ABSTRACT
Spontaneous variation in appearance was studied in bacterial colonies of Serratia marcescens F morphotypeCitation1: (i) A defined array of non-heritable phenotype variations does appear repeatedly; (ii) The presence of colonies of different bacterial species will narrow the variability toward the typical F appearance, as if such an added environmental factor curtailed the capacity of colony morphospace; (iii) Similarly the morphospace becomes reduced by random mutations leading to new, heritable morphotypes—at the same time opening a new array of variations typical for the mutant but not accessible directly from the original F morphospace. Results are discussed in context with biphasic model of early morphogenesis applicable to all multicellular bodies.
Introduction
Microbial colonies represent macroscopic bodies usually originated by coordinated growth from a single cell or a group of cells belonging to a single species (or clone). They are commonplace in basic and applied biological research, and serve, before all, for diagnostics based on the shape, growth on defined media, gene expression, antibiotic resistance, mutagenesis, genetic polymorphism (heritable or not), etc.Citation2-5 And yet, their very existence, morphogenesis, and structure became a matter of interest only to a handful of investigators (e.g. refs. Citation1, 6-15), even if processes in the background are far from being understood in detail. Changes in colony shape were studied, caused by mutations either in mother colony, or upon long-term cultivationCitation16-18
Bacteria typically dwell not in colonies but in multi-species consortia, like biofilms, stromatolites, bowel assemblages, etc., where isolation—the necessary precondition for early morphogenesis—cannot be guaranteed. Obviously, in such ecosystems they are unable to display structures typical for a colony, like symmetry, coloring and/or structural patterning, defined growth pattern, or maintaining its individuality when faced with neighbors.Citation14 Their survival and reproduction under such conditions is thus independent on their capacity of colony-building, and natural selection may be indifferent toward such traits. From this viewpoint, bacterial colonies may be taken for (i) an extremely reduced biofilm, or (ii) a multicellular body with controlled morphogenesis that cannot come to realization when hampered by the presence of partners in the consortium. Our previous work encourages us to prefer the second interpretation.Citation1,13,14 What is important, ontogenesis of multicellular body is uncoupled from natural selection: an ability to build colony is rarely a matter of survival—hence rich patterns and the play of variations.
The colony shape even in well-established bacterial clones exerts an array of variations typical for a “morphospace” of a given clone. Variations may be grounded in stochastic deviations from a “norm” (reversible in the next generation), or caused by mutations that appear with a given frequency—in that case they become irreversible. Here we pay attention to such variability and its constraints, and map the space of possibilities (i.e. the morphospace) available for both kinds of variability. The results suggest that bacterial colonies may become a valuable model for the morphogenesis of multicellular organisms.
Results
Our model organism, Serratia marcescens, morphotype F, gives under defined conditions colonies with finite growth, with typical fountain-like shaping and pigmentation (; for details see ref. Citation1). The pattern is stable in the course of many passages, and develops regardless whether the colony originates from a single cell, bunch of biomass or droplet of dense suspension.
Figure 1. Serratia marcescens clone F characteristics. (A) Scheme of an ideal F colony consisting of central navel, interstitial ring and rim; central navel and outer rim are pigmented, smooth and growing up to approx. 3 mm height, interstitial ring is non-pigmented, rough by texture and grows only to height of approximately 0.5 mm. Time course of colony development, days of growth indicated. (B) Illustration of variability of F pattern (at 7 days); Right: compositional image of the same set. (C) Continuum of growth types from undifferentiated mass to fully developed colonies (day 7); i) fully developed colonies, ii) small red colonies, iii) continuous undifferenced growth–macula. (D) X-structure (broad white rim) formation around F colony in the presence of Serratia rubidaea. Bar = 1 cm.
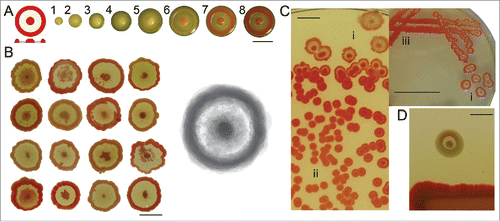
Our perception of ideal shape of the F colony (i.e., symmetrical pattern with red central navel, white interstitial ring and red outer rim) as portrayed in the results from our experience with about 105 colonies. Typical appearance of real F colonies may, however, deviate from the “ideal.” Of course, real shape F will vary around such an ideal type; a composite overlap of real colonies, however gives an image tending to the “true type” ().
Developmental variation of F colonies
Here we describe types of deviation from ideal F shape. Variations are best apparent in the area of interstitial ring (), and in symmetry breaking of the whole colony ().
