 variants display comparable binding activity. Error bars, s.e.m. Statistical significance according to one-way ANOVA, Bonferroni post-test was as follows: * p<0.05, ns, non-significant. (D), The indicated Smaug1 constructs fused to ECFP or V5 were co-transfected in BHK or U2OS cells together with Firefly luciferase reporters carrying wild type or mutant SRE motifs and a Renilla luciferase cDNA as a reference. Plasmids encoding V5 or ECFP were used as a control (Materials and Methods). The ratio of normalized luciferase wt SRE/mut SRE was plotted. Four representative experiments out of 6 are depicted. The two Smaug1 variants display comparable repressor activity whereas a construct lacking the SAM (ΔSAM) shows no effect. Error bars, s.e.m. Statistical significance relative to vector or ΔSAM according to one-way ANOVA, Bonferroni post-test was as follows: *** p<0.0001, ** p<0.001 and * p<0.05, respectively. No significant differences were seen between FL Smaug1 and ΔEIII and between the vector and ΔSAM.
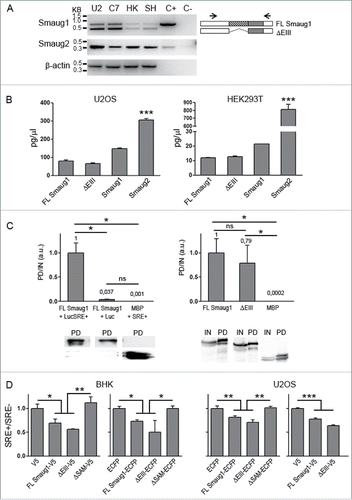
Like the Drosophila counterpart, mammalian Smaug1 regulates SRE-containing messengers, as is the case of CamKII mRNA and of chimeric reporters carrying SREs present in the Drosophila nanos mRNA.Citation2,10 We investigated the RNA binding ability of ΔEIII, which lacks 10 amino acids in the SAM domain involved in the binding to SREs, by performing RNA-pull down assays with luciferase reporters and constructs fused to SBP (streptavidin binding protein). First, we assessed the binding of full length Smaug1 to transcripts carrying or not 3 SREs (described in ref. 10). As expected, we found that the reporter carrying binding sites is significantly recovered in the Smaug1 pull down whereas the recovery of a luciferase reporter without binding sites was negligible (, left panel). We simultaneously tested the binding of the SRE-containing transcript to maltose binding protein (MBP), which does not bind RNA, and found that it was not significantly recovered in the pull down (, left panel). With this experimental design we assessed the binding of the SRE-containing reporter to ΔEIII fused to SBP and found that ΔEIII binds RNA with a strength similar to that of the full length protein (, right panel). Thus, the first 10 amino acids of the SAM domain are dispensable for binding to SREs reporters. Differences in the binding to endogenous transcripts remain to be investigated.
Binding does not directly imply repression and we investigated the repressor activity of ΔEIII. We assessed the repression by the full length molecule and by ΔEIII fused to ECFP or V5 in BHK or U2OS cells using luciferase-based reporters carrying 3 wild type or mutant SREs. As expected, in all cases full length Smaug1 leads to a repression of about 25–40 %, similarly to that reported in other mammalian cell types Citation10 (). Also in Drosophila embryos the overexpression of Smaug leads to a very modest repression of SRE-containing transcripts.Citation16 The effect of the ΔEIII was simultaneously analyzed and we found that 2 different ΔEIII constructs downregulated the expression of SRE-containing luciferase reporters with a strength similar to that of the full length protein () both in BHK and U2OS cells. In contrast, 2 different constructs lacking the SAM domain showed not repression (). Different members of a given family of RNA-binding proteins may compete for their target mRNAs eliciting disparate effects Citation17 and differences in the deadenylation or decay triggered by the multiple mammalian Smaug isoforms cannot be discarded. In addition, the selectivity in the binding to endogenous transcripts remains to be investigated. For example, The CamKII mRNA is positively and negatively regulated by Smaug1 and additional factorsCitation2,18,19 and mammalian nanos1 mRNA is regulated by Smaug2.Citation15 Whether the multiple Smaug 1/2 isoforms differentially affect the fate of their targets is an open question.
