ABSTRACT
Plants have evolved sophisticated surveillance and defense mechanisms against constant attacks by different types of microbial pathogens including viruses. Nucleotide-based RNA silencing and receptor-based innate immunity constitute the primary plant defense to viral infection. To establish their infections, plant viruses have also evolved virulence strategies to suppress host defenses. This ever-lasting co-evolutionary arms race between the plant hosts and viruses has shaped today's multifaceted defense and counter-defense layout. A better understanding of this process will certainly assist in the development of new plant varieties with enhanced durable resistance and novel approaches to control viral pathogens for sustainable agriculture. In this addendum, we briefly review the current knowledge of the plant immune response to viral infection and discuss defense and counter-defense strategies.
Plants are constantly exposed to diverse microorganisms such as nematodes, fungi, oomycetes, bacteria, and viruses, and have evolved sophisticated and efficient mechanisms to protect themselves against pathogen attack.Citation1,2 For non-viral pathogens, plants deploy a complex innate immune system that consists of 2 interconnected tiers of receptors including pattern recognition receptors (PRRs) and nucleotide-binding site leucine-rich repeats (NB-LRR) receptors to modulate plant defense.Citation3,4,5,6 For viral pathogens, in addition to the receptor-based innate immunity system, RNA silencing is another major antiviral cellular pathway.Citation7 To establish their infection, viruses have also evolved counter strategies to suppress or evade plant defense surveillance using the relatively few proteins they encode. In this addendum, we focus on summarizing current knowledge about innate immunity and RNA silencing antiviral defense in plants and viral counter-defense strategies. Due to space constraint, we do not discuss other antiviral strategies such as recessive resistance.Citation8,9
RNA silencing in plants, suppression of RNA silencing by viruses and plant counter strategies
RNA silencing, also called RNA interference (RNAi), is an evolutionary conserved and sequence-specific mechanism that is triggered by double-stranded RNAs (dsRNAs) to induce RNA degradation or inhibit translation.Citation7,10 In virus-infected cells, dsRNAs include replicative intermediates of viral replication.Citation11 dsRNAs may also originate as a result of discrete intramolecular pairing within viral genomic RNA, or through de novo synthesis by the RNA-dependent RNA polymerases (RDRs).Citation12 These dsRNAs are recognized and processed by plant Dice-Like (DCL) proteins, which are type III endoribonucleases, into small 20- to 24-nucleotide (nt) RNA duplexes termed virus-derived short-interfering small RNAs (vsiRNAs).Citation13 The vsiRNAs are then incorporated into Argonaute (AGO) proteins, the core component of RNA-induced silencing complex (RISC) and RNA-induced transcriptional silencing complex (RITS) that directly catalyzes the cleavage of homologous RNAs and suppress the transcription of homologous DNAs via methylation, respectively.Citation14,15 Since knockout of the viral suppressor of RNA silencing (VSR) usually results in the loss of infectivity of the virus, RNA silencing has been recognized as an essential antiviral defense mechanism. For instance, a Turnip mosaic virus (TuMV, a potyvirus) mutant with a defective HC-Pro, which is the major VSR of TuMV, fails to infect Arabidopsis, and the infectivity of this mutant can be restored when the plant RNA silencing pathway is compromised.Citation16 Moreover, the vsiRNA duplexes are motile that enable the plant to establish RNA silencing-based resistance against homologous virus infection in the non-infected tissues.Citation17
In response to host RNA silencing, most plant viruses have evolved one or more VSRs to suppress host RNA silencing.Citation18 For example, most of potyviruses such as TuMV encode 2 VSRs, e.g., HC-Pro and VPg. The VSRs encoded by either the same virus or different viruses share no obvious sequence or structure similarity, suggesting that they have evolved independently. VSRs can suppress RNA silencing virtually at all steps of RNA silencing.Citation19 For example, many VSRs, such as p19 of tombusviruses, NS3 of tenuiviruses and HC-Pro of potyviruses, function by sequestering viral dsRNA or vsiRNA to block the silencing initiation step.Citation16,20,21,22 A number of others, such as VPg of potyviruses, P0 of poleroviruses and P25 of potexviruses subvert the host RNA silencing pathway by destabilizing AGOs or SGS3/RDR6.Citation11,23,24 A few VSRs such as the cucumoviral 2b protein can target multiple steps of the RNA silencing pathway.Citation19
To counteract RNA silencing suppression by VSRs, plants simultaneously activate the bulk expression of resistance (R) genes to increase host resistance to viral pathogens.Citation25 It has been shown that these R genes are regulated post-transcriptionally by DCL-4-depdentdent small interfering RNAs (siRNAs).Citation26,27,28,29,30,31,32 Suppression of these siRNAs by VSRs activates expression of the R genes.Citation26 This has been suggested to be a counter-counter-defense mechanism.Citation25 Since these R proteins are indispensable in plant innate immunity, plant RNA silencing and viral suppression of RNA silencing are interlinked to plant innate immunity.