Figure 2. Deviations from a typical F pattern. (A) Secondary outgrowths in the interstitial ring: (i) near the central navel: (ii) in its mid-part; (iii) a film covering part of it (day 7). (B) Induced growth in the ring: (i) mechanical damage at day 5, one and 3 d after the injury. (ii) Transfer of colony growing 5 d at 35°C, to 27°C: day 7 upon transfer. (C) Deflections of colony symmetry, day 7. Bar = 1 cm.
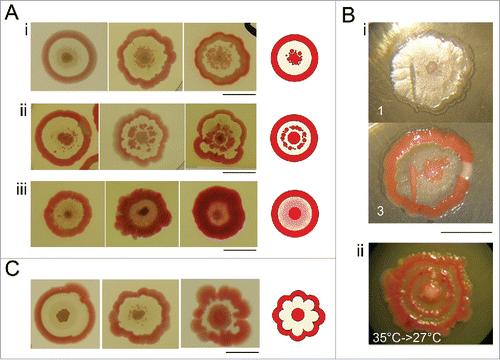
Interstitial ring possesses high potency of secondary outgrowths of either pigmented formations or thin pigmented film. Such secondary outgrowths may appear during both the growth and maturation of the colony. Pigmented formations resemble structure of the central navel and/or outer rim (i.e., pigmented, smooth and elevated; ). Secondary growth may appear in area adjacent to the central navel and even continually fuse with it, or it may appear in the mid-part of interstitial ring and form discontinuous inner circle (). Finally, the entire interstitial ring may be overgrown by smooth, pigmented film ().
If plated on the fresh dish, cells taken from such secondary structures give rise to typical F colonies, indicating that their presence is not a consequence of mutation. Beside these spontaneous changes, interstitial ring bears potency to form secondary growth induced by mechanical injury, or induced by transfer between 2 temperature regimes (35° to 27°C; ).
Symmetry breaking of F colony usually appears as lacy formation of outer rim (). Such pattern may result from incomplete observance of synchronicity of radial growth of the colony.
Sectors
Another type of aberration of typical F colony shape and its symmetry is formation of wedge-shaped sectors ().
Figure 3. Sectored colonies. (A) Sectors may originate from the center (i), interstitial ring (ii), or from the rim of the colony (iii) (day 7). (B) Sectored colony (i) and its progeny: biomass from the F part of sectored colony gives a typical F pattern; colony biomass from the sector forms smooth convex, uniformly pigmented colony; mixed material from the whole colony (ii) gives convex colonies, or typical F colonies (9 cm Petri dish displayed, day 7). (C) Morphogenesis of rdf variant obtained from progeny of smooth red sector of F colony. Development in days 2, 4, and 7, with schemes of profiles. At the beginning, the morphogenesis proceeds similar to F, afterwards it strives toward smooth convex and uniformly pigmented colonies. Bar = 1 cm.
Sectors may originate during growth at different distances from the center (); and apparently represent mutational events: cells from such sectors give rise to non-profiled, uniformly pigmented or non-pigmented colonies. However, there is no hereditary tendency toward generating sectors in F colonies: we were not able to obtain isolates that would give rise to sectors with higher frequencies, neither changes of cultivation parameters led to changes in frequency of mutants. Majority of colonies from a single inoculation either does or do not bear sectors.
As an example of progeny from pigmented sector, shows the stable variant rdf with convex and uniformly pigmented colonies. In early phases of development, its shape resembles a typical F colony, i.e., white colony with elevated center. From the day 3, the growth continues toward smooth unprofiled body with uniform pigmentation.
Prolonged cultivations in liquid medium
Long-term cultivations (weeks to months) of F suspension in liquid media led to a spontaneous appearance of several new and stable phenotypes (). Those new variants may be divided into 2 groups – variants that retain typical F features (K, Kw, D and pF; ) and “unprofiled” variants that grow as convex, uniformly pigmented colonies (rd, M; ).
Figure 4. Variants obtained upon long-term cultivation in liquid media. (A) Profiled variants with secondary growth near central navel: pigmented K and non-pigmented Kw (day 7); variant D with continuous secondary growth in the mid-part of interstitial ring (day 4 and 7); variant pF with its interstitial ring overgrown with pigmented film (day 7). (B) Convex variants: variant rd with convex profile from the beginning of growth (days 2, 4, and 7); variant M (day 7). Bar = 1 cm.
