ABSTRACT
We recently identified the novel persulfide sensor SqrR that functions as a master regulator of sulfide-dependent gene expression in the purple photosynthetic bacterium Rhodobacter capsulatus. SqrR binds to the promoter regions of target genes to repress their expression in the absence of sulfide, and the repressor activity is negated by sulfide treatment. SqrR has 3 cysteine residues, 2 of which are conserved in SqrR homologs from other bacteria: Cys41 and Cys107. SqrR forms an intramolecular tetrasulfide bond between Cys41 and Cys107 when exposed to persulfide, which results in loss of the DNA-binding activity in vitro. Here, we address the mechanism through which these cysteine residues are modified by persulfides. We show that the predicted pKa value of Cys107, as revealed by a putative SqrR structural model, is lower than that of Cys41. Furthermore, C41S SqrR in which Cys41 was changed to serine forms an intermolecular disulfide-bond between Cys107 of 2 SqrRs, suggesting high nucleophilic reactivity of Cy107. These data suggest that Cys107 and Cys41 function as attacking Cys and resolving Cys, respectively; this occurs during tetrasulfide-bond formation of WT SqrR, when it is exposed to persulfide.
Hydrogen sulfide is a toxic chemical that inhibits cellular respiration,Citation1 and also works as the general biological gasotransmitter or signaling molecule in both prokaryotes and eukaryotes.Citation2-5 Although cysteine thiol modifications are likely involved in this sulfide signaling, sulfide is negatively charged and thus will not react with nucleophilic thiolates.Citation6 Recent studies in mammalian cells suggest that more oxidized and highly reactive sulfur-containing species, termed reactive sulfur species (RSS), are the actual sulfide signaling species.Citation7-9 Although persulfidated (S-sulfhydrated) proteins have been identified and a plausible repair pathway(s) elucidated in mammalian cells,Citation7,10-13 the functional impact of persulfidation of proteins and enzymes is as of yet not generally clear.Citation10
We recently identified the novel persulfide-responsive transcriptional repressor SqrR from the purple photosynthetic bacterium R. capsulatus.Citation14 SqrR forms an intramolecular tetrasulfide bond between Cys41 and Cys107 when incubated with the sulfane sulfur donor glutathione persulfide and then loses the repressor activity. Cysteine-dependent enzymes, e.g. thioredoxin (TrxA)Citation15 and peroxide sensor (OxyR),Citation16 which form an intramolecular disulfide crosslink, generally have what are termed attacking cysteine and resolving cysteine. The attacking cysteine initiates the crosslinking catalysis by intramolecular nucleophilic attack, so that its reactivity is higher than that of the resolving Cys. Here, to reveal the functional roles of the 2 conserved Cys residues for tetrasulfide-bond formation of SqrR, we estimated the pKa values of Cys41 and Cys107 that were calculated from a putative SqrR structure. Obtained results are compared with oligomerization characteristics of C41S and C107S SqrRs under oxidized conditions.
We first obtained a SqrR structural model by using the SWISS-MODEL server homology modeling pipeline (http://swissmodel.expasy.org/).Citation17 Homology modeling was performed with the crystal structure of the transcriptional repressor BigR, a SqrR homolog, from Xylella fastidiosa (PDB ID: 3PQJ) as a template. Because the obtained structure was dimer, we deleted one subunit, chain B, using PyMOL (https://www.pymol.org/) to obtain a monomer structure. Then, pKa values of Cys41 and Cys107 at pH7.0 in the SqrR structure were estimated by the web server called PROPKA 3.1 (http://propka.org/).Citation18 By the calculation, pKa values of Cys41 and Cys107 were predicted to be 10.13 and 9.36, respectively. Many hydrophobic amino acids are located around Cys107 (e.g., Leucine, methionine, isoleucine and alanine) that may influence the lower pKa value of Cys107. These results suggest that the nucleophilicity of Cys107 is higher than that of Cys41.