Plant immunity to viral infection and suppression of the immune response by viruses
Plant immunity is another essential defense antiviral mechanism. This concept is adapted from research on the plant immune response to non-viral pathogens. Jones and Dangl summarized research progress in molecular plant–pathogen interactions and proposed a zigzag model.Citation3 The Jones/Dangl model shows that plants use a 2-branched innate immune system against attacks by non-viral pathogens. The extracellular PRRs recognize and respond to evolutionarily conserved pathogen- or microbial-associated molecular patterns (PAMPs or MAMPs) to trigger the first layer of resistance and establish PAMP-triggered immunity (PTI).Citation33 PTI can restrict the majority of potential pathogens. To counteract this defense, some pathogens have evolved to deploy variable pathogen molecules, termed effectors, to interfere with PTI. As a result, these pathogens can establish a successful infection. In response, plants have developed the second branch of defense that is activated by dominant resistance (R) gene-encoded proteins. Most of these R gene- proteins belong to the extremely polymorphic superfamily of NB-LRRs. R proteins can directly or indirectly recognize the effectors of specific pathogens, which function as avirulence (Avr) factors, leading to effector-triggered immunity (ETI). ETI contains the invading pathogen at the infection site by activating much faster and stronger resistant responses than PTI and inducing the hypersensitive response (HR).Citation3,33
Although the relevance of the 2-tier immunity concepts to viral pathogens were not initially included in the Jones/Dangl model,Citation3 several lines of recent evidence suggest that plants also deploy the innate immune system to combat viral pathogens.Citation6,34,35,36,37 Viral infections induce PTI-based antiviral responses in plants.Citation36,37,38 Knock-out of BRI1-ASSOCIATED KINASE 1 (BAK1) in Arabidopsis encoding a central regulator of PTI results in an increased susceptibility to several viruses, suggesting that PTI indeed suppresses viral infection.Citation34 dsRNA, a replicative intermediate of viral replication, has been shown as a PTI elicitor to induce typical PTI responses such as activation of mitogen-activated protein kinases (MAPKs), ethylene synthesis and defense gene expression.Citation38 The dsRNA-mediated PTI signaling requires the pattern recognition co-receptor kinase SERK1, but is independent on DCLs.Citation38 To overcome dsRNA-activated PTI, viruses seem to have also evolved counter-PTI strategies. It has been reported that Plum pox virus (PPV) uses its capsid protein (CP) to suppress PTI.Citation36 Therefore, potyviral CP can function as a virus-encoded innate immunity suppressor (VIIS).
Plants activate ETI against viral infection through direction or indirect interaction between a specific virus-encoded effector and a corresponding R protein, leading to HR.Citation2,37 Like antibacterial and antifungal R proteins, proteins encoded by most of cloned dominant R genes to plant viruses are NB-LRR proteins too.Citation2,6,37 Regardless of these NB-LRR proteins conferring resistance to viral or non-viral pathogens, they share structural similarity. Therefore, it is not surprising that plant antiviral R proteins can activate typical ETI responses.