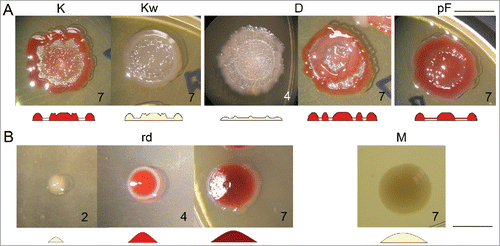
New variants with typical F features (dubbed “profiled” here) are sometimes hardly distinguishable from a typical F pattern (), as they show specific secondary growth in interstitial ring resembling those seen in typical Fs (). However, the pattern is heritable and uniform in subsequent generations. They do not grow beyond typical F diameter.
Sister variants K (pigmented) and Kw (non-pigmented) form secondary structures in the interstitial ring adjacent to central navel. Their growth starts in early phase of morphogenesis and in mature colony the central navel continually blends with those structures and may lose its contours. Pigmentation of both navel and secondary structures takes place in parallel in K.
Double-rim variant D forms additional pigmented circle in mid-part of interstitial ring. Development of this variant, however, differs from development of typical F colonies. It starts as a thin colony without elevated center. Around 3rd day, base of internal circle forms, either as a full circle or as discontinuous dots, that merges as colony maturates. Only after the inner circle becomes visible, center is about to appear, and subsequently the outer rim grows.
Variant pF resembles development of F colony, but from 3rd day on, the ring becomes overgrown by a thin pigmented film. However, fountain-like overall shape remains.
Another type of variants is represented by unprofiled (convex) colony that lose ability to form interstitial ring. The variant rd forms uniformly pigmented smooth convex colony. It resembles rdf variant, but has a smaller diameter with higher elevation in the center. Also its morphogenesis differs from rdf variant, rd grows as a smooth, convex, non-pigmented colony and from 3rd day becomes pigmented.
Variant M was obtained by the procedure differing from those above. Clone F do not grow (but persists) in minimal medium. Variant M appeared after several cycles of cultivation of F clone in minimal medium and plating on nutrient agar; the clone is able to grow in minimal media. Both its colony shape and other properties differ from all other variant descending from F origin.
Manipulating variability
Density of colonies per plate influences F colony development, its final shape (; see also refs. Citation1, Citation13) and also tendency to form deviations. Abovementioned typical development of F colony occurs only when colony density does not exceed 20–30 bodies on 9 cm Petri dish. Otherwise, it is generally true that the more dense plating, the smaller colonies and the faster their development (). When density of colonies exceeds ca 500 per 9 cm Petri dish, only small red colonies without typical profilation will develop (), and stop their growth within 2 or 3 d.
Table 1. Colony development according to density of colonies.
Development of F colonies is thus realized in dependence on area of substrate available, from unprofiled red dots to fully developed F colonies (with a maximal perimeter about 20 mm when plated as a single colony per 9 cm dish). shows situation, where the density of colonies decreases from left to right: as the area available for the colony increases, so does colony size, hand in hand with its profilation (see ref. Citation13). Note that in small colonies, tendency to form deviations from ideal F pattern is less apparent. Also manifestation of sectored pattern seems to diminish in smaller colonies (; ).
Heterospecific body in the vicinity
When F colonies grow in the presence of a heterospecific neighbor (S. rubidaea, E. coli), an outer white rim will appear, dubbed X-structure, around F colonies (; ref. Citation14). This reaction is not caused by newly obtained variants, thus we consider them as homospecific to F. The only exception is the variant M, which acts as if heterospecific, albeit originating from clone F (as confirmed by MALDI-TOF analysisCitation14).
We observed the effect of heterospecific neighbors (S. rubidaea, E. coli and S. marcescens clone M) on the appearance of typical F colonies, with their deviations from ideal shape and sectored pattern (); as well as on newly obtained variants (K, D, pF; ).
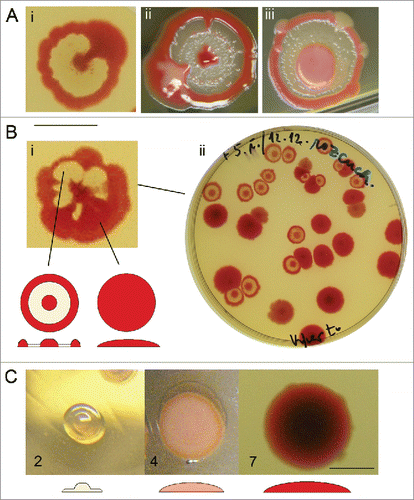
Figure 5. Effect of a heterospecific neighbor on the deviations from the F pattern and on the formation of sectors. (A) Secondary growth in the interstitial ring in uninfluenced control (i) and colonies originating from the same inoculum, but with a S. rubidaea macula neighbor (6th day). (B) Sectored pattern present in uninfluenced controls (i, iii) or controls grown in the presence of homospecific macula – S. marcescens clone F (v); Inocula taken from material in controls (left) were placed in the neighborhood of heterospecific maculae (S. rubidaea – ii; E. coli – iv; S. marcescens clone M – vi) which resulted in strong suppression of sector formation. All photos taken at the 7th day of growth. Bar = 1 cm.