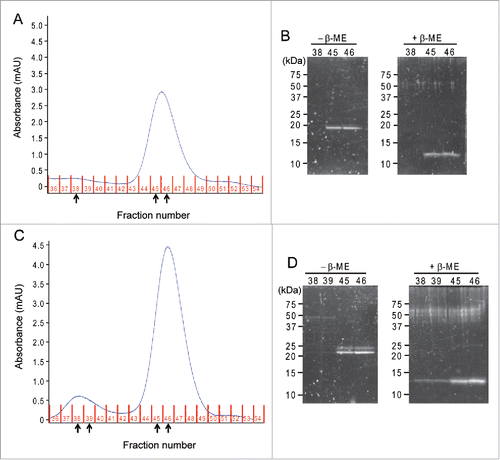
Next, we tested the reactivity of the 2 Cys residues by biochemical analysis. Recombinant C41S and C107S SqrR were expressed by an E. coli overexpression system followed by affinity chromatography as described previously.Citation14 C41S and C107S SqrR were treated with CuCl2 to induce disulfide bond formation and then applied to size-exclusion chromatography (, ). The chromatogram of C41S SqrR displayed a single peak (fraction 44–48) (). On the other hand, C107S SqrR showed 2 peaks; one is similar to that of C41S SqrR (fraction 44–48) and the second peak (fraction 37–40) is unique for C107S (). To determine the oligomerization status of the proteins, each peak fraction was subjected to SDS-PAGE under non-reducing and reducing conditions (, ). In the case of C41S SqrR, a single band estimated to be ≈24 kDa was observed in the fraction 45 and 46 under non-reducing conditions (). Given the molecular weight of SqrR is 12.1 kDa, calculated from its deduced amino-acid sequence, C41S SqrR exists as dimer. On the other hand, no signal was detected above fraction 38 under both conditions (). In the case of C107S SqrR, dimer and tetramer were detected in fractions 45–46 and 38–39, respectively (). Moreover, a double band was observed in each fraction, indicating that there are at least 2 patterns of intermolecular crosslink between cysteine residues (e.g., Cys9-Cys9, Cys41-Cys41 and/or Cys9-Cys41). The pKa value of Cys41 was predicted to be higher than that of Cys107, so that the reactivity of Cys41 might be weaker than that of Cys107. Therefore, Cys41 reactivity is not particularly high and could react with various cysteine residues like Cys9. On the other hand, C41S SqrR only forms an intermolecular disulfide bond between Cys107 of the 2 proteins immediately after induction with CuCl2, indicating that the reactivity of Cys107 is higher than those of Cys9 and Cys41. Indeed, in the case of thioredoxin (TrxA), a strong intermolecular crosslinking between attacking cysteine residues is observed when the resolving Cys was changed to Ser.Citation19 A single band estimated to be ≈12 kDa was observed under reduced conditions in all fractions, indicating that dimeric and tetrameric SqrRs were due to intermolecular disulfide cross-link formation (, ). Overall, these results suggest that Cys107 functions as the redox-active attacking cysteine residue for the disulfide-bind formation.
Figure 1. Redox state of C41S and C107S SqrR. The recombinant R. capsulatus C41S and C107S SqrR were purified from E. coli as described previously.Citation14 Purified SqrRs were treated with 50 µM CuCl2 for 1 hour at room temperature to induce intermolecular disulfide-bond formation and then were loaded onto a Superdex 75 gel filtration column (GE) in 20 mM Tris-HCl (pH 8.0), 500 mM NaCl, 6% glycerol. (A and C) The size-exclusion chromatogram of C41S and C107S SqrR. Expected elution fractions are as follows: dimer, fraction number 44–48; tetramer, fraction number 37–40. Protein elution is monitored by measuring absorbance at 280 nm using an AKTA prime. (B and D) SDS-PAGE analysis of C41S SqrR and C107S SqrR of each fraction indicated by arrows in A and C under oxidizing (−2-mercaptoethanol; β-ME) and reducing (+β-ME) conditions. The gels were stained with SYPRO Ruby stain (Molecular Probes) and imaged using UV-transillumination.
SqrR senses persulfide by tetrasulfide formation between Cys41 and Cys107 based on nucleophilic reaction of thiol of cysteine residues and persulfide.Citation20 This reaction may be undertaken by similar mechanisms of the disulfide formation. Our structural prediction analysis as well as biochemical characterization of Cys mutants strongly suggested that Cys41 and Cys107 function as resolving and attacking residues during tetrasulfide-bond formation; Cys107 is initially modified by persulfide and then forms a tetrasulfide bond by performing an intramolecular nucleophilic attack on Cys41. Further characterization of the redox-dependent reaction of cysteine residues of SqrR should be important for understanding the step-by-step molecular mechanism of persulfide sensing in cells.
Funding
This work was supported by a Rrant-in-Aid for Scientific Research KAKENHI (17H05524, 16H03280).