To protect plants against pathogen attacks, PTI and ETI upregulate a large number of integrated defense responses not only in local leaves but also in non-infected tissues, a phenomenon called systemic acquired resistance (SAR).Citation2,39 Non-expressor of pathogenesis related genes-1 (NPR1) is the master regulator of the plant immunity signaling pathway in local resistance and SAR.Citation39,40,41 NPR1 may be regulated by sumoylation, and sumoylation of NPR1 by SMALL-UBIQUITIN LIKER MODIFIER 3 (SUMO3) activates the NPR1-mediated resistance pathwayCitation42 SUMO3 is one of the 4 functional SUMOs encoded in Arabidopsis,Citation43 and is required to maintain the basal expression level of PATHOGEN-RELATED (PR) genes in plants without pathogen challenging.Citation44 Viral infection upregulates SUMO3 expression, leading to enhanced NPR1-mediated resistance.Citation44 Potyviral NIb is the only viral RNA-dependent RNA polymerase (RdRp) that is absolutely required for viral genome replication. Similar to NPR1, NIb targets the nucleus and is also sumoylated wherein by SUMO3. Sumoylation of NIb by SUMO3 effectively suppresses the host immune response, possibly through inhibition of NPR1 sumoylation.Citation44 Therefore, NIb is another VIIS that suppresses the cellular immune response to promoter viral infection. Interestingly, the NIb protein of several potyviruses, such as Potato virus Y (PVY), Pepper mottle virus (PepMoV), and Pepper severe mosaic virus (PepSMV), has also been shown as an Avr protein that triggers the HR response in plants carrying corresponding R genes.Citation45,46 These data suggest that potyviral NIb catalyzes biosynthesis of the replicative intermediates as a generator of the RNA silencing inducer, promotes viral infection as a VIIS, and inhibits viral infection as an ETI elicitor. Thus, a single viral component can play contrasting roles in viral infection, reflecting the complex and specific virus-plant interactions as a result of the co-evolutionary arms race.
Perspective
In the past 20 years, significant progress has been made in understanding the mechanisms of defense and counter-defense between plants and viruses at the nucleic acid level. We are just beginning to touch plant receptor-based innate immunity to viral infection and viral counter-defense strategies. We summarize what we have discussed above in . However, on the one hand, it still remains unresolved how plants recognize infection by diverse viruses, what the signaling pathways immediately after acceptor recognition are that lead to the activation of antiviral innate immune responses and if the signaling defense pathways are shared in different plants against different viruses. On the other hand, it is yet largely unclear how viruses suppress PTI and ETI to establish their infection in plants and if similar defense suppression strategies are used by distinct viruses. These are certainly future research directions. It is worth mentioning that at the protein level, recently autophagy and unfolded protein response (UPR) pathways have been shown to be involved in viral infection.Citation11,47 These pathways are likely interlinked with the receptor-based innate immunity. Rapid research in these areas may also contribute to elucidation of the molecular events of PTI and ETI pathways.
