ABSTRACT
Cell fusion takes place in all domains of life and contributes greatly to the formation of complex multicellular structures. In particular, many fungi, such as the filamentous Neurospora crassa, rely on conspecific somatic cell fusion to drive the unicellular-to-multicellular transition and formation of the interconnected mycelial syncytium. This can, however, lead to the transmission of infectious elements and deleterious genotypes that have a negative impact on the organismal fitness. Accumulating evidence obtained from natural populations suggests that N. crassa has evolved various self/non-self or allorecognition systems to avoid fusion between genetically non-identical spores or hyphae at all costs. Here we present an overview of the recent advances made in the field of fungal allorecognition, describe its genetic basis, and comment on its evolutionary meaning. These data pinpoint the multilayered complexity of the cooperative social behaviors undertaken by a model eukaryotic microbe.
Cell-cell fusion: a social lifestyle
The classical philosopher Aristotle once wrote that “Man is by nature a social animal” (ca. 328 BC). That is not less true at the cellular level, where social behaviors are typified by the process of cell-cell fusion. Cell fusion occurs in all domains of life and all humans originate from a cell fusion event – an oocyte and a sperm cell fuse to form a zygote and the germinal stage of development ensues. More generally, cell fusion is involved in various stages of development such as syncytiotrophoblast and skeletal muscle formation, bone remodeling, organ sculpting, neuronal development, and tumorigenesis, just to enumerate a few (recently reviewed by [Citation1,Citation2]). In fungi, more specifically in the eukaryotic model Neurospora crassa, somatic cell fusion drives the unicellular-to-multicellular transition and is fundamental for the formation of the interconnected hyphal network known as the mycelium [Citation3].
Cell fusion between genetically non-identical N. crassa cells is controlled by allorecognition systems
The mycelial colony constitutes a polarized syncytium that expands and radiates as a result of tip elongation and cooperative cell fusion, allowing maximal ability to colonize substrates [Citation4]. However, this example of conspecific cooperation comes with a risk. On the one hand, Cell-cell and hyphal fusion within and between colonies potentially enhances the ability to withstand environmental variations and promotes an increase in organismal size due to the public nature of the created goods in a multinuclear syncytium. However, the transition to a multicellular state can result in conflict due to the potential transmission of infectious elements via somatic fusion events and incorporation of deleterious mitochondrial or nuclear genotypes that are detrimental for the overall fitness of the individuals. In this context, evidence from Neurospora and other microbes has shown that the higher the genetic relatedness, the higher the likelihood that cells will undergo cooperative behaviors in order to prevent parasitism and the exploitation of public goods by cheaters, defined as selfish units that receive benefit at the cost of others [Citation5–Citation9].
In N. crassa, the initial stages of multicellular colony establishment are characterized by cell fusion events between germinated unicellular spores (germlings) that are in close proximity. In natural settings, spores encounter numerous other spores with varying degrees of genetic similarity and fusion of genetically dissimilar cells results in cytoplasmic pools that contain a mixture of organelles originating from dissimilar genetic backgrounds, the so-called ‘heterokaryons’. To reduce the risk of detrimental heterokaryon formation, fungi have evolved allorecognition systems that ‘surveil’ genetic identity and impede the process of cell fusion from occurring whenever two partner cells are genetically distant. A well-established example of such a system is triggered during fusion of vegetative hyphae of N. crassa: if fusion between hyphae that are genetically identical at all het loci, a stable heterokaryon is formed; if hyphae have alternate specificity at any het locus, a defense response is elicited leading to cell death. Due to septal plugging, the death reaction is restricted to the heterokaryotic fusion cell and surrounding compartments, sparing the remainder of the colony [Citation10]. This phenomenon, termed heterokaryon incompatibility or vegetative incompatibility, is put in place to prevent somatic parasitism and the transmission of infectious elements and mycoviruses [Citation11–Citation13]. Importantly, het loci typically display features of rapid diversification [Citation14]. All het loci have been identified in hyphal-based experimental setups and het-gene-based allorecognition has not been extensively studied in germlings. Evidence exists that vegetatively incompatible strains can fuse at the germling stage in Colletotrichum lindemuthianum [Citation15], while in N. crassa, germling fusion occurs, although delayed, in situations of heterokaryon incompatibility [Citation16].
