ABSTRACT
The regions at which the ER and mitochondria come into close proximity, known as ER-mitochondria contact sites provide essential platforms for the exchange of molecules between the two organelles and the coordination of various fundamental cellular processes. In addition to the well-established role of ER-mitochondria interfaces in calcium and lipid crosstalk, emerging evidence supports that a proper communication between ER and mitochondria is critical for the regulation of mitochondrial morphology and the initiation of autophagy. Within this context, our recent data indicate that glycogen is targeted to ER-mitochondria contacts through the Stbd1 protein, a proposed autophagy receptor for glycogen. Glycogen-bound Stbd1 influences ER-mitochondria tethering and the morphology of the mitochondrial network. We here suggest possible roles of glycogen recruitment to ER-mitochondria contact sites. Stbd1-mediated targeting of glycogen to ER-mitochondria junctions could represent a mechanism through which glycogen is sequestered into autophagosomes for lysosomal degradation, a process described as glycogen autophagy or glycophagy. Additionally, we discuss a possible mechanism which links the observed effects of Stbd1 on mitochondrial morphology with the previously reported impact of nutrient availability on mitochondrial dynamics. In this model we propose that glycogen-bound Stbd1 signals nutrient status to ER-mitochondria junctions resulting in adaptations in the morphology of the mitochondrial network.
A key feature of eukaryotic cells is the compartmentalization of metabolic reactions in membrane-bound organelles. Moreover, communication between organelles has emerged as an additional level of regulation aiming at the fine tuning of cellular processes within cells. This is achieved through membrane-contact sites (MCS) at which the membranes of two organelles come into close contact.Citation1,2 The most extensively studied MCS are those between the endoplasmic reticulum (ER) and mitochondria, known as ER-mitochondria contact sites. Since their discovery almost 60 years ago,Citation3 a plethora of studies highlighted the importance of this type of inter-organelle communication in a variety of cellular processes such as calcium crosstalk, mitochondrial morphology and dynamics, lipid homeostasis, autophagosome formation, ER stress and apoptosis.Citation2,4,5 Furthermore, increasing evidence suggests that a miscommunication between ER and mitochondria occurs in a number of neurodegenerative and metabolic diseases, including Parkinson's and Alzheimer's disease as well as type 2 diabetes. However, whether distortions in ER-mitochondria crosstalk are the cause or the consequence of the above pathologic conditions still remains elusive.Citation2,6
Most of our current knowledge regarding ER-mitochondria contacts comes from electron microscopy studies which revealed that the two organelles are tethered together through protein bridges and that no membrane fusion takes place.Citation2,7 The mitochondrial surface coming into contact with an adjacent ER membrane was estimated around 5–20% in HeLa cells.Citation8 Similarly, the distance between ER and mitochondria at MCS is not constant but can lie between ∼10 and ∼80 nm.Citation7 Several reports indicate that ER-mitochondria coupling is adjusted depending on the metabolic state of the cell,Citation9-12 supporting the dynamic nature of ER-mitochondria associations.