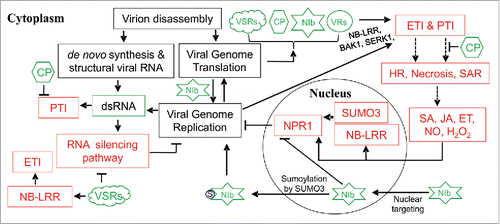
Figure 1. Antiviral responses in plants. As most of plant viruses are positive-sense RNA viruses and potyviruses represent over 30% of known plant viruses, potyviruses are used as a representative. Upon entry into a plant cell, virion disassembly takes place. After virion uncoating, potyviral genomic RNAs are translated to generate 2 polyproteins that are proteolytically processed by 3 protease domains into 11 viral proteins (VRs) including coat protein (CP), nuclear inclusion b (NIb) which is the only viral RNA-dependent RNA polymerase (RdRp), 2 viral suppressors of RNA silencing (VSRs), e.g., HC-Pro and VPg. Viral replicase proteins including NIb recruit host factors to form cellular membrane-associated viral replication complexes (VRCs) that catalyze viral replication. Viral dsRNAs, as replicative intermediates from replication), or generated by de novo synthesis by cellular RdRps using the viral genomic RNA as a template or as a result of internal pairing of viral RNA, trigger RNA silencing to defense against viral infection. VSRs suppress RNA silencing to promote viral infection. VSRs can also suppress biogenesis cellular DCL4-dependent small interference RNAs (siRNAs) that regulate post-transcriptionally resistance (R) genes mainly encoding nucleotide-binding site leucine-rich repeats (NB-LRR) receptors. Upregulated NB-LRR enhances effector-triggered immunity (ETI). Potyviral dsRNAs also function as a pathogen-associated molecular pattern (PAMP) to induce PAMP-triggered immunity (PTI). Potyviral CP may function as a virus-encoded innate immunity suppressor (VIIS) to suppress PTI. Other elements associated with viral genome replication and translation can also induce PTI and ETI. For example, some R proteins recognize potyviral NIb and other viral proteins to activate ETI. Plants may percept viral infection by PTI acceptors BAK1 and SERK1 to activate PTI. Activated PTI and/or ETI lead to the hypersensitive response (HR), necrosis and systemic acquired resistance (SAR). These are associated with changes in salicylic acid (SA), jasmonic acid (JA), ethylene (ET), nitric oxide (NO) and hydrogen peroxide (H2O2) levels. As a result, defense genes including those regulated by non-expressor of pathogenesis related genes-1 (NPR1) are upregulated. SMALL-UBIQUITIN LIKER MODIFIER 3 (SUMO3) is an activator of NPR1. Potyviral NIb targets the nucleus where it is sumopylated by SUMO3, which inhibits sumoylaiton of NPR1. Thus, NIb plays a role as a VIIS to promote viral infection. Sumoylated NIb retargets the cytoplasm for viral replication.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Jamie McNeil for technical support.
Funding
Work in the Wang laboratory was supported in part by a Genomics R&D Initiative (GRDI) grant from Agriculture and Agri-Food Canada (AAFC) and a discovery grant from the Natural Sciences and Engineering Research Council of Canada (NSERC).
References
- Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 2006; 124:803-14; PMID:16497589; https://doi.org/10.1016/j.cell.2006.02.008
- Mandadi KK, Scholthof KB. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013; 25:1489-505; PMID:23709626; https://doi.org/10.1105/tpc.113.111658
- Jones JD, Dangl JL. The plant immune system. Nature 2006; 444:323-9; PMID:17108957; https://doi.org/10.1038/nature05286
- Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat Rev Genet 2010; 11:539-48; PMID:20585331; https://doi.org/10.1038/nrg2812
- Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science 2013; 341:746-51; PMID:23950531; https://doi.org/10.1126/science.1236011
- Nicaise V. Crop immunity against viruses: outcomes and future challenges. Front Plant Sci 2014; 5:1-18; https://doi.org/10.3389/fpls.2014.00660
- Ding SW. RNA-based antiviral immunity. Nat Rev Immunol 2010; 10:632-644; PMID:20706278; https://doi.org/10.1038/nri2824
