ABSTRACT
Professional phagocytes engulf microbial invaders into plasma membrane-derived phagosomes. These mature into microbicidal phagolysosomes, leading to killing of the ingested microbe. Phagosome maturation involves sequential fusion of the phagosome with early endosomes, late endosomes, and the main degradative compartments in cells, lysosomes. Some bacterial pathogens manipulate the phosphoinositide (PIP) composition of phagosome membranes and are not delivered to phagolysosomes, pointing at a role of PIPs in phagosome maturation. This hypothesis is supported by comprehensive microscopic studies. Recently, cell-free reconstitution of fusion between phagosomes and endo(lyso)somes identified phosphatidylinositol 4-phosphate [PI(4)P] and phosphatidylinositol 3-phosphate [PI(3)P] as key regulators of phagolysosome biogenesis. Here, we describe the emerging roles of PIPs in phagosome maturation and we present tools to study PIP involvement in phagosome trafficking using intact cells or purified compartments.
Phagocytosis and phagosome maturation
Phagosomes are compartments which are formed when phagocytic cells ingest microorganisms and which subsequently mature into microbicidal phagolysosomes, leading to oxidative and nonoxidative killing of enclosed microbes.Citation1,2,3 During maturation, phagosomes sequentially fuse with and acquire the characteristics of early endosomes, late endosomes, and lysosomes.Citation3 The interactions between the various phagocytic and endocytic compartments are vectorially ordered in that early phagosomes do not directly fuse with lysosomes.Citation2,4 As with other membranes of the secretory and endocytic pathway, phagosome-endo(lyso)some fusion is governed by compartment-specific Rab (Ras-like protein from rat brain) and SNARE (soluble NSF attachment protein receptor) proteins,Citation1,3 some of which have been identified.Citation5 Rab5 and Rab7 are the best-characterized regulator GTPases of the endocytic and phagocytic pathways.Citation5,6 Similar to maturing endosomes, phagosomes first recruit Rab5 which then is gradually replaced by Rab7. This Rab5-to-Rab7 conversion is presumably regulated by SAND1/Mon1-Ccz1 (SAND1, SAND endocytosis protein family 1; Mon1, monensin sensitivity 1; Ccz1, calcium caffeine zinc sensitivity 1), a Rab5 effector complex which links inactivation and release of Rab5 to Rab7 recruitment and activation.Citation7,8,9 Rab5 promotes fusion of the phagosome with early endosomes Citation10 and Rab7 with late endosomes/lysosomes.Citation2,11 Hence, Rab5-to-Rab7 conversion on phagosomes drives transition of early phagosomes into late phagosomes/phagolysosomes.
As they mature, phagosomes are acidified by the proton-pumping vacuolar ATPase (vATPase), resulting in a luminal pH of 4.5–5.0 in phagolysosomes.Citation12,13 The acidic milieu within phagolysosomes activates lysosomal hydrolases and regulates degradation of the phagosome's contents.Citation14 In addition to acid hydrolases and low pH, the microbicidal arsenal of phagolysosomes includes antimicrobial peptides (e.g., defensins), reactive oxygen (ROS) and nitrogen species (RNS). ROS and RNS are generated by a NADPH-oxidase complex and a NO-synthase, respectively.Citation1
As with the endocytic pathway, membrane constituents to be degraded within or recycled from phagosomes are presumably incorporated into vesicles that either enter the phagosome lumen or bud from the phagosome.Citation5 Intriguingly, the machineries that drive inward (i.e., ESCRT complex, endosomal sorting complex required for transport) or outward budding (i.e., SNXs, sorting nexins) of the phagosome membrane, are required for phagolysosome formation.Citation15,16 Phagolysosome formation, moreover, is regulated by the cytoskeleton,Citation11,17,18 calcium currents,Citation1,5,19 and lipids, especially phosphoinositides (PIPs).Citation20,21
PIPs
“PIPs” is a summary expression for 7 different, mono-, bis-, or tris-phosphorylated derivatives of the glycerophospholipid phosphatidylinositol (PI) that are generated by phosphorylation of the D3-, D4-, and/or D5-hydroxyl groups of the inositol moiety of PI.Citation22 Many of the PIP isomers can be interconverted by PIP kinases and PIP phosphatasesCitation22,23 and subcellular distribution and substrate specificity of these enzymes causes accumulation of PIPs in defined subcellular compartments.Citation24 PI(3)P is enriched in early endosomes/phagosomes, PI(4)P in the Golgi complex, secretory vesicles, and the plasma membrane, PI(3,5)P2 in late endosomes/lysosomes, and PI(4,5)P2 and PI(3,4,5)P3 in the plasma membrane.Citation22,25,26 Notably, each of these lipids can be present in small amounts in other membrane compartments.Citation22 Though PIPs are rare lipids, they regulate many cellular processes, such as membrane remodeling, transport, and fusion in the biosynthetic/secretory and endocytic pathway.
Most often, PIPs exert their functions by recruiting effector proteins to specific subcellular compartments and/or to regulate the activity of these factors.Citation27 PIP effector proteins usually contain conserved PIP-binding modules such as ANTH (AP180 N-terminal homology), C2 (conserved region-2 of protein kinase C), ENTH (epsin N-terminal homology), FERM (Protein 4.1, Ezrin, Radixin, Moesin), FYVE (Fab1, YOTB, Vac1, EEA1), Golph3 (Golgi phosphoprotein 3), GRAM (glycosyltransferases, Rab-like GTPase activators and myotubularins), PDZ (postsynaptic density 95, disk large, zonula occludens), PH (pleckstrin homology), PHD (plant homeodomain), PTB (phosphotyrosine binding), or PX domains (phox homology).Citation27,28 Some of these PIP-binding proteins domains bind to multiple PIPs, some to one PIP isomer exclusively. Often, PIP effectors require additional determinants to bind to membranes or to influence a cellular process. Examples for such additional factors are other lipids (e.g., PIPs), small GTPases and/or presence of certain membrane curvature. These additional affinities of PIP effectors are either encoded in the PIP-binding domain itself or they are mediated by a distinct portion of the effector protein.Citation29 Hence, PIPs function in a process commonly referred to as “coincidence detection.”Citation29
Detection of PIPs in biological samples
The major challenge of analyzing the PIP content of a biological sample is to distinguish between the various PIP isomers. This can be done by either specific PIP-binding proteins or by separating (deacylated) PIPs according to physical properties (i.e. mass, charge, and/or hydrophobicity) using TLC (thin layer chromatography), HPLC (high performance liquid chromatography), or mass spectrometry.Citation27 All of these approaches are suitable to analyze the composition of lipid extractsCitation30,31,32 and can be combined with cell fractionation techniques to determine the subcellular localization of the various PIPs.Citation33 However, the subcellular distribution of PIPs has largely been analyzed using PIP-binding proteins which can be applied to microscopically detect PIPs in (intact) cells or tissue sections.Citation27 The above approaches to analyze PIPs are detailed below:
Detection of PIPs by PIP-binding proteins
Upon expression of epitope tag PIP-binding domain fusion proteins in cells, the subcellular localization of the PIP probe and, thus, of the corresponding lipid can be visualized by immunoelectron or immunofluorescence microscopy. Expression of PIP-binding domains fused to fluorescent proteins (e.g., GFP, green fluorescent protein) even allows for detection of PIPs in live cells. Alternatively, purified PIP-binding domains or PIP-specific antibodies can be used to stain free PIPs in untransfected cells after fixation and permeabilization.Citation27,32,34 As discussed above, some PIP-binding modules bind to more than one PIP isomer and require additional cues for membrane localization. The experimentally perfect PIP sensor is specific for a single PIP and should not depend on additional membrane receptors.Citation35 To reliably assign a PIP to a certain subcellular compartment, it is moreover advisable to use independent sensors against the corresponding lipid.
Aside the use of PIP-binding domains, other methods have been developed to analyze the PIP content of purified compartments. These include the detection of radiolabeled (deacylated) PIPs by TLC or HPLC, or of unlabeled PIPs by HPLC or mass spectrometry:
Detection of radiolabeled (deacylated) PIPs by TLC or HPLC
Here, membrane preparations are incubated in the presence of 32P labeled ATP which is biosynthetically incorporated into PIPs by phosphoinositide kinases (PIKs). Lipids are extracted, separated by TLC or reverse phase HPLC, and radioactive PIPs are quantified.Citation27 Alternatively, PIPs can be deacylated and separated by anion exchange HPLC. PIP isomers are identified by co-migration (TLC) or co-elution (HPLC) with (deacylated) PIP standards. The latter can be used to quantify the amount of a defined PIP in a lipid mixture. These approaches, though highly sensitive, visualize only the turnover of PIPs and exclude dormant PIP pools, such as the pool of PI(3,4,5)P3 in the plasma membrane of resting cells.Citation36,37
Detection of non-radioactive PIPs by HPLC and/or mass spectrometry
Non-radioactive methods for the biochemical detection of free and complexed PIPs in a sample rely on HPLCCitation33 and/or mass spectrometry.Citation38 Detection of unlabeled PIPs by HPLC is less sensitive than by radioactivity-based methods.Citation27 Mass spectrometry detection of unlabeled PIPs is rather sensitive, yet fails to distinguish between the mono- or the bis-phosphorylated PIP species.Citation39 Recently, a combinatorial approach of HPLC separation and subsequent mass spectrometry analysis of deacylated PIPs has been developed, allowing for detection and quantification of all PIP isomers in a complex lipid mixture.Citation36
PIPs in phagosome maturation
Although there is a detailed picture of which PIPs are required for phagosome formation at the plasma membrane,Citation21,40 little is known about the PIPs involved in subsequent phagosome maturation. summarize the current knowledge on the roles of PIPs and PIP-modifying enzymes in phagosome maturation.