More recently, our group has used population genomics to discover cell fusion-associated allorecognition checkpoints that occur in germlings as well as hyphae. Conceptually speaking, cell fusion in germlings and hyphae can be divided into four steps (): 1) intercellular communication leading to chemotropic growth of the two fusing partners; 2) cell wall remodeling that is triggered after cell-cell contact has occurred; 3) plasma membrane merger; and 4) cytoplasmic mixing and the conclusion of the fusion process [Citation17]. Cells that have identical allelic specificity at the determinant of communication loci (doc-1 and doc-2) loci are capable of communicating with each other, while cells encoding doc alleles of different specificities fail to reinforce chemotropic interactions and consequently show a greatly reduced level of cell fusion [Citation18]. The DOC-1 and DOC-2 loci encode proteins lacking annotation or conserved domains and therefore their biochemical function remains unclear. Cells with identical specificity at the doc loci will undergo chemotropic interactions, but a second cell fusion checkpoint can be triggered upon cell contact and adhesion. In this case, the ability to proceed with cell wall dissolution is defined by genetic identity at the cell wall remodeling loci (cwr-1 and cwr-2). If cells have different cwr allelic specificity, adhered cells are unable to switch from chemotropism to cell wall dissolution, as shown by the continued oscillation of chemotropism-associated proteins and accumulation of cell wall material at the zone of contact [Citation19]. CWR-1 is a carbohydrate-binding polysaccharide monooxygenase potentially involved in directly modifying the cell wall while CWR-2 is a poorly conserved protein with several transmembrane domains and thus hypothesized to behave as a receptor at the cell periphery.
Figure 1. The different stages of somatic cell fusion in N. crassa and respective allorecognition checkpoint systems and genetic determinants that have been identified thus far
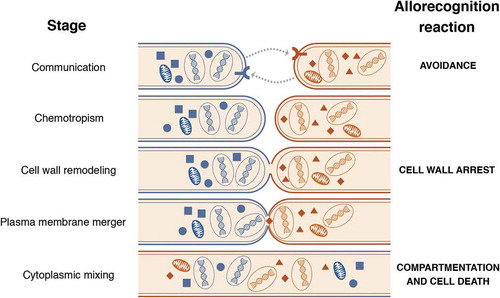
When the doc and cwr checkpoints are crossed, cell fusion and cytoplasmic mixing occur with the cellular constituents becoming shared goods in cytoplasmic pools. However, a third checkpoint controlled by allelic specificity at the sec-9 and plp-1 loci regulates the post-fusion viability of heterokaryons in germlings and hyphae. If cells encode different allelic specificities at sec-9/plp-1, rapid vacuolization and cell death are triggered [Citation20]. PLP-1 functions as a nucleotide-binding domain leucine-rich repeat containing receptor (NLR) in one cell, which is activated by interactions with the soluble N-ethylmaleimide sensitive factor attachment protein receptor (SNARE) SEC-9 in the fusion partner cell. More recently, another cell death-triggering locus was identified in wild N. crassa populations and that causes a phenotype similar to the sec-9/plp-1 reaction; in this case, the allorecognition determinant is the rcd-1 [Citation21]. The rcd-1 gene belongs to a large and uncharacterized family widespread in Ascomycota fungi and is one of the most polymorphic genes in the genomes of wild N. crassa and Neurospora discreta isolates (but not Neurospora tetrasperma) [Citation21]. The evolutionary benefit and/or meaning of the consequences of activation of these cell death-inducing allorecognition systems are intriguing – as they were found in germlings, cell death results in organismal death. However, as with genetic differences at het loci, differences at sec-9/plp-1 and rcd-1 also cause programmed cell death of hyphal fusion compartments [Citation20].