Finally, we investigated whether these previously uncharacterized mammalian Smaug1/2 variants form cytosolic bodies, as described before for the Drosophila molecule and for the full length mammalian Smaug1.Citation2,5,10,20 We found that when transfected in cell lines, the 3 Smaug1/2 isoforms aggregate in discrete foci (). The fraction of cells showing cytosolic bodies was similar (59% ± 8 for Smaug1-ECFP; 58% ± 11 for ΔEIII-ECFP; and 49% ± 11 for Smaug2-Myc). The number and size of the bodies was comparable in all cases and moreover, the 3 molecules colocalized in the same bodies when co-expressed in pairs: 98.8% ± 1.3 of Smaug1-ECFP foci contained Smaug2-Myc and 98% ± 1 of Smaug2-Myc contained Smaug1-ECFP (N of foci: 293); 98.8% ± 1.3 of Smaug1-EYFP foci contained Smaug2-Myc and 98% ± of Smaug2-Myc contained Smaug1-EYFP (N of foci: 230); 97.8% ± 3.3 of Smaug1-EYFP foci contained ΔEIII-ECFP and 98.8% ± 1.3 of ΔEIII-ECFP foci contained Smaug1-EYFP (N of foci: 312); 97.5% ± 3 of Smaug1-V5 foci contained ΔEIII-ECFP and 99.5% ± 2 of ΔEIII-ECFP foci contained Smaug1-V5 (N of foci: 187); finally 97.4% ± 4 of Smaug2-Myc foci contained ΔEIII-ECFP and 98.8% ± 1.3 of ΔEIII-ECFP foci contained Smaug2-Myc (N of foci: 421) ( and unpublished data). We also calculated the Pearson's correlation coefficient (PCC) Citation21 in at least 20 regions of interest (ROIs) randomly taken for each pair of constructs. On average, we found high correlation coefficients (0.71 ± 0.06 for Smaug1-ECFP and Smaug2-Myc; 0.83 ± 0.05 for Smaug1-EYFP and Smaug2-Myc; 0.82 ± 0.08 for ΔEIII-ECFP and Smaug1-EYFP; 0.79 ± 0.11 for ΔEIII-ECFP and Smaug1-V5; and 0.68 ± 0.06 for ΔEIII-ECFP and Smaug2-Myc). It was recently reported that endogenous Smaug molecules form cytosolic bodies in HEK293Tcells Citation5 and cortical neuron precursors Citation15 and our results suggest similar aggregation mechanisms for all known Smaug variants. The possibility of multiple Smaug bodies with different stoichiometry and that may respond to different cellular cues to trigger specific responses remains to be investigated.
Figure 3. Smaug1 and Smaug2 variants form cytosolic bodies. (A) U2OS cells were transfected with constructs for Smaug1, ΔEIII and Smaug2 fused to the indicated tags. The percentage of transfected cells with foci is shown in each case. (B) Smaug1, ΔEIII and Smaug2 constructs were co-transfected in pairs as indicated. The ΔEIII bodies colocalize almost completely with the bodies formed by the full length isoform. Smaug1 and Smaug2, as well as Smaug2 and ΔEIII also colocalize completely (for quantification see the main text). (C) The indicated constructs were co-transfected and endogenous XRN1 was stained with a specific antibody. XRN1 is present in foci (likely PBs), which are largely excluded from the Smaug1/2 bodies. Percentage of Smaug1/2 foci containing endogenous XRN1 is indicated for each construct.
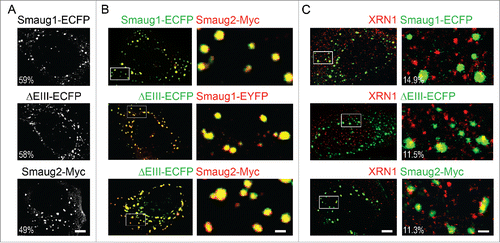
In neurons, Smaug1 and XRN1 form mRNA silencing bodies located at post-synapses and termed S-foci and SX-bodies, respectively. These bodies respond to a number of stimuli with distinct patterns.Citation3 XRN1 and Smaug also co-exist in cell lines, where XRN1 is mostly present in PBs. We investigated whether XRN1 and Smaug1 form different bodies in U2OS. We found that most Smaug1-ECFP, ΔEIII-ECFP and Smaug2-Myc bodies exclude endogenous XRN1, as previously described for neurons.Citation3 We found that 14.9% ± 8.8 of Smaug1-ECFP foci contain XRN1; 11.5% ± 8 of ΔEIII-ECFP foci contain XRN1; and 11.3% ± 10.7 of Smaug2-Myc foci contain XRN1.This relatively low colocalization in bodies is consistent with the PCCs, which are 0.23 ± 0.08 for Smaug1-ECFP and XRN1; 0.2 ± 0.09 for ΔEIII-ECFP and XRN1; and 0.25 ± 0.09 for Smaug2-Myc and XRN1.