- Wang A, Krishnaswamy S. Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol Plant Pathol 2012; 13:795-803; PMID:22379950; https://doi.org/10.1111/j.1364-3703.2012.00791.x
- Wang A. Dissecting the molecular network of virus-plant interactions: The complex roles of host factors. Annu Rev Phytopathol 2015; 53:45-66; PMID:25938276; https://doi.org/10.1146/annurev-phyto-080614-120001
- Baulcombe D. RNA silencing in plants. Nature 2004; 431:356-63; PMID:15372043; https://doi.org/10.1038/nature02874
- Cheng X, Wang A. The potyviral silencing suppressor protein VPg mediates degradation of SGS3 via ubiquitination and autophagy pathways. J Virol 2017; 91:e01478-16; PMID:27795417; https://doi.org/10.1128/JVI.01478-16
- Zhao JH, Hua CL, Fang YY. Guo HS The dual edge of RNA silencing suppressors in the virus-host interactions. Curr Opin Virol 2016; 17:39-44; PMID:26802204; https://doi.org/10.1016/j.coviro.2015.12.002
- Schauer SE, Jacobsen SE, Meinke DW, Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 2002; 7:487-91; PMID:12417148; https://doi.org/10.1016/S1360-1385(02)02355-5
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004; 303:672-6; PMID:14704433; https://doi.org/10.1126/science.1093686
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 2000; 404:293-6; PMID:10749213; https://doi.org/10.1038/35005107
- Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 2010; 328:872-5; PMID:20413459; https://doi.org/10.1126/science.1187959
- Garcia-Ruiz H, Takeda A, Chapman EJ, Sullivan CM, Fahlgren N, Brempelis KJ, Carrington JC. Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip mosaic virus infection. Plant Cell 2010; 22:481-96; PMID:20190077; https://doi.org/10.1105/tpc.109.073056
- Wu Q, Wang X, Ding SW. Viral suppressors of RNA-based viral immunity: host targets. Cell Host Microbe 2010; 8:12-5; PMID:20638637; https://doi.org/10.1016/j.chom.2010.06.009
- Csorba T, Burgyán J. Antiviral silencing and suppression of gene silencing in plants, p. 1–33. In Wang A, Zhou X (ed), Curr Res Top Plant Virol. Springer International Publishing, Cham; 2016.
- Hemmes H, Lakatos L, Goldbach R, Burgyan J, Prins M. The NS3 protein of Rice hoja blanca tenuivirus suppresses RNA silencing in plant and insect hosts by efficiently binding both siRNAs and miRNAs. RNA 2007; 13:1079-89; PMID:17513697; https://doi.org/10.1261/rna.444007
- Havelda Z, Hornyik C, Valoczi A, Burgyan J. Defective interfering RNA hinders the activity of a tombusvirus‐encoded posttranscriptional gene silencing suppressor. J Virol 2005; 79:450-7; PMID:15596838; https://doi.org/10.1128/JVI.79.1.450-457.2005
- Lakatos L, Csorba T, Pantaleo V, Chapman EJ, Carrington JC, Liu YP, Dolja VV, Calvino LF, Lopez-Moya JJ, Burgyan J. Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J 2006; 25:2768-80; PMID:16724105; https://doi.org/10.1038/sj.emboj.7601164
- Chiu MH, Chen IH, Baulcombe DC, Tsai CH. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol Plant Pathol 2010; 11:641-9; PMID:20696002
- Hendelman A, Kravchik M, Stav P, Zik M, Lugassi N, Arazi T. The developmental outcomes of P0-mediated ARGONAUTE destabilization in tomato. Planta 2013; 237:363-77; PMID:23080016; https://doi.org/10.1007/s00425-012-1778-8
- Pumplin N, Voinnet O. RNA silencing suppression by plant pathogens: defence, counter-defence and counter-counter-defence. Nat Microbiol 2013; 11:745-760; https://doi.org/10.1038/nrmicro3120
- Zhai JX, Jeong DH, De Paoli E, Park S, Rosen BD, Li YP, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 2011; 25:2540-53; PMID:22156213; https://doi.org/10.1101/gad.177527.111
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B. MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 2012; 109:1790-5; PMID:22307647; https://doi.org/10.1073/pnas.1118282109
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012; 24:859-74; PMID:22408077; https://doi.org/10.1105/tpc.111.095380