Table 1. PIPs and their roles in phagosome maturation.
Table 2. PIP-modifying enzymes with a role in phagosome maturation.
PIPs in phagosome maturation – lessons learned from intracellular pathogens
Selection pressure from the host can lead to the evolution of pathogen structures, molecules, and strategies which protect the pathogen from being delivered to and killed within phagolysosomes. Such pathogens (i) disrupt the phagosome membrane and escape into the cytosol (e.g., Listeria monocytogenes), (ii) block phagosome maturation at a pre-phagolysosome stage (e.g., Salmonella enterica, Mycobacterium tuberculosis), (iii) they transform the phagosomes in which they reside, into non-endocytic compartments (e.g., Legionella pneumophila), or (iv) are adapted to the acidic and degradative milieu within phagolysosomes (e.g., Coxiella burnetii).Citation1,41 Some of these “intracellular” pathogens manipulate the PIP metabolism in their host cells, resulting in altered maturation of pathogen-containing phagosomes.Citation42 Pathogen strategies impacting on or exploiting host cell PIP metabolism include (i) production of PIP-binding effector proteins that use PIPs as a membrane anchor, (ii) introduction of lipid and protein factors that activate, inactivate, or recruit host cell PIP-metabolizing enzymes, or of (iii) PIP-metabolizing enzymes that directly modify host cell PIPs.Citation41 For excellent reviews on PIP-targeting effector proteins of bacterial pathogens, we refer the reader to references 41 and 42. Here, we confine our presentation to pathogenic bacteria which block phagolysosome formation by secreting phosphatases which directly dephosphorylate PIPs:
S. enterica
Salmonellae secrete effector proteins into host cells using a dedicated bacterial type III secretion system. One of the effector proteins translocated in this way is SopB (Salmonella outer protein B) which dephosphorylates PI(3,5)P2, PI(3,4,5)P3,Citation43 and PI(4,5)P2.Citation44 SopB is required for the S. enterica phagosome maturation arrest, as knockout strains lacking SopB are more efficiently delivered to phagolysosomes.Citation44
M. tuberculosis
Phagosomes containing viable M. tuberculosis can be devoid of PI(3)P, a feature likely caused by the M. tuberculosis secreted PI(3)P phosphatase SapM (secreted acid phosphatase of M. tuberculosis).Citation45 Phagosomes containing SapM-deficient M. tuberculosis acidifyCitation46 and fuse with lysosomes,Citation47 yet it has not been tested when and how much PI(3)P they acquire.
Another M. tuberculosis PIP phosphatase, MptpB (M. tuberculosis protein tyrosine phosphatase), dephosphorylates PI(3)P, PI(4)P, PI(5)P, and PI(3,5)P2 in vitro.Citation48 Mutant strains lacking MptpB are killed by activated macrophages in guinea pigs.Citation49 Possibly, MptpB deletion mutants are more readily delivered to and killed within phagolysosomes which, however, remains to be tested. Moreover, it is unclear, whether MptpB acts by dephosphorylating PIPs and/or proteins in vivo.
L. pneumophila
The Legionella effector protein SidP (substrate of Icm/Dot transporter P) secreted through a type IV secretion system dephosphorylates PI(3)P and PI(3,5)P2,Citation50 whereas SidF is a secreted PI(3,4)P2 and PI(3,4,5)P3 phosphatase.Citation51 Knock-out strains lacking either of these effector proteins have not been tested for interference with phagosome maturation.
The above observations point to an essential role of PIPs in phagosome maturation. To truly understand how manipulation of PIPs can contribute to altered trafficking of phagosomes, it is necessary to define which PIPs are required for each step of the default maturation of phagosomes into phagolysosomes.
Analysis of PIP involvement in phagosome maturation using intact cells or purified compartments
Whether and which PIPs are required for phagolysosome formation can be determined using a combination of techniques to track the progression of phagosome maturation, to specifically visualize PIP isomers, and to manipulate the PIP composition of phagosomes.
As detailed below, in whole cells, the progression of phagosome maturation can be analyzed by visualizing marker proteins or lipids that specify early phagosomes, late phagosomes, or phagolysosomes, and phagosome PIPs can be detected by ectopically expressed fluorescent protein- or epitope-tagged lipid-binding domains. The impact of PIPs on phagosome maturation can be assessed by overexpression of PIP-binding domains to sequester defined PIP species or by manipulating the PIP composition of phagosomes using inhibition, silencing, depletion, and/or overexpression of PIP-modifying enzymes. Moreover, polyamine carrier-complexed exogenous PIPs or membrane-permeable PIP analogs can be incorporated into subcellular membranes, including phagosomes. Alternatively, sub-reactions of phagosome maturation (e.g., phagosome-lysosome fusion) can be reconstituted in vitro with purified compartments. In such cell-free assays, phagosome/endosome PIPs can be detected by PIP-binding domains or antibodies, TLC, HPLC, and/or mass spectrometry and PIP-sequestering protein domains or PIP-modifying enzymes can be used to identify PIPs relevant to the sub-reaction of phagosome maturation studied.
Analysis of PIP requirements of phagosome maturation in whole cells
As they mature, phagosomes pick up and lose endocytic marker molecules following a characteristic temporal pattern. Accordingly, different-aged phagosomes vary in marker molecule composition: early phagosomes contain Rab5, the transferrin receptor (TfR), and syntaxin 13 (Stx13); late phagosomes lack early endocytic proteins and possess Rab7, lysosomal hydrolases (e.g., cathepsins), and LAMPs (lysosome-associated membrane proteins).Citation1,3 The composition of phagolysosomes is very similar to that of late phagosomes. Differentiation between late phagosomes and phagolysosomes is possible in that the latter acquire fluid phase tracers (e.g., fluorochrome-conjugated dextrans) preloaded into lysosomes,Citation3 although even after very long chase periods, some of the tracer will still be in late endosomes.Citation52 Visualization of endocytic marker molecules on phagosomes at different times post phagocytosis allows to track the progression of phagosome maturation and to reveal altered maturation of phagosomes in experimentally manipulated or pathogen-infected cells.
The use of lipid-binding proteins to visualize PIPs on maturing phagosomes in intact cells
An important step toward the understanding of how PIPs govern phagosome maturation was to determine which PIPs occur on phagosomes at defined maturation stages. To this end, numerous studies have analyzed association of overexpressed fluorescent protein-tagged PIP-binding domains with nascent and/or maturing phagosomes. This approach has provided a detailed picture of the PIP dynamics at sites of phagocytosisCitation40 and on early phagosomes, yet it only occasionally detected PIPs on late phagocytic compartments.Citation53,54
Notably, overexpression of PIP-binding protein domains can compete with membrane recruitment of the relevant, authentic PIP effectors and, hence, inhibit PIP-dependent cellular processes.Citation27,55 For instance, overexpression of PI(3)P-binding probes (i.e., 2xFYVE domain of EEA1 or PX domain of p40phox) to visualize PI(3)P blocks phagosome maturation at an early stage,Citation15 making this approach unsuitable to analyze the PI(3)P content of late phagosomes or phagolysosomes.
To circumvent alterations of phagosome maturation due to ectopic expression of lipid-binding probes, the experimenter can use epitope-tagged and/or fluorochrome-conjugated PIP probes or PIP-specific antibodies with fixed and permeabilized, untransfected cells.Citation27,55 For example, Alexa633-conjugated 2xFYVE domain of Hrs has been used to reveal the presence of PI(3)P on latex bead phagosomes (LBPs) and its absence from M. tuberculosis phagosomes.Citation32 However useful for solving certain questions, this approach fails to detect PIPs which were already complexed with downstream effector proteins.Citation27 In sum, combining low expression levels of genetically encoded fluorescent PIP sensors and high sensitivity imaging techniques might be the key to detection of PIPs on maturing phagosomes without affecting phagosome maturation and to visualize PIPs on late phagocytic compartments, too.
Recombinant PIP probes or antibodies for the detection of all PIP isomers are available. Importantly, however, the suitability of ML1Nx2 (2 copies of residues 1–68 of TRPML1) as one of a very few selective PI(3,5)P2 probes is disputed.Citation26,56 Moreover, specific PI(5)P-binding protein domains have not yet been used to visualize this PIP in cells.
Presence of a given PIP in phagosomes suggests that it may be required for the progression of phagosome maturation, yet it does not provide proof. Hence, to identify which PIPs are actually relevant to phagosome maturation, PIPs or the enzymes involved in their turnover need to be manipulated, as detailed below.