Collectively, these observations indicate that the repertoire of Smaug repressors present in mammalian cells include at least 3 molecules, all them with comparable ability to aggregate. All mammalian Smaug isoforms and splicing variants described up to date are present in neurons and non-neuronal cell lines. Functional differences are expected to be discovered between the different Smaug proteins, related to their redundancy or specificity in the repression of different transcripts, and to their responses upon synapse activation or specific cellular stimuli. These observations help to stablish the molecular groundwork for future research on Smaug-mediated post-transcriptional control in mammals, which we anticipate will be highly relevant to multiple cellular process, as Smaug regulates thousands of different messengers and is linked to synapse plasticity, muscle physiology and mitochondrial function in health and disease.Citation5-7,14,16,22,23
Materials and methods
Hippocampal cultures were prepared as described previously.Citation2,3 All cell lines were obtained from the American Tissue Culture Collection and grown and maintained as indicated. Cell lines were transfected with Jet Prime (Polyplus Transfection) according to manufacturer's instructions. Most used constructs were previously described.Citation2,10
pSmaug1-ΔEIII-ECFP was constructed by S.O.E. PCR using pSmaug1-ECFP as a template and the following primers: forward 5′-GCACAAGTACAAATGTTCCAGCCTG-3′, reverse 5′-AGGCTGGAACATTTGTACTTGTGCTG-3′. PCR product was inserted using HindIII/BamHI in pECFP-N1 plasmid.
SBP (streptavidin binding protein) constructs were obtained by subcloning Smaug1 or ΔEIII cDNA sequence from EGFP containing plasmids into a pT7-V5-SBP vector.
For RT-PCR and qRT-PCR, total RNA was isolated by using TRIzol reagent (Invitrogen). First-strand cDNA was synthesized from 1 µg of total RNA using a random hexamer and expand reverse transcriptase (Roche Diagnostics). The cDNA was used as a template for PCR or quantitative PCR. Quantitative PCR was performed using Syber Green reagent (Applied Biosystem) and the amount of total cDNA was analyzed using specific primers for rat β-actin.
Primers used in PCR were: rat full lenght Smaug1: forward 5′-CGACCATAACCAGCACATTG3′-, reverse 5′- GTTTCAAAGGGCTGTGGTGT-3′ ratSAM_Rv GAATGCTGATGACGATTTTGTG ; rat ΔEIII Smaug1: forward 5′-CGACCATAACCAGCACATTG-3′, reverse 5′-ACCAAGCTGGAACATTCGTA-3′ ; human Smaug1 forward 5′-TCGACCAGCTTTGGTGGCCA-3′ reverse 5′-GAATACTGATGACAATTTTGTG-3; human Smaug2: forward 5′-ACCCCTACTGCCAAGGATG -3′, reverse 5′-AGCCTCATGAGTCAGGCACT-3′; human β-actin: forward 5′-CTCTTCCAGCCTTCCTTCCT-3′, reverse 5′-AAAGCCATGCCAATCTCATC-3′.
Primers used for quantitative PCR were: rat Smaug1: forward 5′-ATTCTGGGATTTGCATCAGC-3′; reverse, 5′-GTTTCAAAGGGCTGTGGTGT-3′ ; rat Smaug2: forward 5′-CCCTCTGATAGCAGTGAGCC-3′, reverse 5′-GAATGTCCGGAGGAGTTTCA-3′ ; rat βIII-tubulin: forward 5′-CCTGGAACCATGGACAGCGTTCG-3′, reverse 5′-CGTTGTAGGGCTCTACCACGGTG-3′; rat β-actin: forward 5′-TGTCACCAACTGGGACGATA-3′, forward 5′-GGGGTGTTGAAGGTCTCAAA-3′; human Smaug1: forward 5′-ACCAGTGGATTCGTCAGCTC-3′; reverse 5′-GTTGGCAAACCACAGGTTCT-3′, full length Smaug1: forward 5′-ACAAAAGCATGGGGTGTGAG-3′; reverse 5′-CCTGGCCTGAGAGAATTGTA-3′, ΔEIII Smaug1: forward 5′-ACAAAAGCATGGGGTGTGAG-3′, reverse 5′-GCCAGGCTGGAACATTTGTA -3′; human Smaug2: forward 5′-ACACGGGTGATGGGCAAAGT-3′, reverse 5′-AGCCTCATGAGTCAGGCACT -3′.