For all three germling fusion checkpoints, the corresponding alleles are highly polymorphic, segregate into discrete haplotypes, and show trans-species polymorphisms and evidence of balancing selecting acting on these loci. A simple calculation that takes into account the three germling checkpoints, the het hyphal checkpoints, and the number of haplotypes identified for each of them results in an estimated 2,211,840 incompatibility groups, that is, sets of strains that can only fuse successfully amongst themselves (). We speculate that it is highly possible that the addition of more isolates from various geographic provenances would result in the discovery of several more haplotypes, particularly in the case of the het loci, for which, in some cases, only two incompatible haplogroups have been described. Dozens of additional het loci have been recently identified [Citation22] and experimental testing of their role in heterokaryon incompatibility will likely expand the number of self/non-self recognition systems. Moreover, we have preliminary evidence that additional allorecognition checkpoints are activated during germling cell fusion.
Table 1. Summary of allorecognition systems in N. crassa. The right column shows the cumulative calculation of the total number of incompatible groups identified so far. The estimated total of incompatible genotypes is indicated in bold
A number of evolutionary pressures might have contributed to the high number of allorecognition systems in N. crassa. First of all, despite being important for multicellularity, cell fusion also provides an opportunity for cheater individuals to take advantage of shared cellular goods without participating in their production, weakening the fitness of the fungal colony [Citation6]. As a consequence, multicellular cooperation is enhanced in highly related individuals 6. Secondly, allorecognition presents an effective mechanism to prevent the accumulation of mycoviruses, as shown in Cryphonectria parasitica [Citation13], or mitochondrial defects, as shown in N. crassa [Citation12] and Neurospora intermedia [Citation23]. It seems puzzling that such a large number of incompatibility groups have been selected during evolution only for the purpose of avoidance of conspecific cell fusion, and therefore, additional research is required to fully comprehend the range of functions of these genetic identity-conferring molecules.
Avoiding fusion at all costs
Taken together, our findings strongly suggest that N. crassa has evolved to avoid conspecific somatic cell fusion with genetically non-identical cells at all costs. This has likely been driven by the detrimental effects caused by the incorporation of cheaters and infectious elements into the mycelium, which in turn impairs organismal fitness [Citation6,Citation11–Citation13,Citation17]. Pre-fusion allorecognition (based on doc and cwr loci) could function as a tool to avoid the drastic cost of the downstream cell death-associated non-self recognition systems.
Altogether, these studies have shown a series of multilayered and intricate non-self recognition systems that surveil genetic identity whenever cell fusion is imminent (). This phenomenon is a good illustration of the high level of complexity of the social interactions that can take place in the microbial world by seemingly simple organisms.
Acknowledgments
We would like to thank all the members of the Glass Lab at the University of California, Berkeley who have made important contributions to the field of fungal allorecognition over the last 25 years.
Disclosure statement
The authors declare no competing interests.
Additional information
Funding
References
- Brukman NG , Uygur B , Podbilewicz B , et al. How cells fuse. J Cell Biol. 2019;218:1436–1451.
- Platt JL , Cascalho M. Cell fusion in malignancy: a cause or consequence? A provocateur or cure? Cells. 2019;8:587.
- Fischer MS , Glass NL. Communicate and fuse: how filamentous fungi establish and maintain an interconnected mycelial network. Front Microbiol. 2019;10:619.
- Riquelme M , Aguirre J , Bartnicki-Garcia S , et al. Fungal morphogenesis, from the polarized growth of hyphae to complex reproduction and infection structures. Microbiol Mol Biol Rev. 2018;82:e00068–17.
- Aanen DK , Debets AJ , de Visser JA , et al. The social evolution of somatic fusion. BioEssays. 2008;30:1193–1203.