The use of lipid-binding proteins to sequester defined PIP isomers in intact cells
As discussed above, competitive inhibition of phagosome maturation by ectopically expressed PIP-binding domains can preclude visualization of PIPs on late phagosomes or phagolysosomes. However, it can also be exploited to identify PIPs relevant to phagosome maturation. For instance, sequestration of PI(3)P by overexpression of a high-avidity PI(3)P probe (5 copies of FYVE domain of Hrs [hepatocyte growth factor regulated tyrosine kinase substrate]) blocked actin polymerization on early phagosomes and concomitantly early-to-late phagosome transition.Citation57
Since specific binding probes are available for each PIP isomer, the sequestration approach can be used to assign roles in phagosome maturation to the various PIPs, with the following limitations: (i) Phagocytosis depends on PI(4,5)P2 which is generated from PI(4)P by phosphatidylinositol 4-phosphate 5-kinase Iα. Sequestration of PI(4)P or PI(4,5)P2 will delay phagocytosis,Citation58 which could be mistaken for inhibition of phagosome maturation. This problem could be circumvented using live cell imaging and analysis of individual phagosomes after completion of phagocytosis. (ii) The various subreactions of phagosome maturation are interdependent. Thus, if phagosome maturation required a given PIP in more than one step, the sequestration approach would block the first step depending on this PIP, thereby obscuring the impact of PIP sequestration on the following step(s).
Inhibition, silencing, or depletion of PIP-modifying enzymes in living cells
Inhibitors of PIP-modifying enzymes
Inhibitors to specifically block the formation and/or consumption of a given PIP by PIP kinases or PIP phosphatases are available and some of these agents have already been used to analyze the interplay of PIP metabolism and phagosome maturation:
Pharmacological inhibitors Phosphatidylinositol 3-kinase (PI3K) inhibitors
Mammals possess 3 classes of PI3K which differ in structure, regulation, and substrate preference and generate PI(3)P, PI(3,4)P2, or PI(3,4,5)P3 by phosphorylation of the D3-hydroxyl group of PI, PI(4)P, or PI(4,5)P2.Citation59
The panspecific PI3K inhibitor wortmannin (WM) has been widely used to analyze the contribution of PI3Ks to phagosome maturation. Phagosomes in WM-treated cells get stuck at an early maturation stage; they contain less PI(3)P and EEA1, do not release Rab5, and do not acquire Rab7, LAMP1, or a fluid phase tracer preloaded into lysosomes.Citation4,60-62 Microinjection into phagocytes of an antibody which specifically inhibits class III PI3K Vps34 Citation63 also decreases contents of phagosomes in PI(3)P, EEA1, and LAMP1.Citation61,62 Thus, Vps34 is the major target of WM here, although another WM-sensitive PI3K, i.e. class IA PI3K p110α, is also involved in phagosome maturation.Citation64 Recently, more isoform-specific PI3K inhibitors have been identified, which may, in the future become useful in phagosome research. These include inhibitors of class I PI3K isoforms (e.g., YM024 (p110α), TGX221 (p110β), and AS252424 (p110δ))Citation39 and of class III PI3K Vps34 (e.g., PIK-III,Citation65 SAR405,Citation66 and Vps34-IN1 Citation67).
Phosphatidylinositol 4-kinase (PI4K) inhibitors
Mammals possess 4 different PI4Ks, i.e., PI4KIIα, PI4KIIβ, PI4KIIIα, and PI4KIIIβ. Isoform-specific inhibitors of class II PI4Ks are not available. Class III PI4Ks, by contrast, are sensitive to A1 (PI4KIIIαCitation68) or Pik-93 (PI4KIIIβCitation69). These inhibitors, however, also target PI3Ks, limiting their use in analysis of phagosome maturation which requires both class IA and class III PI3K activity.
Phosphatidylinositol 5-kinase (PI5K) inhibitors
The pharmacological inhibitors of the phosphatidylinositol 3-phosphate 5-kinase (PI3,5K) PikFYVE, namely MF-4, apilimod, and YM201636, block phagosome maturation before the late phagosome/phagolysosome stage, suggesting that PikFYVE and its lipid product PI(3,5)P2 regulate phagosome maturation.Citation70 As PikFYVE also generates PI(5)P from PI,Citation71 it remains to be tested whether phagosome maturation requires PI(5)P, PI(3,5)P2, or the both of these lipids.
Further PIK inhibitors and PIP phosphatase inhibitors
Type II phosphatidylinositol 5-phosphate 4-kinases (PI5,4KIIs) generate PI(4,5)P2 from PI(5)P. PI5,4KII isoforms β and γ are selectively blocked by SAR088Citation72 and NIH-12848,Citation73 respectively. Selective inhibitors of phosphatidylinositol 4-phosphate 5-kinases (PI4,5Ks) have not yet been identified.
Pharmacological inhibitors of PIP phosphatases include VO-OHpic inhibiting PTEN (phosphatase and tensin homolog),Citation74 YU142670 inhibiting OCRL (oculocerebrorenal syndrome of Lowe) and Inpp5b (inositol polyphosphate 5-phosphatase),Citation75 and AS1949490 inhibiting SHIP2 (SH2 domain containing inositol 5-phosphatase 2).Citation76 Although PTEN,Citation77 OCRL, and Inpp5bCitation78 manipulate the PIP composition of phagosomes, the corresponding inhibitors have not yet been tested for interference with phagosome maturation.
PIK-inhibiting antibodies
Inhibitory antibodies against PI3Ks Vps34 and p110α,Citation63 and class II PI4KsCitation79 have been generated. Class IA PI3K p110α- or class II PI4K-inhibiting antibodies have not been tested for interference with phagosome maturation in whole cells. By contrast, introduction of a Vps34-inhibiting antibody into macrophages blocks phagosome maturation at the early phagosome stage.Citation61,62 A major obstacle to the in vivo use of such antibodies is that immunoglobulins do not permeate membranes and hence need to be introduced into cells by microinjection or proteofection.
Silencing or depletion PIP-modifying enzymes
To assign certain PIPs a role in phagosome maturation, some studies have used siRNA-mediated reduction of expression of PIP-modifying enzymes and revealed that PI3,5K PikFYVE and phosphatidylinositol 3-phosphatase PTEN remove PI(3)P from early phagosomesCitation77 and that phagolysosome formation requires class IA PI3K p110α Citation64 and PikFYVE.Citation70 Moreover, in macrophages from mice lacking the PI(3,4,5)P3 5-phosphatase SHIP1, phagosomes were decreased in PI(3,4)P2 and in NADPH oxidase activity.Citation80
A related approach to test the impact of a PIP-modifying enzyme on a given cellular process is to ectopically express enzymatically inactive versions of the respective enzyme. These mutated enzymes often act in a dominant-negative fashion through binding to PI or PIPs without being able to perform the proper function and, hence, essentially function like isolated PI or PIP-binding domains (s. above). Their overexpression often reproduces phenotypes seen with expression knock-down or pharmacological inhibition of the corresponding enzymes.
Inhibition or siRNA-mediated knock down of PIP effector proteins in whole cells
PIPs usually are ‘white collar’ compounds which recruit downstream effector proteins to perform the actual job. The identification of PIP effectors is therefore an important step toward the understanding of how PIPs govern a cellular process. PIP-binding proteins which are recruited to maturing phagosomes are good candidate regulators of phagolysosome biogenesis. Putative phagosome PIP effectors can be tested for involvement in phagosome maturation by selective inhibitors –if available– or by siRNA-mediated expression silencing. For instance, microinjection into phagocytes of antibodies against the PI(3)P effector EEA1 blocks phagosome maturation,Citation62 as does expression knock down of PI(3)P effectors Hrs,Citation15 SNX1, SNX6, SNX9,Citation16 FYCO-1 (FYVE and coiled-coil domain containing 1)Citation81 or of the PI(3,5)P2 effector TRPML1 (transient receptor potential cation channel, mucolipin subfamily, member 1).Citation19
Acute manipulation of PIPs in living cells
Ectopic expression or silencing of PIP-modifying enzymes in (phagocytic) cells takes days. Inhibitory antibodies against PIP kinases or PIP phosphatases act more quickly, yet still need to be introduced into cells hours before phagocytosis.Citation61,63
Pharmacological inhibitors allow for more acute manipulations of cellular PIP metabolism; e.g., PI3K inhibitor WM blocks phagosome maturation even if administered after completion of phagocytosis.Citation4,57,62 Addition of pharmacological inhibitors at different times post phagocytosis hence can be used to analyze late subreactions of phagosome maturation detached from preceding maturation events. Using this approach, we showed that late phagosome-to-lysosome fusion in vivo is WM-sensitive and PI3K-dependent.Citation4
Recently, a technique has been developed to acutely deplete a defined PIP from a given subcellular compartment in living cells. This technique relies on the rapamycin-induced heterodimerization of FKBP12 and the FRB (FKBP12-rapamycin binding) domain of mTOR (mechanistic target of rapamycin). A PIP-modifying enzyme of interest is fused to FRB domain of mTOR and a membrane receptor of choice to FKBP12. Upon coexpression of the resulting fusion proteins in cells, addition of rapamycin to the culture medium makes the PIP-modifying enzyme bind to and convert PIPs in vicinity of the corresponding membrane receptor. Basically, this heterodimerization approach is suitable to increase or reduce the amount of a PIP in any defined subcellular compartment (e.g., phagosomes, endosomes, Golgi) within minutes. Yet, importantly, membrane receptor and PIP-modifying enzyme need to be carefully chosen: Ideally, the membrane receptor is a cytosol-exposed transmembrane protein unique to the compartment to be analyzed and the PIP-modifying enzyme targets just one of the various PIP isomers.