References
- Cooper CE, Brown GC. The inhibition of mitochondrial cytochrome oxidase by the gases carbon monoxide, nitric oxide, hydrogen cyanide and hydrogen sulfide: Chemical mechanism and physiological significance. J Bioenerg Biomembr 2008; 40:533-9; PMID:18839291; https://doi.org/10.1007/s10863-008-9166-6
- Kaneko Y, Kimura Y, Kimura H, Niki I. L-cysteine inhibits insulin release from the pancreatic beta-cell: Possible involvement of metabolic production of hydrogen sulfide, a novel gasotransmitter. Diabetes 2006; 55:1391-7; PMID:16644696; https://doi.org/10.2337/db05-1082
- Kimura H. Hydrogen sulfide: From brain to gut. Antioxid Redox Signal 2010; 12:1111-23; PMID:19803743; https://doi.org/10.1089/ars.2009.2919
- Wang R. Physiological implications of hydrogen sulfide: A whiff exploration that blossomed. Physiol Rev 2012; 92:791-896; PMID:22535897; https://doi.org/10.1152/physrev.00017.2011
- Shatalin K, Shatalina E, Mironov A, Nudler E. H2S: A universal defense against antibiotics in bacteria. Science 2011; 334:986-90; PMID:22096201; https://doi.org/10.1126/science.1209855
- Pan J, Carroll KS. Persulfide reactivity in the detection of protein S-sulfhydration. ACS Chem Biol 2013; 8:1110-6; PMID:23557648; https://doi.org/10.1021/cb4001052
- Ida T, Sawa T, Ihara H, Tsuchiya Y, Watanabe Y, Kumagai Y, Suematsu M, Motohashi H, Fujii S, Matsunaga T, et al. Reactive cysteine persulfides and S-polythiolation regulate oxidative stress and redox signaling. Proc Natl Acad Sci U S A 2014; 111:7606-11; PMID:24733942; https://doi.org/10.1073/pnas.1321232111
- Cuevasanta E, Lange M, Bonanata J, Coitiño EL, Ferrer-Sueta G, Filipovic MR, Alvarez B. Reaction of hydrogen sulfide with disulfide and sulfenic acid to form the strongly nucleophilic persulfide. J Biol Chem 2015; 290:26866-80; PMID:26269587; https://doi.org/10.1074/jbc.M115.672816
- Yadav PK, Martinov M, Vitvitsky V, Seravalli J, Wedmann R, Filipovic MR, Banerjee R. Biosynthesis and reactivity of cysteine persulfides in signaling. J Am Chem Soc 2016; 138:289-99; PMID:26667407; https://doi.org/10.1021/jacs.5b10494
- Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Sadia K, Barrow RK, Yang G, Wang R, Snyder SH, et al. H2S signals through protein S-sulfhydration. Sci Signal 2009; 2:ra72; PMID:19903941; https://doi.org/10.1126/scisignal.2000464
- Gao XH, Krokowski D, Guan BJ, Bederman I, Majumder M, Parisien M, Diatchenko L, Kabil O, Willard B, Banerjee R, et al. Quantitative H2S-mediated protein sulfhydration reveals metabolic reprogramming during the integrated stress response. Elife 2015; 4:e10067; PMID:26595448; https://doi.org/10.7554/eLife.10067
- Dóka É, Pader I, Bíró A, Johansson K, Cheng Q, Ballagó K, Prigge JR, Pastor-Flores D, Dick TP, Schmidt EE, et al. A novel persulfide detection method reveals protein persulfide- and polysulfide-reducing functions of thioredoxin and glutathione systems. Sci Adv 2016; 2:e1500968; PMID:26844296; https://doi.org/10.1126/sciadv.1500968
- Wedmann R, Onderka C, Wei S, András Szijártó I, Miljkovic J, Femic A, Lange M, Savitsky S, Kumar P, Torregrossa R, et al. Improved tag-switch method reveals that thioredoxin acts as depersulfidase and controls the intracellular levels of protein persulfidation. Chem Sci 2016; 7(5):3414-26; PMID:27170841; https://doi.org/10.1039/C5SC04818D
- Shimizu T, Shen J, Fang M, Zhang Y, Hori K, Trinidad JC, Bauer CE, Giedroc DP, Masuda S. SqrR functions as a master regulator of sulfide-dependent photosynthesis. Proc Natl Acad Sci U S A 2017; 114:2355-60; PMID:28196888; https://doi.org/10.1073/pnas.1614133114
- Chivers PT, Prehoda KE, Volkman BF, Kim B, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli Thioredoxin. Biochemistry 1997; 36:14985-91; PMID:9398223; https://doi.org/10.1021/bi970071j
- Jo I, Chung IY, Bae HW, Kim JS, Song S, Cho YH, Ha NC. Structural details of the OxyR peroxide-sensing mechanism. Proc Natl Acad Sci U S A 2015; 112:6443-8; PMID:25931525; https://doi.org/10.1073/pnas.1424495112
- Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011; 27:343-50; PMID:21134891; https://doi.org/10.1093/bioinformatics/btq662
- Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J Chem Theory Comput 2011; 7:525-37; PMID:26596171; https://doi.org/10.1021/ct100578z
- Kouwen TRHM, Andréll J, Schrijver R, Dubois JYF, Maher MJ, Iwata S, Carpenter EP, van Dijl JM. Thioredoxin A active-site mutants form mixed disulfide dimers that resemble enzyme-substrate reaction intermediates. J Mol Biol 2008; 379:520-34; PMID:18455736; https://doi.org/10.1016/j.jmb.2008.03.077
- Mishanina T V, Libiad M, Banerjee R. Biogenesis of reactive sulfur species for signaling by hydrogen sulfide oxidation pathways. Nat Chem Biol 2015; 11(7):457-64; PMID:26083070; https://doi.org/10.1038/nchembio.1834