The identification and characterization of the protein complexes mediating the tethering between ER and mitochondria has been the subject of active research, yet we are still far from a complete understanding of the mechanisms governing ER-mitochondria association. The picture is clearer in yeast, where two protein complexes known as the ER-mitochondria encounter structure (ERMES)Citation13 and the ER membrane complex (EMC)Citation14 were found to be responsible for ER-mitochondria tethering. Analysis of a differential centrifugation fraction consisting of ER membranes co-purified with mitochondria, known as mitochondria-associated membranes (MAMs), resulted in the identification of a number of protein complexes that are proposed to tether ER with mitochondria in mammalian cells. Protein-protein interactions mediating ER-mitochondria association in mammalian cells include those between the ER-targeted IP3R and VDAC1 at the outer mitochondrial membrane (OMM) via the chaperone Grp75. The IP3R/VDAC1/Grp75 ternary complex is also responsible for the transport of calcium from ER to mitochondria.Citation15,16 A number of studies reported contradicting findings regarding the role of the mitochondrial fusion protein Mfn2 in ER-mitochondria association.Citation17-19 Nevertheless, the function of Mfn2 as a bona fide ER-mitochondria tether has been recently reconfirmed.Citation20 In particular, tethering is achieved through the formation of homo- and heterodimers between ER-targeted Mfn2 and Mfn2 or Mfn1 located on the OMM, respectively.Citation17,20 Moreover, the ER-targeted protein VAPB was shown to interact with the OMM PTPIP51 protein and regulate both structural and functional features of ER-mitochondria contact sites.Citation21,22 An additional tethering complex results from the interaction between Fis1 on the OMM and the Bap31 protein at the ER, comprising a platform for the transport of apoptotic signals from mitochondria to ER.Citation23
In addition to the well-established functions of ER-mitochondria interfaces (e. g. calcium flux, lipid homeostasis), new roles for these inter-organelle junctions are emerging. ER-mitochondria contacts were shown to be the site of the formation of double membrane vesicles, known as autophagosomes during the initial stages of the autophagic process.Citation24 Autophagy is an evolutionarily conserved catabolic process which involves the sequestration of cytoplasmic material into autophagosomes and their delivery to lysosomes for degradation.Citation25 Within this context, we have recently identified that Starch binding domain-containing protein 1 (Stbd1), a proposed selective autophagy receptor for glycogen thought to mediate its trafficking to lysosomes through an autophagy-like process,Citation26,27 is targeted in addition to bulk ER, to ER-mitochondria contact sites.Citation28 Our findings further demonstrated that Stbd1 undergoes a specific co-translational lipid modification, known as N-myristoylation, which together with glycogen determine the subcellular targeting of the protein to the ER or ER-mitochondria contact sites. Specifically, targeting of Stbd1 to ER-mitochondria interfaces is promoted by the lack of N-myristoylation and the concomitant binding of the protein to glycogen.Citation28 At ER-mitochondria contact sites, Stbd1 was shown to influence ER-mitochondria tethering and mitochondrial morphology. Particularly, we showed that overexpression of MAM-targeted Stbd1 increased ER-mitochondrial coupling and also induced mitochondrial fragmentation and clustering. On the other hand, shRNA-mediated Stbd1 silencing resulted in a weakening of ER-mitochondria tethering and an elongated morphology of the mitochondrial network.Citation28
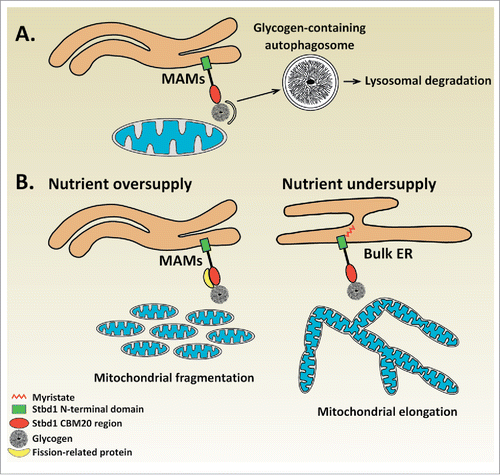
With regards to the biological significance of the Stbd1-mediated recruitment of glycogen to ER-mitochondria contact sites, we can only speculate at the current state of knowledge. Given the proposed role of Stbd1 as a selective autophagy receptor for glycogen,Citation26,27 and the identification of ER-mitochondria contacts as platforms for autophagosome formation,Citation24 it is conceivable that Stbd1-mediated glycogen targeting to ER-mitochondria junctions may serve the sequestration of glycogen into autophagosomes for lysosomal degradation (). Alternative scenarios may exist, which could explain the observed effects of Stbd1 on mitochondrial morphology in relation to glycogen recruitment at ER-mitochondria contact sites. ER-mitochondria junctions play an important role in the regulation of mitochondrial dynamics which involves mitochondrial fusion (elongation),Citation17,20 and fission (fragmentation). Importantly, ER-mitochondria contact sites were shown to mark the sites of mitochondrial fission.Citation29 A number of studies established a functional connection between the availability of nutrients and changes in mitochondrial morphology. Specifically, nutrient oversupply was found to correlate with a fragmented mitochondrial morphology