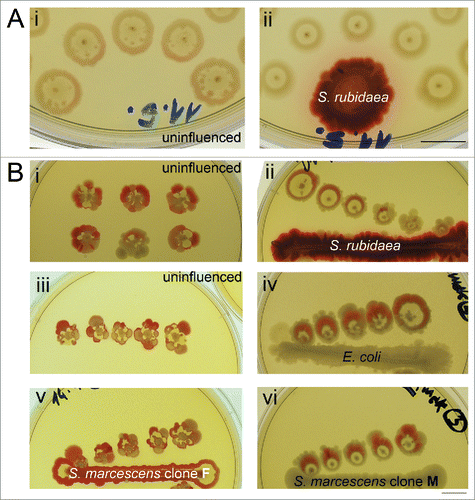
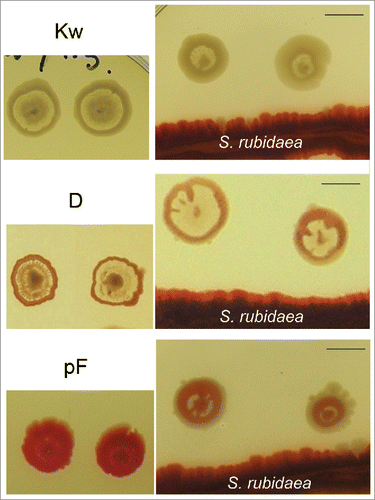
Figure 6. Influence of a heterospecific neighbor on profiled variants. Left: colonies of Kw, D and pF; right, the same variants growing in the presence of S. rubidaea – as a result, typical features of interstitial ring are suppressed (day 7). Bar = 1 cm.
The heterospecific neighbor has a profound influence on both colony symmetry and secondary growth in the interstitial ring. The presence of macula of S. rubidaea nearby developing F colonies leads to loss of the secondary growth in interstitial ring, compared with uninfluenced colonies (). Moreover, the presence of the heterospecific neighbor (S. rubidaea, E. coli and S. marcescens clone M) led to suppression of the sector formation compared with uninfluenced colonies from single inoculum (). The progeny of such “disciplined” colonies is also uniformly F, without the presence of any mutated pattern.
Similarly, variants K, D and pF exposed to heterospecific neighbor (), secondary growth in the interstitial ring typical for these variants was substantially suppressed, strongly forcing the colony pattern toward forms resembling standard F colony. Variants rd and rdf growing in the presence of heterospecific neighbor develop X-structures as in former cases, their colonies, however, remain convex (not-shown).
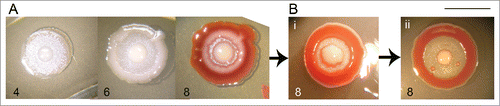
Progeny of heterospecifically treated K, D and pF variants is again K, D and pF phenotype. Only in one single case, a new variant with altered phenotype was obtained as a progeny of treated D colony (). This variant called wpr forms fountain-profiled colony with broad non-pigmented center, narrow interstitial ring and broad rim, which is not pigmented on inner side. Morphogenesis of this phenotype resembles that of F colony until the 6th day. This variant is however not stable: after further passages it tends more to F pattern, but without pigmentation of the center.
Figure 7. Variant wpr – example of variability fading away. (A) First generation, days 4, 6, and 8. (B) Second (i) and third generation (ii), day 8. In subsequent passages, the rim becomes thinner and the interstitial ring broader to an extent comparable to typicalF. Bar = 1 cm.
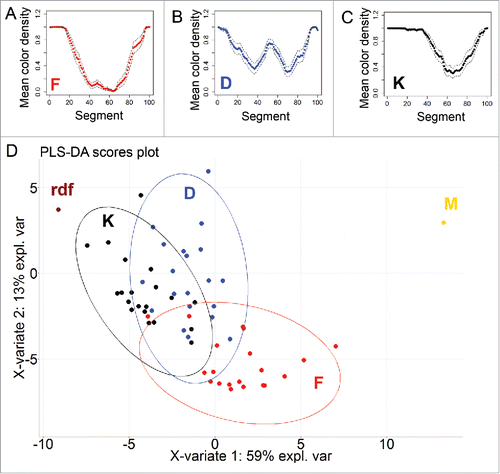
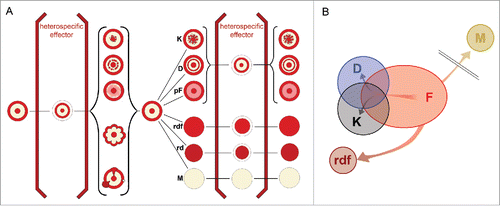
shows the normalized extent of normalized variants in the morphotype F and 4 mutant morphotypes derived from it. Whereas the morphospace of M and rdf collapsed to a moonotonous pattern beyond the set available to F, mutants D and K retain the subset of original variations exerted by F, expanding at the same time to new state spaces not available to F. The overlapping areas of the 3 morphospaces represent the “kernel” of variants that are accessible to both mother morphotype and its derivatives.