The above technique has already been used to target PIP-modifying enzymes to phagocytic or endocytic compartments: Bohdanowicz and coworkers targeted a phosphatidylinositol 4-phosphate 5-kinase to nascent/early phagosomes, leading to an accumulation of PI(4,5)P2 in these compartments.Citation57 Hammond et al. targeted PI(4)P phosphatase Sac1p to late endocytic, Rab7-positive compartments which resulted in a decrease of late endosomes in PI(4)P.Citation35
To study PIP involvement in phagosome maturation, one could use stage-specific marker proteins of early and late endosomes/phagosomes as membrane receptors for PIP-modifying enzymes. However, as marker molecules unique to lysosomes have not yet been identified,Citation3 there is no membrane receptor available to specifically deliver PIP-modifying enzymes to (phago)lysosomes. Hence, the impact of PIPs on phagosome-lysosome fusion is difficult to analyze by the above method.
Incorporation of exogenous PIPs into subcellular compartment membranes of live cells
Aside the aforementioned approaches to manipulate endogenous PIPs, methods to incorporate exogenically added PIPs into subcellular compartment membranes of live cells have been developed. These methods rely on either membrane-permeable PIP analogsCitation39,82 or polyamine carriers that complex PIPs and guide them through membranes.Citation83 In both cases, the negative charges of the PIP headgroup are masked either by chemical modification or by binding of a suitable, positively charged polyamine.
Although PIPs are unselectively incorporated into subcellular compartment membranesCitation39,82 using these methods, addition to cells of a membrane-permeable PI(3)P analog specifically stimulated homotypic early endosome fusion,Citation84 a PI(3)P-dependent process.Citation85 Moreover, intracellular delivery of PI(4)P or PI(5)P by polyamine carriers recently assigned these PIPs roles in autophagosome-lysosome fusionCitation86 and autophagosome formation,Citation87 respectively.
Introduction of exogenous PIPs into live cells, for instance, could be useful to clarify whether the phagosome maturation block caused by WMCitation60,64 is due to decreased levels of PI(3)P, PI(3,4)P2, and/or PI(3,4,5)P3. To this end, it could be tested whether intracellular delivery of the above D3-PIPs alone or in combinations can revert inhibition of phagolysosome formation in phagocytes treated with WM.
Cell-free analysis of PIP requirements of phagosome maturation
The cell-free reconstitution of membrane fusion pathways has proven to be well-suited to identify fusion-relevant factors, as masterfully exemplified by the purification of N-ethylmaleimide-sensitive fusion factor (NSF) by Block and coworkers in 1988.Citation88 Moreover, it can be used to order fusion sub-reactions (i.e., priming, tethering, docking, [hemi]fusion) and has led to identification of the function of NSF as a SNARE chaperoneCitation89 and of SNAREs as fusion catalysts.Citation90 Cell-free fusion assays further are compatible with addition of membrane-impermeable inhibitory agents, such as inhibitory antibodies or recombinant proteins, or with inhibitors toxic to the cell.
Phagosome maturation comprises a number of hierarchically ordered sub-reactions which impedes analysis of this process in intact cells: If a factor were required for early and late subreactions, then its inhibition would block phagosome maturation early and obscure the effects of such treatment on later stages. This issue can be circumvented by reconstituting phagosome maturation sub-reactions in cell-free systems. To this end, phagosomes and endosomes are purified at different times post phagocytosis or endocytosis yielding phagocytic and endocytic compartments of different maturation stages. These compartments are mixed in a test tube with cytosolic proteins, an ATP-regenerating system, and ions. Incubation at physiological temperature leads to membrane fusion which can be quantified by various methods: Fusion assays between phagosomes and endosomes are based either on the formation of an avidin-biotin HRP (horseradish peroxidase) complex, on dequenching of a fluorescent lipid analog introduced into lysosomes, or on colocalization of phagosomes with a fluorescent tracer preloaded into endocytic compartments (for a review see ref. Citation91).
In cell-free fusion experiments, the compartments to be fused are incubated under conditions designed to mimick the cytoplasmic situation. Nevertheless, not all aspects of phagosome maturation are readily reconstructed in vitro: For instance microtubules are required for phagolysosome formation in whole cells,Citation92 yet dispensable for reconstituted late phagosome-lysosome fusion.Citation2 Hence, observations made with cell-free fusion assays always need to be validated, as good as possible, in experiments using intact cells. But, of course, experiments with complete, live cells have their potential drawbacks, too. E.g., knock down of expression of an important gene may result in uncontrolled additional changes in the same cell.
PIP involvement in vitro is studied by adding PIP-sequestering protein domains to fusion reactions. If a given PIP were required for fusion, sequestration of this PIP would inhibit the reconstituted reaction. This approach has been used for various reconstituted membrane fusion reactions and has identified PI(3)P, PI(4)P, and PI(4,5)P2 as required for yeast vacuole fusion,Citation93,94 PI(4)P as required for COPII vesicle-Golgi fusion,Citation95 and PI(3)P as required for homotypic early endosome fusion.Citation96,97 Importantly, PIP-binding domains specific for each of the various PIP isomers have been identified, so that each PIP can be analyzed for involvement in a given fusion reaction.
Additional to the experimental approach of masking PIPs by specific protein domains, dephosphorylating them can also reveal their requirement in membrane fusion. For instance, reconstituted fusion between yeast vacuoles and between phagosomes and lysosomes depend on PI(3)P and hence are blocked by addition of recombinant PI(3)P phosphatase MTM1 (myotubularin).Citation4,93
We recently used this approach to identify PIPs relevant for phagosome maturation and showed that PI(3)P is required for fusion of early phagosomes with early endosomes and of late phagosomes with lysosomes, whereas PI(4)P is selectively required for late phagosome-lysosome fusionCitation4 ().
Figure 1. Regulation of phagolysosome formation by phosphatidylinositol 3-phosphate and phosphatidylinositol 4-phosphate. During phagocytosis, phagocytes ingest large particles (e.g. microorganisms) into plasma membrane-derived phagosomes. These mature into microbicidal and degradative phagolysosomes (‘PLYS’), leading to killing and destruction of the enclosed cargo. Solutes and small particles (< 400 nm in diameter) are taken up by endocytosis. The resulting endosomes deliver their cargo to lysosomes (‘LYS’) where it is degraded. Three different compartments have been defined for both, the phagocytic and the endocytic pathway: Early phagosomes/endosomes (‘EP’/‘EE’), late phagosomes/endosomes (‘LP’/‘LE’), and phagolysosomes/lysosomes. Note that the definition of 3 different phagocytic or endocytic compartments substantially simplifies the diversity of organelles along the phagocytic and endocytic pathway. As phagosomes mature, they sequentially fuse with and acquire the characteristics of early endosomes, late endosomes, and lysosomes. Blue arrows mark the progression of phagosome maturation. The different phagosome-endo(lyso)some fusion events are indicated by black, double-sided arrows. Dependence of the reconstituted phagosome-endo(lyso)some fusion on PI(3)P (‘3’) or PI(4)P (‘4’), as judged by its sensitivity to PI(3)P- or PI(4)P-sequestering protein domains, is indicated: ‘-’, no inhibition; ‘+’, >25% inhibition; ‘++’, >50% inhibition; ‘+++’, >75% inhibition. In summary, PI(3)P regulates phagosome maturation at early and late stages, whereas PI(4)P is selectively required late in the pathway.
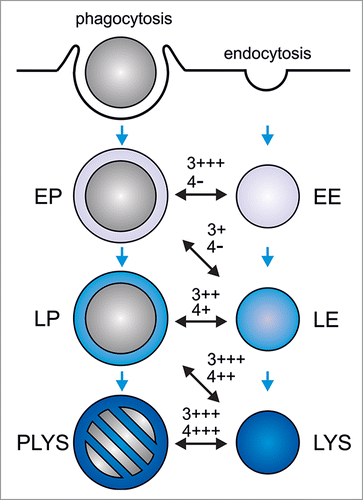
The likely role of PI(3)P in early phagosome-early endosome fusion is to recruit and/or anchor EEA1 to early endocytic and phagocytic compartments, a tethering factor which is required for homotypic early endosome fusionCitation97 and, presumably, early phagosome-early endosome fusion.Citation3 How PI(3)P governs phagosome-lysosome fusion is unclear, yet it might serve to recruit fusion-relevant proteins to late phagosomes/endosomes, such as the “priming” ATPase NSF,Citation98 the “tethering” factors, HOPSCitation99 or FYCO-1,Citation100 and/or the “docking” late endocytic R-SNARE Vamp8.Citation101
As with PI(3)P, PI(4)P might serve to recruit fusion factors to late phagosomes/endosomes, such as NSFCitation98 and HOPS.Citation99 The recent observation that PI(4)P-generating PI4KIIα interacts with the late endocytic R-SNARE Vamp7 which had been implicated in phagosome-lysosome fusion earlier,Citation2 supports the hypothesis that PI4Ks, PI4KIIIβ and PI4KIIα, and their common lipid product PI(4)P are required for late phagosome/endosome maturation.Citation102,103
Analysis of PIP kinase activities associated with phagosomes
We recently identified the kinases responsible for the formation of PI(3)P and PI(4)P in phagolysosomes. To this end, we incubated purified latex bead phagolysosomes under fusion assay conditions and visualized phagosomal PI(3)P or PI(4)P using 2xFYVE domain and a PI(4)P-specific antibody, respectively. Addition of inhibitory antibodies against class III PI3K Vps34 or class II PI4Ks strongly decreased levels of PI(3)P and PI(4)P in phagosomes.Citation4 This approach would hardly have been possible with intact cells, as the use of inhibitory antibodies requires introduction of these molecules into cells which is difficult to achieve. Moreover, in the case of Vps34 inhibition, phagosome maturation would have been blocked early, impeding analysis of PI3K activities associated with phagolysosomes.Citation60,61
In general, this assay is applicable to each purified organelle, as exemplified in our recent publication for different-aged latex bead phagosomes and paramagnetically labeled early endosomes, late endosomes, or lysosomes.Citation4 Moreover, the method can be adapted to analyze activities of other PIP-modifying enzymes, because specific probes are available for each PIP isomer. For instance, using the PH domain of PLCδ1 (phospholipase C δ) as a probe for PI(4,5)P2, we observed that during in vitro incubation phagolysosome PI(4)P is constantly converted to PI(4,5)P2Citation4 as described previously.Citation104
Outlook
Fifteen years ago, Vieira and coworkers observed that class III PI3K Vps34 and its lipid product PI(3)P are required for phagosome maturation.Citation61 Since then, it has been shown for all PIP isomers, except for PI(5)P and PI(3,5)P2, that they regulate the development of newly formed phagosomes into microbicidal phagolysosomes (). Moreover, we now know that some intracellular pathogens possess PIP-modifying effector proteins which, upon introduction into host cells, manipulate the PIP composition of pathogen-containing phagosomesCitation41,42 and help to inhibit their fusion with microbicidal lysosomes. Yet, to truly understand how manipulation of PIPs can contribute to altered trafficking of phagosomes, it is necessary to systematically identify the PIPs required for each step of the default maturation of phagosomes into phagolysosomes. This can be done using cell-free reconstitution of fusion between phagosomes and endosomes of different maturation stages and PIP-sequestering protein domains, an approach which recently identified PI(3)P as needed for fusion of phagosomes with early endosomes or lysosomes and PI(4)P as selectively required for phagosome-lysosome fusion.Citation4 Briefly, comprehensive mapping of the PIP requirements along the phagosome maturation sequence, as exemplified for PI(3)P and PI(4)P in is an important task for the future.