whereas undersupply of nutrients was found to induce mitochondrial elongation.Citation30-32 This dynamic change in the morphology of the mitochondrial network represents a cellular adaptation response which enables cells to regulate their energy levels according to the availability of nutrients, since mitochondrial fragmentation is associated with reduced whereas mitochondrial elongation with increased ATP production, respectively.Citation31,32 Additionally, high glucose levels induce mitochondrial fragmentation accompanied by reduced ER-mitochondria association in liver cells, suggesting that glucose is a metabolic regulator of ER-mitochondria contact sites.Citation12 The finding that Stbd1 is targeted to ER-mitochondria contacts in glycogen-bound form along with the observed effects of Stbd1 on mitochondrial dynamics, raise the hypothesis that Stbd1-mediated recruitment of glycogen at ER-mitochondria junctions acts as a nutrient sensing mechanism implicated in the maintenance of cellular energy homeostasis by modulating mitochondrial morphology. Our proposed model suggests that under nutrient-rich conditions, cellular glycogen levels rise leading to the targeting of non-myristoylated, glycogen-bound Stbd1 to ER-mitochondria contacts where it may recruit proteins involved in mitochondrial fission (e.g. Drp1), resulting in fragmentation of the mitochondrial network. On the other hand, nutrient scarcity is expected to result in the degradation of intracellular glycogen which would hence prevent Stbd1 targeting to ER-mitochondria contact sites. Consequently, the failure to recruit proteins mediating mitochondrial fission to ER-mitochondrial junctions would cause a shift of the equilibrium between mitochondrial fusion and fission toward fusion resulting in fused, elongated mitochondria ().
Figure 1. Proposed role for Stbd1-mediated recruitment of glycogen to ER-mitochondria contact sites. (A) Glycogen autophagy (Glycophagy) | Binding of glycogen to non-myristoylated Stbd1 promotes its targeting to ER-mitochondria contact sites. The recruitment of glycogen to ER-mitochondria junctions which have been identified as the sites of autophagosome formation could serve its sequestration into autophagosomes for lysosomal degradation. (B) Nutrient sensing mechanism | Stbd1-mediated recruitment of glycogen to ER-mitochondria contacts may represent a mechanism for nutrient sensing that influences mitochondrial morphology. Upon nutrient oversupply, intracellular glycogen levels increase leading to the targeting of glycogen-bound, non-myristoylated Stbd1 to ER-mitochondria contacts. Stbd1 may interact at ER-mitochondria contact sites with proteins involved in mitochondrial fission such as Drp1 or its known adaptors resulting in the fragmentation of the mitochondrial network. On the other hand, under conditions of nutrient undersupply, cellular glycogen is degraded and hence Stbd1 is preferentially targeted to bulk ER instead of ER-mitochondria contact sites. Failure of recruiting mitochondrial fission proteins through Stbd1 at ER-mitochondria interfaces would consequently result in the formation of fused, elongated mitochondria. This model is in accordance with the observed effects of Stbd1 on mitochondrial morphology, i.e. overexpression of non-myristoylated Stbd1 causes fragmentation whereas Stbd1 knockdown elongation of the mitochondrial network, and the available data on the correlation between nutrient availability and morphology of the mitochondrial network.
Since its discovery as the major carbohydrate storage form in animals, glycogen has intrigued biologists for over a century and a half. It is becoming clear that glycogen is not only a glucose polymer which covers energy needs of cells and tissuesCitation33 but also serves additional, more complex regulatory functions. The finding that glycogen is targeted through Stbd1 to ER-mitochondria contact sites adds more to this complexity and opens a new perspective regarding potentially important structural, metabolic and regulatory roles for glycogen at these interorganellar contact sites.
Abbreviations
| Bap31 | = | B-cell receptor-associated protein 31 |
| Drp1 | = | dynamin-related protein 1 |
| EMC | = | ER membrane complex |
| ER | = | endoplasmic reticulum |
| ERMES | = | ER-mitochondria encounter structure |
| Fis1 | = | fission 1 protein |
| Grp75 | = | glucose-regulated protein 75 |
| IP3R | = | inositol 1,4,5-triphosphate receptor |
| MAMs | = | mitochondria-associated membranes |
| MCS | = | membrane-contact sites |
| Mfn1 | = | mitofusin 1 |
| Mfn2 | = | mitofusin 2 |
| OMM | = | outer mitochondrial membrane |
| PTPIP51 | = | protein tyrosine phosphatase-interacting protein 51 |
| Stbd1 | = | starch binding domain-containing protein 1 |
| VAPB | = | vesicle-associated membrane protein-associated protein B |
| VDAC1 | = | voltage-dependent anion channel 1 |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Cyprus Telethon (#33173116) and Cancer Research UK (Program Foundation Award C29637/A20183).