RNA-binding was analyzed by RNA-pull down assay. Briefly, HEK293T cells were co-transfected with SBP-tagged constructs and LUC-SRE+ plasmids. Cells were resuspended in lysis buffer (150 mM NaCl, 25 mM Tris pH7.4, 5 mM EDTA, 0.5 mM DTT, 1% NP40 and 5% glycerol) using mild sonication and RNPs were pulled down with streptavidin sepharose beads (GE Health Care Lifesciences). The total RNA from inputs and pull down fractions was isolated and reverse transcribed. The cDNA was treated with DNase I and subsequently used for real-time PCR analysis of luciferase transcript using specific primers forward 5′-CCAGGGATTTCAGTCGATGT-3′, reverse 5′-AATCTCACGCAGGCAGTTCT-3′.
Luciferase assays were performed as previously described.Citation10
Immunofluorescence of cultured cells was performed after fixation, permeabilization, and blocking as usual.Citation2,3,24,25 Primary rabbit anti-XRN1 (Bethyl Laboratories) and rabbit anti-Myc (Cell Signaling), were diluted 1:100 and 1:200 respectively.
Pearson's correlation coefficients were calculated in random ROIs, using the Coloc2 plug-in of the Fiji (ImageJ) Software.
Abbreviations
| ΔEIII | = | Delta exon III |
| CaMKII | = | Ca2+/CaM dependent protein kinase II |
| DM1 | = | Myotonic Dystrophy Type 1 |
| PB | = | processing body |
| SAM | = | sterile alpha motif |
| SG | = | stress granule |
| S-foci | = | Smaug1/Samd4a foci |
| SRE | = | Smaug recognition element |
| SX-bodies | = | Synaptic XRN1 bodies |
| SyAS | = | Synaptic activity regulated mRNA-silencing foci |
| XRN1 | = | 5′-3′ exoribonuclease 1 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Boccaccio's lab members for their constant support and fruitful discussions. We are grateful to Gianluca Amadei from Dr.Freda Miller Laboratory, University of Toronto, Canada, for providing the pSmaug2-Myc.
Funding
This work was supported by the following grants: PICT 2013–3280, PICT 2011–1301 and PICT 2010–1850 to GLB, PICT 2010–2339, PICT 2012–2493 and PIP 205-2011-2013 to MGT. AJFA, MLP and GLB are investigators from the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Argentina.
References
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal 2011; 23:324-34; PMID:20813183; http://dx.doi.org/10.1016/j.cellsig.2010.08.011
- Baez MV, Luchelli L, Maschi D, Habif M, Pascual M, Thomas MG, Boccaccio GL. Smaug1 mRNA-silencing foci respond to NMDA and modulate synapse formation. J Cell Biol 2011; 195:1141-57; PMID:22201125; http://dx.doi.org/10.1083/jcb.201108159
- Luchelli L, Thomas MG, Boccaccio GL. Synaptic control of mRNA translation by reversible assembly of XRN1 bodies. J Cell Sci 2015; 128:1542-54; PMID:25736288; http://dx.doi.org/10.1242/jcs.163295
- Pascual ML, Luchelli L, Habif M, Boccaccio GL. Synaptic activity regulated mRNA-silencing foci for the fine tuning of local protein synthesis at the synapse. Commun Integr Biol 2012; 5:388-92; PMID:23060966; http://dx.doi.org/10.4161/cib.20257
- Chartier A, Klein P, Pierson S, Barbezier N, Gidaro T, Casas F, Carberry S, Dowling P, Maynadier L, Bellec M, et al. Mitochondrial dysfunction reveals the role of mRNA poly(A) tail regulation in oculopharyngeal muscular dystrophy pathogenesis. PLoS Genet 2015; 11:e1005092; PMID:25816335; http://dx.doi.org/10.1371/journal.pgen.1005092
- de Haro M, Al-Ramahi I, Jones KR, Holth JK, Timchenko LT, Botas J. Smaug/SAMD4A restores translational activity of CUGBP1 and suppresses CUG-induced myopathy. PLoS Genet 2013; 9:e1003445; PMID:23637619; http://dx.doi.org/10.1371/journal.pgen.1003445
- Chen Z, Holland W, Shelton JM, Ali A, Zhan X, Won S, Tomisato W, Liu C, Li X, Moresco EM, et al. Mutation of mouse Samd4 causes leanness, myopathy, uncoupled mitochondrial respiration, and dysregulated mTORC1 signaling. Proc Natl Acad Sci U S A 2014; 111:7367-72; PMID:24799716; http://dx.doi.org/10.1073/pnas.1406511111