Since PIPs use effector proteins as mediators of their downstream functions, the identification of PIP effectors is a crucial step toward the understanding of how PIPs govern a cellular process. Phagosomal/endosomal PIP effector proteins can be purified from detergent lysates of isolated phagosomes/endosomes using PIP-coated beads or PIP-containing liposomes. After mass spectrometric identification, putative PIP effectors can be tested for involvement in phagolysosome formation using the various approaches described in this article.
Once PIPs and the corresponding effectors relevant to a given step of phagosome maturation have been identified, cell-free fusion assays can be used to determine whether a PIP or a PIP effector is required for the priming, tethering, docking, and/or (hemi)fusion sub-reaction of phagosome-endosome fusion. Such dissection of fusion sub-reactions has especially been done for reconstituted homotypic yeast vacuole fusion,Citation105 yet, presumably is readily adapted to also study phagosome-endosome fusion.
Reconstituted fusion between different-aged phagosomes and endosomes will allow to assign bacterial PIP-modifying effector proteins to a given sub-reaction of phagosome maturation. This will enrich our understanding of how the phagosome's fate is reprogrammed by pathogens: Recently, it has been reported that Legionella effector LegC3 and Vibrio parahaemolyticus effector VopQ block homotypic yeast vacuole fusion.Citation106,107 These observations underline the wide range of eukaryotic species against which pathogen effectors can act, likely because they are usually targeted to central regulatory components which are often highly conserved. However, as yeast is not a natural host of Legionella nor of Vibrio, inhibitory actions of these effectors need be also tested with purified compartments from mammalian cells. Importantly, using our fusion assay setup, we have reproduced inhibition of phagosome-lysosome fusion seen with S. enterica-containing phagosomes in intact cells.Citation2 Hence, cell-free fusion analysis can be used to analyze both, interference with fusion by pathogen factors and identification of factors that force pathogen phagosomes to fuse with lysosomes, e.g., after macrophage activation.
Given the currently big interest into PIPs and into how they regulate cellular processes, we will likely soon have a detailed map of how PIPs govern regular phagolysosome formation. Such map will help to understand how pathogen infection and other pathological events alter host cell PIP metabolism and will offer new approaches to disease intervention.
Abbreviations
| ANTH | = | AP180 N-terminal homology |
| C2 | = | conserved region-2 of protein kinase C |
| COPII | = | coat protein complex II |
| EEA1 | = | early endosome antigen 1 |
| ENTH | = | epsin N-terminal homology |
| ESCRT | = | endosomal sorting complex required for transport |
| FERM | = | Protein 4.1, Ezrin, Radixin Moesin |
| FRB | = | FKBP12-rapamycin binding domain of mTOR |
| FYCO-1 | = | FYVE and coiled-coil domain containing 1 |
| FYVE | = | Fab1, YOTB, Vac1, EEA1 |
| GFP | = | green fluorescent protein |
| Golph3 | = | golgi phosphoprotein 3 |
| GRAM | = | glycosyltransferases, Rab-like GTPase activators and myotubularins |
| HOPS | = | homotypic fusion and vacuole protein sorting |
| HPLC | = | high performance liquid chromatography |
| HRP | = | horseradish peroxidase |
| Hrs | = | hepatocyte growth factor regulated tyrosine kinase substrate |
| Inpp5b | = | inositol polyphosphate 5-phosphatase |
| LAMPs | = | lysosome-associated membrane proteins |
| LBP | = | latex bead phagosome |
| MptpB | = | M. tuberculosis protein tyrosine phosphatase |
| MTM1 | = | myotubularin |
| mTOR | = | mechanistic target of rapamycin |
| NSF | = | N-ethylmaleimide-sensitive fusion factor |
| OCRL | = | oculocerebrorenal syndrome of Lowe |
| PDZ | = | postsynaptic density 95, disk large, zonula occludens |
| PH | = | pleckstrin homology |
| PHD | = | plant homeodomain |
| PI(3)P | = | phosphatidylinositol 3-phosphate |
| PI(3,4,5)P3 | = | phosphatidylinositol 3,4,5-trisphosphate |
| PI(3,4)P2 | = | phosphatidylinositol 3,4-bisphosphate |
| PI(3,5)P2 | = | phosphatidylinositol 3,5-bisphosphate |
| PI(4)P | = | phosphatidylinositol 4-phosphate |
| PI(4,5)P2 | = | phosphatidylinositol 4,5-bisphosphate |
| PI(5)P | = | phosphatidylinositol 5-phosphate |
| PI | = | phosphatidylinositol |
| PI3K | = | phosphatidylinositol 3-kinase |
| PI4,5K | = | phosphatidylinositol 4-phosphate 5-kinase |
| PI4K | = | phosphatidylinositol 4-kinase |
| PI5,4KII | = | type II phosphatidylinositol 5-phosphate 4-kinase |
| PI5K | = | phosphatidylinositol 5-kinase |
| PIK | = | phosphoinositide kinase |
| PIP | = | phosphoinositide |
| PLCδ1 | = | phospholipase Cδ1 |
| PTB | = | phosphotyrosine binding |
| PTEN | = | phosphatase and tensin homolog |
| PX | = | phox homology |
| Rab | = | Ras-like protein fom rat brain |
| RNS | = | reactive nitrogen species |
| ROS | = | reactive oxygen species |
| SAND1/Mon1-Ccz1 | = | SAND1, SAND endocytosis protein family 1 |
| Mon1 | = | monensin sensitivity 1 |
| Ccz1 | = | calcium caffeine zinc sensitivity 1 |
| SapM | = | secreted acid phosphatase of M. tuberculosis |
| SHIP1 | = | SH2 domain containing inositol 5-phosphatase 1 |
| SHIP2 | = | SH2 domain containing inositol 5-phosphatase 2 |
| SidF | = | substrate of Icm/Dot transporter F |
| SidP | = | substrate of Icm/Dot transporter P |
| SNARE | = | soluble NSF attachment protein receptor |
| SNXs | = | sorting nexins |
| SopB | = | Salmonella outer protein B |
| Stx13 | = | syntaxin 13 |
| TfR | = | Transferrin receptor |
| TLC | = | thin layer chromatography |
| TRPML1 | = | transient receptor potential cation channel, mucolipin subfamily, member 1 |
| vATPase | = | vacuolar ATPase |
| Vps34 | = | vacuolar protein sorting 34 |
| WM | = | wortmannin |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Haas A. The phagosome: compartment with a license to kill. Traffic 2007; 8:311-330; PMID:17274798; http://dx.doi.org/10.1111/j.1600-0854.2006.00531.x
- Becken U, Jeschke A, Veltman K, Haas A. Cell-free fusion of bacteria-containing phagosomes with endocytic compartments. Proc Natl Acad Sci U S A 2010; 107:20726-20731; PMID:21071675; http://dx.doi.org/10.1073/pnas.1007295107
- Vieira OV, Botelho RJ, Grinstein S. Phagosome maturation: aging gracefully. Biochem J 2002; 366:689-704; PMID:12061891; http://dx.doi.org/10.1042/bj20020691
- Jeschke A, Zehethofer N, Lindner B, Krupp J, Schwudke D, Haneburger I, Jovic M, Backer JM, Balla T, Hilbi H, et al. Phosphatidylinositol 4-phosphate and phosphatidylinositol 3-phosphate regulate phagolysosome biogenesis. Proc Natl Acad Sci U S A 2015; 112:4636-4641; PMID:25825728; http://dx.doi.org/10.1073/pnas.1423456112
- Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol 2012; 33:397-405; PMID:22560866; http://dx.doi.org/10.1016/j.it.2012.03.003
- Huotari J, Helenius A. Endosome maturation. EMBO J 2011; 30:3481-3500; PMID:21878991; http://dx.doi.org/10.1038/emboj.2011.286