Figure 8. Statistical evaluation of differences between colonies of different variants. (A-C) Standardized profiles of 3 different variant (F, D and K) based on colony pigmentation, dashed lines denotes 95% confidence interval. (D) PLS-DA clustering based on pigmentation as viewed above, while rdf variant is taken as pigmented over the whole area and M variant as entirely non-pigmented; ellipses indicate 95% confidence region.
Discussion
In previous papers, we studied the variability of a selected clone F and its malleability by the environment: its area available to growth; effects of medium composition; density of the substrate; temperature; the presence of neighbors, homospecific or not; signals carried by both medium and atmosphere etc.Citation1,12-15,20 It is self-evident that we cared for the constancy of the clone: it was stored deep frozen and periodically renewed after a couple of transmissions. Rare spontaneous deviations from the typical colony appearance (that always appear in cultivation) were not studied further and were excluded from experiments—yet meticulously documented.
In contrast, in the present paper we focused our attention to so frequent and mostly unwelcome variability and tried to uncover some regularities (if any) in pattern distribution. We analyzed protocols accumulated over 10 y and established new experiments, in attempts to induce and/or increase both frequency and repertoire of variants. The results can be divided into 3 groups:
(i) Small random, nonheritable deviations from the standard F pattern (), apparently originating in imperfect synchronization of growth.Citation15,21 (ii) Red or white sectors appearing at random in otherwise standard colony () represent mutant clones that lost the original variation potential of the mother (F) morphotype, giving rise to heritable smooth red or white colonies. (iii) Heritable and stable, i.e., mutant, morphotypes were isolated also upon a prolonged cultivation in liquid medium (). A single exception is the morphotype wpr () tended toward the F morphotype after several generations.
Comparison of groups (i) and (iii) (see cartoon in ) reveals that both produce similar variants; the set (i), however, is much richer in such patterns than the set (iii). What is also common to both sets is their “disciplining” in the presence of a heterospecific neighbor(s). This is especially apparent when a foreign body enters the game: the play of variants becomes reduced to the original motif F surrounded by an X-structure, without producing baroque structural “excesses.” All profiled forms apparently remember their F-origin, whereas convex smooth mutants either escaped the boundaries of the variation space available to F, or they may represent a reversal to some more ancestral forms older than F.
Figure 9. Summary of effects of heterospecific neighbor on both hereditary and non-hereditary variants of the F colony. The left side of the scheme shows examples of non-hereditary variability, like disturbances of the ring, or deviations of symmetry (), or productions of sectors (Fig. 3). All such forms can be “brought into line” when a heterospecific macula is present in the common substrate; such a disciplining is accompanied by the formation of X-structure (). The progeny of such colonies give standard F colony. The right side of the diagram gives examples of hereditary variability: rimmed variants K, D, and pF (), and rimless forms rdf (), rd and M(). Only rimmed variants react to the presence of heterospecific influence by a reversal toward the form F, accompanied by the X-structure (). The progeny of such disciplined forms is again—in contrast to non-hereditary variants—the default hereditary rimmed form. (B) Schematic projection of the distribution of state spaces of variations in 5 morphotypes analyzed in .
Back to the long-term cultivations (iii): by such—quite drastic—treatment we obtained forms similar to the set (i) above, but stable. They, however, all belonged to a subset of the much broader array of forms observed in non-hereditary variations of the mother morphotype F: mutants (with an exception of M) did not escape from the manifold of variability typical for F, they represent rather limited but fixed subset from it. Such a narrowing of possibilities, however, enables the mutant to explore a new space of morphological possibilities – beyond that available to F, as documented in this and in previous publications (refs. Citation1, 12-15, 20). Whereas the “basic” appearance of such mutants is tuned to the mother phenotype F, it will produce a manifold of new possibilities not available to F.