References
- Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci 2011; 36:616-23; PMID:21958688; https://doi.org/10.1016/j.tibs.2011.08.004
- Vance JE. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim Biophys Acta 2014; 1841:595-609; PMID:24316057; https://doi.org/10.1016/j.bbalip.2013.11.014
- Copeland DE, Dalton AJ. An association between mitochondria and the endoplasmic reticulum in cells of the pseudobranch gland of a teleost. J Biophys Biochem Cytol 1959; 5:393-6; PMID:13664679; https://doi.org/10.1083/jcb.5.3.393
- de Brito OM, Scorrano L. An intimate liaison: spatial organization of the endoplasmic reticulum-mitochondria relationship. EMBO J 2010; 29:2715-23; PMID:20717141; https://doi.org/10.1038/emboj.2010.177
- Marchi S, Patergnani S, Pinton P. The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim Biophys Acta 2014; 1837:461-9; PMID:24211533; https://doi.org/10.1016/j.bbabio.2013.10.015
- Paillusson S, Stoica R, Gomez-Suaga P, Lau DH, Mueller S, Miller T, Miller CC. There's something wrong with my MAM; the ER-Mitochondria axis and neurodegenerative diseases. Trends Neurosci 2016; 39:146-57; PMID:26899735; https://doi.org/10.1016/j.tins.2016.01.008
- Giacomello M, Pellegrini L. The coming of age of the mitochondria-ER contact: a matter of thickness. Cell Death Differ 2016; 23(9):1417-27; PMID:27341186; https://doi.org/10.1038/cdd.2016.52
- Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 1998; 280:1763-6; PMID:9624056; https://doi.org/10.1126/science.280.5370.1763
- Bravo R, Vicencio JM, Parra V, Troncoso R, Munoz JP, Bui M, Quiroga C, Rodriguez AE, Verdejo HE, Ferreira J, et al. Increased ER-mitochondrial coupling promotes mitochondrial respiration and bioenergetics during early phases of ER stress. J Cell Sci 2011; 124:2143-52; PMID:21628424; https://doi.org/10.1242/jcs.095455 10.1242/jcs.080762
- Csordas G, Renken C, Varnai P, Walter L, Weaver D, Buttle KF, Balla T, Mannella CA, Hajnoczky G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J Cell Biol 2006; 174:915-21; PMID:16982799; https://doi.org/10.1083/jcb.200604016
- Sood A, Jeyaraju DV, Prudent J, Caron A, Lemieux P, McBride HM, Laplante M, Toth K, Pellegrini L. A Mitofusin-2-dependent inactivating cleavage of Opa1 links changes in mitochondria cristae and ER contacts in the postprandial liver. Proc Natl Acad Sci U S A 2014; 111:16017-22; PMID:25352671; https://doi.org/10.1073/pnas.1408061111
- Theurey P, Tubbs E, Vial G, Jacquemetton J, Bendridi N, Chauvin MA, Alam MR, Le Romancer M, Vidal H, Rieusset J. Mitochondria-associated endoplasmic reticulum membranes allow adaptation of mitochondrial metabolism to glucose availability in the liver. J Mol Cell Biol 2016; 8:129-43; PMID:26892023; https://doi.org/10.1093/jmcb/mjw004
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 2009; 325:477-81; PMID:19556461; https://doi.org/10.1126/science.1175088
- Lahiri S, Chao JT, Tavassoli S, Wong AK, Choudhary V, Young BP, Loewen CJ, Prinz WA. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol 2014; 12:e1001969; PMID:25313861; https://doi.org/10.1371/journal.pbio.1001969
- Szabadkai G, Bianchi K, Varnai P, De Stefani D, Wieckowski MR, Cavagna D, Nagy AI, Balla T, Rizzuto R. Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J Cell Biol 2006; 175:901-11; PMID:17178908; https://doi.org/10.1083/jcb.200608073
- De Stefani D, Bononi A, Romagnoli A, Messina A, De Pinto V, Pinton P, Rizzuto R. VDAC1 selectively transfers apoptotic Ca2+ signals to mitochondria. Cell Death Differ 2012; 19:267-73; PMID:21720385; https://doi.org/10.1038/cdd.2011.92