- Kinchen JM, Ravichandran KS. Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 2010; 464:778-782; PMID:20305638; http://dx.doi.org/10.1038/nature08853
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell 2010; 141:497-508; PMID:20434987; http://dx.doi.org/10.1016/j.cell.2010.03.011
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 2010; 20:1654-1659; PMID:20797862; http://dx.doi.org/10.1016/j.cub.2010.08.002
- Alvarez-Dominguez C, Barbieri AM, Berón W, Wandinger-Ness A, Stahl PD. Phagocytosed live Listeria monocytogenes influences Rab5-regulated in vitro phagosome-endosome fusion. J Biol Chem 1996; 271:13834-13843; PMID:8662791; http://dx.doi.org/10.1074/jbc.271.23.13834
- Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol 2003; 23:6494-6506; PMID:12944476; http://dx.doi.org/10.1128/MCB.23.18.6494-6506.2003
- Hackam DJ, Rotstein OD, Zhang WJ, Demaurex N, Woodside M, Tsai O, Grinstein S. Regulation of phagosomal acidification. Differential targeting of Na+/H+ exchangers, Na+/K+-ATPases, and vacuolar-type H+-atpases. J Biol Chem 1997; 272:29810-29820; PMID:9368053; http://dx.doi.org/10.1074/jbc.272.47.29810
- Flannagan RS, Jaumouillé V, Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol 2012; 7:61-98; PMID:21910624; http://dx.doi.org/10.1146/annurev-pathol-011811-132445
- Geisow MJ, D'Arcy Hart P, Young MR. Temporal changes of lysosome and phagosome pH during phagolysosome formation in macrophages: studies by fluorescence spectroscopy. J Cell Biol 1981; 89:645-652; PMID:6166620; http://dx.doi.org/10.1083/jcb.89.3.645
- Vieira OV, Harrison RE, Scott CC, Stenmark H, Alexander D, Liu J, Gruenberg J, Schreiber AD, Grinstein S. Acquisition of Hrs, an essential component of phagosomal maturation, is impaired by mycobacteria. Mol Cell Biol 2004; 24:4593-4604; PMID:15121875; http://dx.doi.org/10.1128/MCB.24.10.4593-4604.2004
- Lu N, Shen Q, Mahoney TR, Liu X, Zhou Z. Three sorting nexins drive the degradation of apoptotic cells in response to PtdIns(3)P signaling. Mol Biol Cell 2011; 22:354-374; PMID:21148288; http://dx.doi.org/10.1091/mbc.E10-09-0756
- Marion S, Hoffmann E, Holzer D, Le Clainche C, Martin M, Sachse M, Ganeva I, Mangeat P, Griffiths G. Ezrin promotes actin assembly at the phagosome membrane and regulates phago-lysosomal fusion. Traffic 2011; 12:421-437; PMID:21210911; http://dx.doi.org/10.1111/j.1600-0854.2011.01158.x
- Jahraus A, Egeberg M, Hinner B, Habermann A, Sackman E, Pralle A, Faulstich H, Rybin V, Defacque H, Griffiths G. ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol Biol Cell 2001; 12:155-170; PMID:11160830; http://dx.doi.org/10.1091/mbc.12.1.155
- Dayam RM, Saric A, Shilliday RE, Botelho RJ. The Phosphoinositide-Gated Lysosomal Ca(2+) Channel, TRPML1, Is Required for Phagosome Maturation. Traffic 2015; 16:1010-1026; PMID:26010303; http://dx.doi.org/10.1111/tra.12303
- Steinberg BE, Grinstein S. Pathogen destruction versus intracellular survival: the role of lipids as phagosomal fate determinants. J Clin Invest 2008; 118:2002-2011; PMID:18523652; http://dx.doi.org/10.1172/JCI35433
- Bohdanowicz M, Grinstein S. Role of phospholipids in endocytosis, phagocytosis, and macropinocytosis. Physiol Rev 2013; 93:69-106; PMID:23303906; http://dx.doi.org/10.1152/physrev.00002.2012
- Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 2013; 93:1019-1137; PMID:23899561; http://dx.doi.org/10.1152/physrev.00028.2012
- Sasaki T, Takasuga S, Sasaki J, Kofuji S, Eguchi S, Yamazaki M, Suzuki A. Mammalian phosphoinositide kinases and phosphatases. Prog Lipid Res 2009; 48:307-343; PMID:19580826; http://dx.doi.org/10.1016/j.plipres.2009.06.001
- De Matteis MA, Godi A. PI-loting membrane traffic. Nat Cell Biol 2004; 6:487-492; PMID:15170460; http://dx.doi.org/10.1038/ncb0604-487
- Jean S, Kiger AA. Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol 2012; 13:463-470; PMID:22722608; http://dx.doi.org/10.1038/nrm3379
- Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RGW, Weisman LS, Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci U S A 2013; 110:21165-21170; PMID:24324172; http://dx.doi.org/10.1073/pnas.1311864110
- Rusten TE, Stenmark H. Analyzing phosphoinositides and their interacting proteins. Nat Methods 2006; 3:251-258; PMID:16554828; http://dx.doi.org/10.1038/nmeth867
- Kutateladze TG. Translation of the phosphoinositide code by PI effectors. Nat Chem Biol 2010; 6:507-513; PMID:20559318; http://dx.doi.org/10.1038/nchembio.390
- Carlton JG, Cullen PJ. Coincidence detection in phosphoinositide signaling. Trends Cell Biol 2005; 15:540-547; PMID:16139503; http://dx.doi.org/10.1016/j.tcb.2005.08.005
- Kim Y, Shanta SR, Zhou LH, Kim KP. Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: a brief review. Exp Mol Med 2010; 42:1-11; PMID:19887898; http://dx.doi.org/10.3858/emm.2010.42.1.001
- Furutani M, Itoh T, Ijuin T, Tsujita K, Takenawa T. Thin layer chromatography-blotting, a novel method for the detection of phosphoinositides. J Biochem 2006; 139:663-670; PMID:16672266; http://dx.doi.org/10.1093/jb/mvj076
- Purdy GE, Owens RM, Bennett L, Russell DG, Butcher BA. Kinetics of phosphatidylinositol-3-phosphate acquisition differ between IgG bead-containing phagosomes and Mycobacterium tuberculosis-containing phagosomes. Cell Microbiol 2005; 7:1627-1634; PMID:16207249; http://dx.doi.org/10.1111/j.1462-5822.2005.00580.x
- Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J 2010; 428:375-384; PMID:20370717; http://dx.doi.org/10.1042/BJ20100129
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19:4577-4588; PMID:10970851; http://dx.doi.org/10.1093/emboj/19.17.4577
- Hammond GRV, Machner MP, Balla T. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol 2014; 205:113-126; PMID:24711504; http://dx.doi.org/10.1083/jcb.201312072
- Kiefer S, Rogger J, Melone A, Mertz AC, Koryakina A, Hamburger M, Küenzi P. Separation and detection of all phosphoinositide isomers by ESI-MS. J Pharm Biomed Anal 2010; 53:552-558; PMID:20399587; http://dx.doi.org/10.1016/j.jpba.2010.03.029
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006; 443:651-657; PMID:17035995; http://dx.doi.org/10.1038/nature05185
- Wenk MR, Lucast L, Paolo GD, Romanelli AJ, Suchy SF, Nussbaum RL, Cline GW, Shulman GI, McMurray W, Camilli PD. Phosphoinositide profiling in complex lipid mixtures using electrospray ionization mass spectrometry. Nat Biotechnol 2003; 21:813-817; PMID:12808461; http://dx.doi.org/10.1038/nbt837
- Idevall-Hagren O, De Camilli P. Detection and manipulation of phosphoinositides. Biochim Biophys Acta 2015; 1851:736-745; PMID:25514766; http://dx.doi.org/10.1016/j.bbalip.2014.12.008
- Levin R, Grinstein S, Schlam D. Phosphoinositides in phagocytosis and macropinocytosis. Biochim Biophys Acta 2014; 1851:805-823.
- Weber SS, Ragaz C, Hilbi H. Pathogen trafficking pathways and host phosphoinositide metabolism. Mol Microbiol 2009; 71:1341-1352; PMID:19208094; http://dx.doi.org/10.1111/j.1365-2958.2009.06608.x
- Pizarro-Cerda J, Kuhbacher A, Cossart P. Phosphoinositides and host-pathogen interactions. Biochim Biophys Acta 2015;1851:911-918.