- Kallman T, Chen J, Gyllenstrand N, Lagercrantz U. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol 2013; 162:741-54; PMID:23580593; https://doi.org/10.1104/pp.113.214643
- Yi H, Richards EJ. A cluster of disease resistance genes in Arabidopsis is coordinately regulated by transcriptional activation and RNA silencing. Plant Cell 2007; 19:2929-39; PMID:17890374; https://doi.org/10.1105/tpc.107.051821
- Burch-Smith TM, Schiff M, Caplan JL, Tsao J, Czymmek K, Dinesh-Kumar SP. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 2007; 5:e68; PMID:17298188; https://doi.org/10.1371/journal.pbio.0050068
- Li HW, Lucy AP, Guo HS, Li WX, Ji LH, Wong SM, Ding SW. Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J 1999; 18:2683-91; PMID:10329615; https://doi.org/10.1093/emboj/18.10.2683
- Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 2009; 324:742-3; PMID:19423812; https://doi.org/10.1126/science.1171647
- Kørner CJ, Klauser D, Niehl A, Dom ınguez-Ferreras A, Chinchilla D, Boller T, Heinlein M, Hann DR. The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol Plant Microbe Interact 2013; 26:1271-80; PMID:23902263; https://doi.org/10.1094/MPMI-06-13-0179-R
- Chen H, Arsovski AA, Yu K, Wang A. Deep sequencing leads to the identification of eukaryotic translation initiation factor 5A as a key element in Rsv1-mediated lethal systemic hypersensitive response to Soybean mosaic virus infection in soybean. Mol Plant Pathol 2017; 18:391-404; PMID:27019403; https://doi.org/10.1111/mpp.12407
- Nicaise V, Candresse T. Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol Plant Pathol 2016; PMID:27301551; https://doi.org/10.1111/mpp.12447
- Gouveia BC, Calil IP, Machado JP, Santos AA, Fontes EP. Immune receptors and co-receptors in antiviral innate immunity in plants. Front Microbiol 2017; 7:2139; PMID:28105028; https://doi.org/10.3389/fmicb.2016.02139
- Niehl A, Wyrsch I, Boller T, Heinlein M. Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol 2016; 211:1008-19; PMID:27030513; https://doi.org/10.1111/nph.13944
- Fu ZQ, Dong X. Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 2013; 64:839-63; PMID:23373699; https://doi.org/10.1146/annurev-arplant-042811-105606
- Withers J, Dong X. Posttranslational modifications of NPR1: a single protein playing multiple roles in plant immunity and physiology. PLoS Pathog 2016; 12:e1005707; PMID:27513560; https://doi.org/10.1371/journal.ppat.1005707
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, Dong X. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012; 486:228-32; PMID:22699612
- Saleh A, Withers J, Mohan R, Marques J, Gu Y, Yan S, Zavaliev R, Nomoto M, Tada Y, Dong X. Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 2015; 18:169-82; PMID:26269953; https://doi.org/10.1016/j.chom.2015.07.005
- van den Burg HA, Kini RK, Schuurink RC, Takken FLW. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 2010; 22:1998-2016; PMID:20525853; https://doi.org/10.1105/tpc.109.070961
- Cheng X, Xiong R, Li Y, Li F, Zhou X, Wang A. Sumoylation of Turnip mosaic virus RNA polymerase promotes viral infection by counteracting the host NPR1-mediated immune response. Plant Cell 2017; 29(3):508-25; PMID:28223439; https://doi.org/10.1105/tpc.16.00774
- Fellers JP, Tremblay D, Handest MF, Lommel SA. The Potato virus Y M(S)N(R) NIb-replicase is the elicitor of a veinal necrosis-hypersensitive response in root knot nematode resistant tobacco. Mol Plant Pathol 2002; 3:145-52; PMID:20569320; https://doi.org/10.1046/j.1364-3703.2002.00106.x
- Kim SB, Lee HY, Seo S, Lee JH, Choi D. RNA-dependent RNA polymerase (NIb) of the potyviruses is an avirulence factor for the broad-spectrum resistance gene Pvr4 in Capsicum annuum cv. CM334. PLoS One 2015; 10(3):e0119639; PMID:25760376; https://doi.org/10.1371/journal.pone.0119639
- Zhang L, Chen H, Brandizzi F, Verchot J, Wang A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet 2015; 11:e1005164; PMID:25875739; https://doi.org/10.1371/journal.pgen.1005164