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008; 456:605-10; PMID:19052620; https://doi.org/10.1038/nature07534
- Cosson P, Marchetti A, Ravazzola M, Orci L. Mitofusin-2 independent juxtaposition of endoplasmic reticulum and mitochondria: an ultrastructural study. PLoS One 2012; 7:e46293; PMID:23029466; https://doi.org/10.1371/journal.pone.0046293
- Filadi R, Greotti E, Turacchio G, Luini A, Pozzan T, Pizzo P. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc Natl Acad Sci U S A 2015; 112:E2174-81; PMID:25870285; https://doi.org/10.1073/pnas.1504880112
- Naon D, Zaninello M, Giacomello M, Varanita T, Grespi F, Lakshminaranayan S, Serafini A, Semenzato M, Herkenne S, Hernandez-Alvarez MI, et al. Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proc Natl Acad Sci U S A 2016; 113:11249-54; PMID:27647893; https://doi.org/10.1073/pnas.1606786113
- De Vos KJ, Morotz GM, Stoica R, Tudor EL, Lau KF, Ackerley S, Warley A, Shaw CE, Miller CC. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum Mol Genet 2012; 21:1299-311; PMID:22131369; https://doi.org/10.1093/hmg/ddr559
- Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau KF, Vizcay-Barrena G, Lin WL, Xu YF, Lewis J, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun 2014; 5:3996; PMID:24893131; https://doi.org/10.1038/ncomms4996
- Iwasawa R, Mahul-Mellier AL, Datler C, Pazarentzos E, Grimm S. Fis1 and Bap31 bridge the mitochondria-ER interface to establish a platform for apoptosis induction. EMBO J 2011; 30:556-68; PMID:21183955; https://doi.org/10.1038/emboj.2010.346
- Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, Oomori H, Noda T, Haraguchi T, Hiraoka Y, et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389-93; PMID:23455425; https://doi.org/10.1038/nature11910
- Kaur J, Debnath J. Autophagy at the crossroads of catabolism and anabolism. Nat Rev Mol Cell Biol 2015; 16:461-72; PMID:26177004; https://doi.org/10.1038/nrm4024
- Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, Roach PJ. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J Biol Chem 2010; 285:34960-71; PMID:20810658; https://doi.org/10.1074/jbc.M110.150839
- Jiang S, Wells CD, Roach PJ. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem Biophys Res Commun 2011; 413:420-5; PMID:21893048; https://doi.org/10.1016/j.bbrc.2011.08.106
- Demetriadou A, Morales-Sanfrutos J, Nearchou M, Baba O, Kyriacou K, Tate EW, Drousiotou A, Petrou PP. Mouse Stbd1 is N-myristoylated and affects ER-mitochondria association and mitochondrial morphology. J Cell Sci 2017; 130:903-15; PMID:28137759; https://doi.org/10.1242/jcs.195263
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science 2011; 334:358-62; PMID:21885730; https://doi.org/10.1126/science.1207385
- Molina AJ, Wikstrom JD, Stiles L, Las G, Mohamed H, Elorza A, Walzer G, Twig G, Katz S, Corkey BE, et al. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes 2009; 58:2303-15; PMID:19581419; https://doi.org/10.2337/db07-1781
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 2011; 13:589-98; PMID:21478857; https://doi.org/10.1038/ncb2220
- Jheng HF, Tsai PJ, Guo SM, Kuo LH, Chang CS, Su IJ, Chang CR, Tsai YS. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol 2012; 32:309-19; PMID:22083962; https://doi.org/10.1128/MCB.05603-11
- Adeva-Andany MM, Gonzalez-Lucan M, Donapetry-Garcia C, Fernandez-Fernandez C, Ameneiros-Rodriguez E. Glycogen metabolism in humans. BBA Clin 2016; 5:85-100; PMID:27051594; https://doi.org/10.1016/j.bbacli.2016.02.001