- Marcus SL, Wenk MR, Steele-Mortimer O, Finlay BB. A synaptojanin-homologous region of Salmonella typhimurium SigD is essential for inositol phosphatase activity and Akt activation. FEBS Lett 2001; 494:201-207; PMID:11311241; http://dx.doi.org/10.1016/S0014-5793(01)02356-0
- Bakowski MA, Braun V, Lam GY, Yeung T, Heo WD, Meyer T, Finlay BB, Grinstein S, Brumell JH. The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe 2010; 7:453-462; PMID:20542249; http://dx.doi.org/10.1016/j.chom.2010.05.011
- Vergne I, Chua J, Lee HH, Lucas M, Belisle J, Deretic V. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 2005; 102:4033-4038; PMID:15753315; http://dx.doi.org/10.1073/pnas.0409716102
- Puri RV, Reddy PV, Tyagi AK. Secreted acid phosphatase (SapM) of Mycobacterium tuberculosis is indispensable for arresting phagosomal maturation and growth of the pathogen in guinea pig tissues. PLoS One 2013; 8:e70514; PMID:23923000; http://dx.doi.org/10.1371/journal.pone.0070514
- Saikolappan S, Estrella J, Sasindran SJ, Khan A, Armitige LY, Jagannath C, Dhandayuthapani S. The fbpA/sapM double knock out strain of Mycobacterium tuberculosis is highly attenuated and immunogenic in macrophages. PLoS One 2012; 7:e36198; PMID:22574140; http://dx.doi.org/10.1371/journal.pone.0036198
- Beresford N, Patel S, Armstrong J, Szoor B, Fordham-Skelton AP, Tabernero L. MptpB, a virulence factor from Mycobacterium tuberculosis, exhibits triple-specificity phosphatase activity. Biochem J 2007; 406:13-18; PMID:17584180; http://dx.doi.org/10.1042/BJ20070670
- Singh R, Rao V, Shakila H, Gupta R, Khera A, Dhar N, Singh A, Koul A, Singh Y, Naseema M, et al. Disruption of mptpB impairs the ability of Mycobacterium tuberculosis to survive in guinea pigs. Mol Microbiol 2003; 50:751-762; PMID:14617138; http://dx.doi.org/10.1046/j.1365-2958.2003.03712.x
- Toulabi L, Wu X, Cheng Y, Mao Y. Identification and structural characterization of a Legionella phosphoinositide phosphatase. J Biol Chem 2013; 288:24518-24527; PMID:23843460; http://dx.doi.org/10.1074/jbc.M113.474239
- Hsu F, Zhu W, Brennan L, Tao L, Luo ZQ, Mao Y. Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase. Proc Natl Acad Sci U S A 2012; 109:13567-13572; PMID:22872863; http://dx.doi.org/10.1073/pnas.1207903109
- Jahraus A, Storrie B, Griffiths G, Desjardins M. Evidence for retrograde traffic between terminal lysosomes and the prelysosomal/late endosome compartment. J Cell Sci 1994; 107:145-157; PMID:8175904.
- Lu N, Shen Q, Mahoney TR, Neukomm LJ, Wang Y, Zhou Z. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol 2012; 10:e1001245; PMID:22272187; http://dx.doi.org/10.1371/journal.pbio.1001245
- Chua J, Deretic V. Mycobacterium tuberculosis reprograms waves of phosphatidylinositol 3-phosphate on phagosomal organelles. J Biol Chem 2004; 279:36982-36992; PMID:15210698; http://dx.doi.org/10.1074/jbc.M405082200
- Hammond GRV, Balla T. Polyphosphoinositide binding domains: Key to inositol lipid biology. Biochim Biophys Acta 2015; 1851:746-758.
- Hammond GRV, Takasuga S, Sasaki T, Balla T. The ML1Nx2 Phosphatidylinositol 3,5-Bisphosphate Probe Shows Poor Selectivity in Cells. PLoS One 2015; 10:e0139957; PMID:26460749; http://dx.doi.org/10.1371/journal.pone.0139957
- Bohdanowicz M, Cosío G, Backer JM, Grinstein S. Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes. J Cell Biol 2010; 191:999-1012; PMID:21115805; http://dx.doi.org/10.1083/jcb.201004005
- Botelho RJ, Teruel M, Dierckman R, Anderson R, Wells A, York JD, Meyer T, Grinstein S. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol 2000; 151:1353-1368; PMID:11134066; http://dx.doi.org/10.1083/jcb.151.7.1353
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010; 11:329-341; PMID:20379207; http://dx.doi.org/10.1038/nrm2882
- Vieira OV, Bucci C, Harrison RE, Trimble WS, Lanzetti L, Gruenberg J, Schreiber AD, Stahl PD, Grinstein S. Modulation of Rab5 and Rab7 recruitment to phagosomes by phosphatidylinositol 3-kinase. Mol Cell Biol 2003; 23:2501-2514; PMID:12640132; http://dx.doi.org/10.1128/MCB.23.7.2501-2514.2003
- Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J Cell Biol 2001; 155:19-25; PMID:11581283; http://dx.doi.org/10.1083/jcb.200107069
- Fratti RA, Backer JM, Gruenberg J, Corvera S, Deretic V. Role of phosphatidylinositol 3-kinase and Rab5 effectors in phagosomal biogenesis and mycobacterial phagosome maturation arrest. J Cell Biol 2001; 154:631-644; PMID:11489920; http://dx.doi.org/10.1083/jcb.200106049
- Siddhanta U, McIlroy J, Shah A, Zhang Y, Backer JM. Distinct roles for the p110alpha and hVPS34 phosphatidylinositol 3′-kinases in vesicular trafficking, regulation of the actin cytoskeleton, and mitogenesis. J Cell Biol 1998; 143:1647-1659; PMID:9852157; http://dx.doi.org/10.1083/jcb.143.6.1647
- Thi EP, Lambertz U, Reiner NE. Class IA phosphatidylinositol 3-kinase p110a regulates phagosome maturation. PLoS One 2012; 7:e43668; PMID:22928013; http://dx.doi.org/10.1371/journal.pone.0043668
- Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 2014; 16:1069-1079; PMID:25327288; http://dx.doi.org/10.1038/ncb3053
- Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol 2014; 10:1013-1019; PMID:25326666; http://dx.doi.org/10.1038/nchembio.1681
- Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shapiro N, Ward R, Cross D, Ganley IG, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J 2014; 463:413-427; PMID:25177796; http://dx.doi.org/10.1042/BJ20140889
- Bojjireddy N, Botyanszki J, Hammond G, Creech D, Peterson R, Kemp DC, Snead M, Brown R, Morrison A, Wilson S, et al. Pharmacological and genetic targeting of the PI4KA enzyme reveals its important role in maintaining plasma membrane phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate levels. J Biol Chem 2014; 289:6120-6132; PMID:24415756; http://dx.doi.org/10.1074/jbc.M113.531426
- Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell 2006; 125:733-747; PMID:16647110; http://dx.doi.org/10.1016/j.cell.2006.03.035
- Kim GHE, Dayam RM, Prashar A, Terebiznik M, Botelho RJ. PIKfyve inhibition interferes with phagosome and endosome maturation in macrophages. Traffic 2014; 15:1143-1163; PMID:25041080; http://dx.doi.org/10.1111/tra.12199
- Sbrissa D, Ikonomov OC, Shisheva A. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J Biol Chem 1999; 274:21589-21597; PMID:10419465; http://dx.doi.org/10.1074/jbc.274.31.21589
- Voss MD, Czechtizky W, Li Z, Rudolph C, Petry S, Brummerhop H, Langer T, Schiffer A, Schaefer HL. Discovery and pharmacological characterization of a novel small molecule inhibitor of phosphatidylinositol-5-phosphate 4-kinase, type II, beta. Biochem Biophys Res Commun 2014; 449:327-331; PMID:24845568; http://dx.doi.org/10.1016/j.bbrc.2014.05.024
- Clarke JH, Giudici ML, Burke JE, Williams RL, Maloney DJ, Marugan J, Irvine RF. The function of phosphatidylinositol 5-phosphate 4-kinase g explored using a specific inhibitor that targets the PI5P-binding site. Biochem J 2015; 466:359-367; PMID:25495341; http://dx.doi.org/10.1042/BJ20141333
- Mak LH, Vilar R, Woscholski R. Characterisation of the PTEN inhibitor VO-OHpic. J Chem Biol 2010; 3:157-163; PMID:21643420; http://dx.doi.org/10.1007/s12154-010-0041-7
- Pirruccello M, Nandez R, Idevall-Hagren O, Alcazar-Roman A, Abriola L, Berwick SA, Lucast L, Morel D, De Camilli P. Identification of inhibitors of inositol 5-phosphatases through multiple screening strategies. ACS Chem Biol 2014; 9:1359-1368; PMID:24742366; http://dx.doi.org/10.1021/cb500161z
- Suwa A, Yamamoto T, Sawada A, Minoura K, Hosogai N, Tahara A, Kurama T, Shimokawa T, Aramori I. Discovery and functional characterization of a novel small molecule inhibitor of the intracellular phosphatase, SHIP2. Br J Pharmacol 2009; 158:879-887; PMID:19694723; http://dx.doi.org/10.1111/j.1476-5381.2009.00358.x
- Hazeki K, Nigorikawa K, Takaba Y, Segawa T, Nukuda A, Masuda A, Ishikawa Y, Kubota K, Takasuga S, Hazeki O. Essential roles of PIKfyve and PTEN on phagosomal phosphatidylinositol 3-phosphate dynamics. FEBS Lett 2012; 586:4010-4015; PMID:23068606; http://dx.doi.org/10.1016/j.febslet.2012.09.043