Colony shaping
In ref. Citation14 we speculatively divide the early morphogenesis of multicellular bodies (including bacterial colonies) into 2 phases. In the first stage the newly established germ must remain insulated from the rest of biosphere. In contrast, the second phase is characterized by establishing manifold of liaisons with other dwellers in the biosphere. If the early phase is disturbed by other life, morphogenesis is aborted: this is what happens to bacteria living in multi-species consortia. In contrast, if it remains insulated during the second phase (germ-free or gnotobiotic organisms), its further development becomes usually flawed. In case of bacteria, however, isolation in the second phase will not lead to a collapse. On the contrary, the colony as if became freed from continuous ecological negotiations, and only at such rare occasions it can display its rich morphogenetic potential. This is why fully developed morphotype F (and its variants) will appear only if the colony is freed from biospheric interactions, i.e., is allowed to grow far from any disturbing neighbors (as is the case in consortia), with no or limited interaction between colonies. The colony therefore represents an analogy of “germ-free” organisms as described in animals and plants, and interactions between colonies are, in turn, similar to gnotobiotic arrangement.
The fragility of such a “germ-free germ” body is apparent if neighbor bodies (homo- or heterospecific) enter the game and influence (reciprocally) the “typical” morphogenesis. If densely sowed, the colony development is slowed down or even frozen (as a population of very small flat smooth colonies, ); even denser population leads to an undifferentiated cell mass – macula (). In the presence of heterospecific colonies in the neighborhood the development may get steered toward some structures—even new ones, as in case of structure X (). Interaction also may be demanding for resources—hence reduced play of variations in interacting bodies.
Why variability?
Variations in colony appearance may be caused by several factors:
| (i) | Random environmental fluctuations. Under experimental conditions used, most of environmental constraints fade away. Any random, minimal fluctuation in physical parameters, nutrients, or signals, may shift the colony appearance within the realm of space of possibilities. As such shifts do not influence the survival, changes do not matter. | ||||
| (ii) | Releasing the brakes of variability. Experimenting with variations may be costly or even dangerous. However, under released conditions mentioned above, internal potential toward self-manifestation (sensu PortmannCitation22) will become released. Again, from the point of survival the changes are unimportant, but in contrast to previous point, they do matter. It seems that any free surface can be assigned a function of such “semantic organ”Citation23 Bacterial colonies released from survival duties may represent non plus ultra semantic organs of a kind. | ||||
| (iii) | Natural selection may, however, be also a decisive factor for variation. Here, bacteria that do not produce dormant stages, must ensure their successful dispersal: attractivity, e.g., to bacteriovorous insects (like Drosophila) by color ornaments and olfactory cocktails.Citation20,24 may play a decisive role, similar to the games played between flowers and pollinators. Such a normative selection (reduction of variations) may take place in the presence of heterospecific bacterial bodies. | ||||
Materials and methods
The strain S. marcescens CNCTS 5965 was obtained from the Czech National Institute of Health. All morphotypes of S. marcescens displayed here are derived from F morphotype.Citation1 The identity of F and M strains was confirmed by MALDI - TOF method, using Bruker Daltonik MALDI Biotyper (performed by A. Nemec, National Health Institute, Prague); the scores assigned to particular strains S. marcescens (F = 2.151, M = 2.168) indicate very high probability of correct determination.Citation14 The strain of S. rubidaea and E. coli strain 281 were obtained from the collection of the Department of Genetics and Microbiology, Faculty of Sciences, Charles University.
Bacterial stocks have been maintained at −80°C as described previously.Citation1
Bacteria have been grown under previously described standard conditionsCitation1 on Nutrient Agar No2 (Imuna Pharm a.s., Order No T 382100001020) supplemented with 0.5% glucose (27 mM), or on a medium obtained by solidifying Nutrient broth No2 (Imuna Pharm a.s., Order No V 382100000098) by addition of 1.5% agar, supplemented with 0.5% glucose (27 mM), with the same results.
For long-term cultivation in liquid broth, Nutrient broth No2 (Imuna Pharm a.s., Order No V 382100000098), supplemented with 0.5% glucose (27mM) was used.
Minimal medium for used in experiment where M clone was isolated: 21 mM KH2PO4, 48 mM Na2HPO4, 8 mM NaCl, 18 mM NH4Cl, 3.9 mM MgSO4, 27 mM glucose.
New colonies were initiated as follows: (1) as clones from single cells, by classical sowing of bacterial suspension; (2) planted by dropping dense suspension (108/ml) on a defined place (diameter about 2 mm); (3) planted by dotting from material taken by a sterile needle from an older body; (4) by smearing (to grow maculae): 30 μl of bacterial suspension (approx. 108 cells) was applied to a line of approx. 5 cm.
Mechanical disruption of interstitial ring was performed by sterile needle, disrupting only bacterial mass without damaging underlying agar.