- Smibert CA, Wilson JE, Kerr K, Macdonald PM. smaug protein represses translation of unlocalized nanos mRNA in the Drosophila embryo. Genes Dev 1996; 10:2600-9; PMID:8895661; http://dx.doi.org/10.1101/gad.10.20.2600
- Smibert CA, Lie YS, Shillinglaw W, Henzel WJ, Macdonald PM. Smaug, a novel and conserved protein, contributes to repression of nanos mRNA translation in vitro. Rna 1999; 5:1535-47; PMID:10606265; http://dx.doi.org/10.1017/S1355838299991392
- Baez MV, Boccaccio GL. Mammalian Smaug is a translational repressor that forms cytoplasmic foci similar to stress granules. J Biol Chem 2005; 280:43131-40; PMID:16221671; http://dx.doi.org/10.1074/jbc.M508374200
- Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One 2012; 7:e51954; PMID:23251661; http://dx.doi.org/10.1371/journal.pone.0051954
- Ong BA, Li J, McDonough JM, Wei Z, Kim C, Chiavacci R, Mentch F, Caboot JB, Spergel J, Allen JL, et al. Gene network analysis in a pediatric cohort identifies novel lung function genes. PLoS One 2013; 8:e72899; PMID:24023788; http://dx.doi.org/10.1371/journal.pone.0072899
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 2003; 10:614-21; PMID:12858164; http://dx.doi.org/10.1038/nsb956
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell 2007; 12:143-55; PMID:17199047; http://dx.doi.org/10.1016/j.devcel.2006.10.005
- Amadei G, Zander MA, Yang G, Dumelie JG, Vessey JP, Lipshitz HD, Smibert CA, Kaplan DR, Miller FD. A Smaug2-based translational repression complex determines the balance between precursor maintenance versus differentiation during mammalian neurogenesis. J Neurosci 2015; 35:15666-81; PMID:26609159; http://dx.doi.org/10.1523/JNEUROSCI.2172-15.2015
- Benoit B, He CH, Zhang F, Votruba SM, Tadros W, Westwood JT, Smibert CA, Lipshitz HD, Theurkauf WE. An essential role for the RNA-binding protein Smaug during the Drosophila maternal-to-zygotic transition. Development 2009; 136:923-32; PMID:19234062; http://dx.doi.org/10.1242/dev.031815
- Kim HH, Lee SJ, Gardiner AS, Perrone-Bizzozero NI, Yoo S. Different motif requirements for the localization zipcode element of beta-actin mRNA binding by HuD and ZBP1. Nucleic Acids Res 2015; 43:7432-46; PMID:26152301; http://dx.doi.org/10.1093/nar/gkv699
- Sosanya NM, Huang PP, Cacheaux LP, Chen CJ, Nguyen K, Perrone-Bizzozero NI, Raab-Graham KF. Degradation of high affinity HuD targets releases Kv1.1 mRNA from miR-129 repression by mTORC1. J Cell Biol 2013; 202:53-69; PMID:23836929; http://dx.doi.org/10.1083/jcb.201212089
- Sosanya NM, Cacheaux LP, Workman ER, Niere F, Perrone-Bizzozero NI, Raab-Graham KF. Mammalian target of Rapamycin (mTOR) tagging promotes dendritic branch variability through the capture of Ca2+/Calmodulin-dependent protein kinase II alpha (CaMKIIalpha) mRNAs by the RNA-binding Protein HuD. J Biol Chem 2015; 290:16357-71; PMID:25944900; http://dx.doi.org/10.1074/jbc.M114.599399
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 2006; 133:4573-83; PMID:17050620; http://dx.doi.org/10.1242/dev.02649
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol 2011; 300:C723-42; PMID:21209361; http://dx.doi.org/10.1152/ajpcell.00462.2010
- Chen L, Dumelie JG, Li X, Cheng MH, Yang Z, Laver JD, Siddiqui NU, Westwood JT, Morris Q, Lipshitz HD, et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol 2014; 15:R4; PMID:24393533; http://dx.doi.org/10.1186/gb-2014-15-1-r4
- Laver JD, Marsolais AJ, Smibert CA, Lipshitz HD. Regulation and function of maternal gene products during the maternal-to-zygotic transition in drosophila. Curr Top Dev Biol 2015; 113:43-84; PMID:26358870; http://dx.doi.org/10.1016/bs.ctdb.2015.06.007
- Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J Cell Sci 2009; 122:563-73; PMID:19193871; http://dx.doi.org/10.1242/jcs.038208
- Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol Biol Cell 2005; 16:405-20; PMID:15525674; http://dx.doi.org/10.1091/mbc.E04-06-0516