- Bohdanowicz M, Balkin DM, De Camilli P, Grinstein S. Recruitment of OCRL and Inpp5B to phagosomes by Rab5 and APPL1 depletes phosphoinositides and attenuates Akt signaling. Mol Biol Cell 2012; 23:176-187; PMID:22072788; http://dx.doi.org/10.1091/mbc.E11-06-0489
- Endemann GC, Graziani A, Cantley LC. A monoclonal antibody distinguishes two types of phosphatidylinositol 4-kinase. Biochem J 1991; 273:63-66; PMID:1846531; http://dx.doi.org/10.1042/bj2730063
- Kamen LA, Levinsohn J, Cadwallader A, Tridandapani S, Swanson JA. SHIP-1 increases early oxidative burst and regulates phagosome maturation in macrophages. J Immunol 2008; 180:7497-7505; PMID:18490750; http://dx.doi.org/10.4049/jimmunol.180.11.7497
- Ma J, Becker C, Reyes C, Underhill DM. Cutting edge: FYCO1 recruitment to dectin-1 phagosomes is accelerated by light chain 3 protein and regulates phagosome maturation and reactive oxygen production. J Immunol 2014; 192:1356-1360; PMID:24442442; http://dx.doi.org/10.4049/jimmunol.1302835
- Hoeglinger D, Nadler A, Schultz C. Caged lipids as tools for investigating cellular signaling. Biochim Biophys Acta 2014; 1841:1085-1096; PMID:24713581; http://dx.doi.org/10.1016/j.bbalip.2014.03.012
- Ozaki S, DeWald DB, Shope JC, Chen J, Prestwich GD. Intracellular delivery of phosphoinositides and inositol phosphates using polyamine carriers. Proc Natl Acad Sci U S A 2000; 97:11286-11291; PMID:11005844; http://dx.doi.org/10.1073/pnas.210197897
- Subramanian D, Laketa V, Müller R, Tischer C, Zarbakhsh S, Pepperkok R, Schultz C. Activation of membrane-permeant caged PtdIns(3)P induces endosomal fusion in cells. Nat Chem Biol 2010; 6:324-326; PMID:20364126; http://dx.doi.org/10.1038/nchembio.348
- Raiborg C, Schink KO, Stenmark H. Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J 2013; 280:2730-2742; PMID:23289851; http://dx.doi.org/10.1111/febs.12116
- Wang H, Sun HQ, Zhu X, Zhang L, Albanesi J, Levine B, Yin H. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci U S A 2015; 112:7015-7020; PMID:26038556; http://dx.doi.org/10.1073/pnas.1507263112
- Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. PI(5)P regulates autophagosome biogenesis. Mol Cell 2015; 57:219-234; PMID:25578879; http://dx.doi.org/10.1016/j.molcel.2014.12.007
- Block MR, Glick BS, Wilcox CA, Wieland FT, Rothman JE. Purification of an N-ethylmaleimide-sensitive protein catalyzing vesicular transport. Proc Natl Acad Sci U S A 1988; 85:7852-7856; PMID:3186695; http://dx.doi.org/10.1073/pnas.85.21.7852
- Mayer A, Wickner W, Haas A. Sec18p (NSF)-driven release of Sec17p (alpha-SNAP) can precede docking and fusion of yeast vacuoles. Cell 1996; 85:83-94; PMID:8620540; http://dx.doi.org/10.1016/S0092-8674(00)81084-3
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell 1998; 92:759-772; PMID:9529252; http://dx.doi.org/10.1016/S0092-8674(00)81404-X
- Becken U, Haas A. In vitro fusion asssays with phagosomes; Intracellular Niches of Microbes, WILEY-VHC Press, Weinheim, Germany 2009; 95-105.
- Blocker A, Severin FF, Habermann A, Hyman AA, Griffiths G, Burkhardt JK. Microtubule-associated protein-dependent binding of phagosomes to microtubules. J Biol Chem 1996; 271:3803-3811; PMID:8631997; http://dx.doi.org/10.1074/jbc.271.7.3803
- Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol 2004; 167:1087-1098; PMID:15611334; http://dx.doi.org/10.1083/jcb.200409068
- Mayer A, Scheglmann D, Dove S, Glatz A, Wickner W, Haas A. Phosphatidylinositol 4,5-bisphosphate regulates two steps of homotypic vacuole fusion. Mol Biol Cell 2000; 11:807-817; PMID:10712501; http://dx.doi.org/10.1091/mbc.11.3.807
- Lorente-Rodríguez A, Barlowe C. Requirement for Golgi-localized PI(4)P in fusion of COPII vesicles with Golgi compartments. Mol Biol Cell 2011; 22:216-229; http://dx.doi.org/10.1091/mbc.E10-04-0317
- Petiot A, Faure J, Stenmark H, Gruenberg J. PI3P signaling regulates receptor sorting but not transport in the endosomal pathway. J Cell Biol 2003; 162:971-979; PMID:12975344; http://dx.doi.org/10.1083/jcb.200303018
- Mills IG, Jones AT, Clague MJ. Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr Biol 1998; 8:881-884; PMID:9705936; http://dx.doi.org/10.1016/S0960-9822(07)00351-X
- Shiozawa K, Goda N, Shimizu T, Mizuguchi K, Kondo N, Shimozawa N, Shirakawa M, Hiroaki H. The common phospholipid-binding activity of the N-terminal domains of PEX1 and VCP/p97. FEBS J 2006; 273:4959-4971; PMID:17018057; http://dx.doi.org/10.1111/j.1742-4658.2006.05494.x
- Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J 2006; 25:1579-1589; PMID:16601699; http://dx.doi.org/10.1038/sj.emboj.7601051
- Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjørkøy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 2010; 188:253-269; PMID:20100911; http://dx.doi.org/10.1083/jcb.200907015
- Dai S, Zhang Y, Weimbs T, Yaffe MB, Zhou D. Bacteria-generated PtdIns(3)P recruits VAMP8 to facilitate phagocytosis. Traffic 2007; 8:1365-1374; PMID:17645435; http://dx.doi.org/10.1111/j.1600-0854.2007.00613.x
- Jovic M, Kean MJ, Szentpetery Z, Polevoy G, Gingras AC, Brill JA, Balla T. Two phosphatidylinositol 4-kinases control lysosomal delivery of the Gaucher disease enzyme, b–glucocerebrosidase. Mol Biol Cell 2012; 23:1533-1545; PMID:22337770; http://dx.doi.org/10.1091/mbc.E11-06-0553
- Sridhar S, Patel B, Aphkhazava D, Macian F, Santambrogio L, Shields D, Cuervo AM. The lipid kinase PI4KIIIb preserves lysosomal identity. EMBO J 2013; 32:324-339; PMID:23258225; http://dx.doi.org/10.1038/emboj.2012.341
- Defacque H, Bos E, Garvalov B, Barret C, Roy C, Mangeat P, Shin HW, Rybin V, Griffiths G. Phosphoinositides regulate membrane-dependent actin assembly by latex bead phagosomes. Mol Biol Cell 2002; 13:1190-1202; PMID:11950931; http://dx.doi.org/10.1091/mbc.01-06-0314
- Wickner W. Membrane fusion: five lipids, four SNAREs, three chaperones, two nucleotides, and a Rab, all dancing in a ring on yeast vacuoles. Annu Rev Cell Dev Biol 2010; 26:115-136; PMID:20521906; http://dx.doi.org/10.1146/annurev-cellbio-100109-104131
- Bennett TL, Kraft SM, Reaves BJ, Mima J, O'Brien KM, Starai VJ. LegC3, an effector protein from Legionella pneumophila, inhibits homotypic yeast vacuole fusion in vivo and in vitro. PLoS One 2013; 8:e56798; PMID:23437241; http://dx.doi.org/10.1371/journal.pone.0056798
- Sreelatha A, Bennett TL, Carpinone EM, O'Brien KM, Jordan KD, Burdette DL, Orth K, Starai VJ. Vibrio effector protein VopQ inhibits fusion of V-ATPase-containing membranes. Proc Natl Acad Sci U S A 2015; 112:100-105; PMID:25453092; http://dx.doi.org/10.1073/pnas.1413764111
- Ellson CD, Anderson KE, Morgan G, Chilvers ER, Lipp P, Stephens LR, Hawkins PT. Phosphatidylinositol 3-phosphate is generated in phagosomal membranes. Curr Biol 2001; 11:1631-1635; PMID:11676926; http://dx.doi.org/10.1016/S0960-9822(01)00447-X
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature 1999; 397:621-625; PMID:10050856; http://dx.doi.org/10.1038/17618
- Lawe DC, Chawla A, Merithew E, Dumas J, Carrington W, Fogarty K, Lifshitz L, Tuft R, Lambright D, Corvera S. Sequential roles for phosphatidylinositol 3-phosphate and Rab5 in tethering and fusion of early endosomes via their interaction with EEA1. J Biol Chem 2002; 277:8611-8617; PMID:11602609; http://dx.doi.org/10.1074/jbc.M109239200
- Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol 151:601-612; PMID:11062261; http://dx.doi.org/10.1083/jcb.151.3.601
- Pizarro-Cerdá J, Payrastre B, Wang YJ, Veiga E, Yin HL, Cossart P. Type II phosphatidylinositol 4-kinases promote Listeria monocytogenes entry into target cells. Cell Microbiol 2007; 9:2381-2390; http://dx.doi.org/10.1111/j.1462-5822.2007.00967.x
- Shin HW, Hayashi M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, Modregger J, Uttenweiler-Joseph S, Wilm M, Nystuen A, et al. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol 2005; 170:607-618; PMID:16103228; http://dx.doi.org/10.1083/jcb.200505128
- Segawa T, Hazeki K, Nigorikawa K, Morioka S, Guo Y, Takasuga S, Asanuma K, Hazeki O. Inpp5e increases the Rab5 association and phosphatidylinositol 3-phosphate accumulation at the phagosome through an interaction with Rab20. Biochem J 2014; 464:365-375; PMID:25269936; http://dx.doi.org/10.1042/BJ20140916