Composite image was crafted in Adobe Photoshop by adjusting all images of full grown colonies in a manner that all pigmented area were automatically set to black by color channel tool. Interstitial ring was left uncolored. All colonies were centered by the central navel; 16 colonies were used, each as separate layer with 6.25% opacity. Plates were photographed in situ using an Olympus C-5050ZOOM digital camera under ambient or penetrating light (Fomei, LP-400 light panel, cold cathode light) or under magnification using a binocular magnifier. Figuress shown were selected from an extensive collection of primary photos from several repetitions of each experiment. Photoshop software was used to assemble the plates but no image doctoring was performed except automatic adjustment of brightness and contrast in some cases.
To perform statistical evaluation, photograph of a colony was first analyzed by ImageJ software.Citation25 Image was turned to 8-bit image type and then threshold function was used to adjust all pigmented areas as black and non-pigmented area as white. Line approximately 1 mm thick was laid over the radius of colony from center to margin in random direction and plot profile function was used to generate data of color presence along this line. Subsequently, this absolute data was fitted to the length of 100 segments (“approx” function in RCitation26) and color density was adjusted ranging from 0 to 1.
For each variant (F, D, K), means for each segment were plotted and 95% confidence interval indicated. These data was also used to perform clustering by PLS-DA method (“plsda” function in R, mixOmics packageCitation27), that is widely used as discrimination method in chemometricsCitation28 or to reduce dimensions in highly correlated data e.g reflectance data.Citation29 Additionally, 2 other clones were considered: rdf variant, what is taken as pigmented over the whole area and M variant as entirely non-pigmented.
Conclusions
Three points emerge from the results presented: (1) Under controlled environment, the colony morphotype exerts an easily recognizable (and probably final) set of variations that will appear repeatedly, provided that the morphotype does not accumulate mutations; (2) If such mutations do occur, they result in fixation of some appearance out of the original extent of patterns and, at the same time, curbing of the spread of original variations; (3) the mutant subsequently explores the morphospace available from its new position – such a morphospace of patterns occupies a different field of possibilities than the original (F) morphotype.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Czech Science Foundation 13–24275S (AM) and the Internal Grant Agency of the Czech University of Life Sciences CIGA, grant no. 20164306 (JČ).
References
- Rieger T, Neubauer Z, Blahuskova A, Cvrčková F, Markos A. Bacterial body plans: colony ontogeny in Serratia marcescens. Commun Integr Biol 2008; 1:78-87; PMID:19513204; http://dx.doi.org/10.4161/cib.1.1.6547
- Buzalewicz I, Kujawińska M, Krauze W, Podbielska H. Novel Perspectives on the Characterization of Species-Dependent Optical Signatures of Bacterial Colonies by Digital Holography. PLoS One 2016; 11:e0150449; PMID:26943121; http://dx.doi.org/10.1371/journal.pone.0150449
- von Eiff C, Peters G, Becker K. The small colony variant (SCV) concept—the role of staphylococcal SCVs in persistent infections. Injury 2006; 37:S26-33; PMID:16651068; http://dx.doi.org/10.1016/j.injury.2006.04.006
- Sousa AM, Pereira MO, Lourenço A. MorphoCol: An ontology-based knowledgebase for the characterisation of clinically significant bacterial colony morphologies. J Biomed Inform 2015; 55:55-63; PMID:25817920; http://dx.doi.org/10.1016/j.jbi.2015.03.007
- Sousa AM, Machado I, Nicolau A, Pereira MO. Improvements on colony morphology identification towards bacterial profiling. J Microbiol Methods 2013; 95:327-35; PMID:24121049; http://dx.doi.org/10.1016/j.mimet.2013.09.020
- Shapiro JA. The significances of bacterial colony patterns. Bioessays 1995; 17:597-607; PMID:7646482; http://dx.doi.org/10.1002/bies.950170706
- Ben-Jacob E, Cohen I, Gutnick DL. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol 1998; 52:779-806; PMID:9891813; http://dx.doi.org/10.1146/annurev.micro.52.1.779
- Di Franco C, Beccari E, Santini T, Pisaneschi G, Tecce G. Colony shape as a genetic trait in the pattern-forming Bacillus mycoides. BMC Microbiol 2002; 2:1; PMID: 11846887; http://dx.doi.org/10.1186/1471-2180-2-33
- Mamou G, Mohan GBM, Rouvinski A, Rosenberg A, Ben-Yehuda S. Early Developmental Program Shapes Colony Morphology in Bacteria. Cell Rep 2016; 14:1850-7; PMID:26904951; http://dx.doi.org/10.1016/j.celrep.2016.01.071
- Shapiro JA. Organization of developing Escherichia coli colonies viewed by scanning electron microscopy. J Bacteriol 1987; 169:142-56; PMID:3539913; http://dx.doi.org/10.1128/jb.169.1.142-156.1987
- Pipe LZ, Grimson MJ. Spatial-temporal modelling of bacterial colony growth on solid media. Mol Biosyst 2008; 4:192-8; PMID:18437261; http://dx.doi.org/10.1039/b708241j
- Čepl J, Blahůšková A, Cvrčková F, Markoš A. Ammonia produced by bacterial colonies promotes growth of ampicillin sensitive Serratia sp. by means of antibiotic inactivation. FEMS Microbiol Lett 2014; 354:126-32; http://dx.doi.org/10.1111/1574-6968.12442
- Čepl JJ, Pátková I, Blahůšková A, Cvrčková F, Markoš A. Patterning of mutually interacting bacterial bodies: close contacts and airborne signals. BMC Microbiol 2010; 10:139; PMID:20462411; http://dx.doi.org/10.1186/1471-2180-10-139
- Pátková I, Čepl JJ, Rieger T, Blahůšková A, Neubauer Z, Markoš A. Developmental plasticity of bacterial colonies and consortia in germ-free and gnotobiotic settings. BMC Microbiol 2012; 12:178; PMID:22894147; http://dx.doi.org/10.1186/1471-2180-12-178
- Čepl J, Scholtz V, Scholtzová J. The fitness change and the diversity maintenance in the growing mixed colony of two Serratia rubidaea clones. Arch Microbiol 2015; 198(3):301-6
- Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci 1999; 96:4023-7; PMID:10097156; http://dx.doi.org/10.1073/pnas.96.7.4023
- Flohr RCE, Blom CJ, Rainey PB, Beaumont HJE. Founder niche constrains evolutionary adaptive radiation. Proc Natl Acad Sci 2013; 110:20663-8; PMID:24306929; http://dx.doi.org/10.1073/pnas.1310310110
- Bunting MI, Ingraham LJ. The distribution of color variants in ageing broth cultures of Serratia marcescens# 274. J Bacteriol 1942; 43:585; PMID:16560523.
- Ben-Jacob E, Cohen I, Shochet O, Tenenbaum A, Czirók A, Vicsek T. Cooperative formation of chiral patterns during growth of bacterial colonies. Phys Rev Lett 1995; 75(15):2899; PMID:10059433; http://dx.doi.org/10.1103/PhysRevLett.75.2899
- Sovova K, Cepl J, Markos A, Spanel P. Real time monitoring of population dynamics in concurrent bacterial growth using SIFT-MS quantification of volatile metabolites. Analyst 2013; 138:4795-4801; http://dx.doi.org/10.1039/c3an00472d
- Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci 2007; 104:19926-30; PMID:18056799; http://dx.doi.org/10.1073/pnas.0710150104
- Portmann A. Neue Wege der Biologie. R. Piper, München, 1960.
- Kleisner K, Markoš A. Mutual understanding and misunderstanding in biological systems mediated by self-representational meaning of organisms. Σημϵιωτκή-Sign Syst Stud 2009; 1-2:299-310.
- Shestivska V, Španěl P, Dryahina K, Sovová K, Smith D, Musílek M, Nemec A. Variability in the concentrations of volatile metabolites emitted by genotypically different strains of Pseudomonas aeruginosa. J Appl Microbiol 2012; 113:701-13; PMID:22726261; http://dx.doi.org/10.1111/j.1365-2672.2012.05370.x
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat methods 2012; 9:671-5; PMID:22930834; http://dx.doi.org/10.1038/nmeth.2089
- Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2015. URL http.www.R-project.org 2016.
- Lê Cao K-A, González I, Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics 2009; 25:2855-6; http://dx.doi.org/10.1093/bioinformatics/btp515
- Hilvo M, Gade S, Hyötyläinen T, Nekljudova V, Seppänen-Laakso T, Sysi-Aho M, Untch M, Huober J, Minckwitz G, Denkert C. Monounsaturated fatty acids in serum triacylglycerols are associated with response to neoadjuvant chemotherapy in breast cancer patients. Int J Cancer 2014; 134:1725-33; PMID:24114462; http://dx.doi.org/10.1002/ijc.28491
- Oussama A, Elabadi F, Platikanov S, Kzaiber F, Tauler R. Detection of olive oil adulteration using FT-IR spectroscopy and PLS with variable importance of projection (VIP) scores. J Am Oil Chem Soc 2012; 89:1807-12; http://dx.doi.org/10.1007/s11746-012-2091-1