- Bastiaans E , Debets AJ , Aanen DK . Experimental evolution reveals that high relatedness protects multicellular cooperation from cheaters. Nat Commun. 2016;7:11435.
- Foster KR , Parkinson K , Thompson CR . What can microbial genetics teach sociobiology? Trends Genet. 2007;23:74–80.
- Kuzdzal-Fick JJ , Fox SA , Strassmann JE , et al. High relatedness is necessary and sufficient to maintain multicellularity in Dictyostelium . Science. 2011;334:1548–1551.
- Wloch-Salamon DM . Sociobiology of the budding yeast. J Biosci. 2014;39:225–236.
- Gonçalves AP , Heller J , Daskalov A , et al. Regulated forms of cell death in fungi. Front Microbiol. 2017;8:1837.
- Biella S , Smith ML , Aist JR , et al. Programmed cell death correlates with virus transmission in a filamentous fungus. Proc Biol Sci. 2002;269:2269–2276.
- Debets F , Yang X , Griffiths AJ . Vegetative incompatibility in Neurospora: its effect on horizontal transfer of mitochondrial plasmids and senescence in natural populations. Curr Genet. 1994;26:113–119.
- Zhang DX , Spiering MJ , Dawe AL , et al. Vegetative incompatibility loci with dedicated roles in allorecognition restrict mycovirus transmission in chestnut blight fungus. Genetics. 2014;197:701–714.
- Paoletti M , Saupe SJ . Fungal incompatibility: evolutionary origin in pathogen defense? BioEssays. 2009;31:1201–1210.
- Ishikawa FH , Souza EA , Shoji JY , et al. Heterokaryon incompatibility is suppressed following conidial anastomosis tube fusion in a fungal plant pathogen. PloS One. 2012;7:e31175.
- Gabriela Roca M , Read ND , Wheals AE . Conidial anastomosis tubes in filamentous fungi. FEMS Microbiol Lett. 2005;249:191–198.
- Hickey PC , Jacobson D , Read ND , et al. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa . Fung Genet Biol. 2002;37: 109–119.
- Heller J , Zhao J , Rosenfield G , et al. Characterization of greenbeard genes involved in long-distance kind discrimination in a microbial eukaryote. PLoS Biol. 2016;14:e1002431.
- Gonçalves AP , Heller J , Span EA , et al. Allorecognition upon fungal cell-cell contact determines social cooperation and impacts the acquisition of multicellularity. Curr Biol. 2019;29:3006–17 e3.
- Heller J , Clave C , Gladieux P , et al. NLR surveillance of essential SEC-9 SNARE proteins induces programmed cell death upon allorecognition in filamentous fungi. Proc Natl Acad Sci USA. 2018;115:E2292–E301.
- Daskalov A , Gladieux P , Heller J , et al. Programmed cell death in Neurospora crassa is controlled by the allorecognition determinant rcd-1. Genetics. 2019;213(4):1387–1400.
- Zhao J , Gladieux P , Hutchison E , et al. Identification of allorecognition Loci in Neurospora crassa by genomics and evolutionary approaches. Mol Biol Evol. 2015;32:2417–2432.
- Bastiaans E , Aanen DK , Debets AJ , et al. Regular bottlenecks and restrictions to somatic fusion prevent the accumulation of mitochondrial defects in Neurospora . Philos Trans R Soc London, Ser B. 2014;369:20130448.
- Hall C , Welch J , Kowbel DJ , et al. Evolution and diversity of a fungal self/nonself recognition locus. PloS One. 2010;5:e14055.
- Shiu PK , Glass NL . Molecular characterization of tol, a mediator of mating-type-associated vegetative incompatibility in Neurospora crassa . Genetics. 1999;151:545–555.
- Glass NL , Dementhon K . Non-self recognition and programmed cell death in filamentous fungi. Curr Opin Microbiol. 2006;9:553–558.
- Perkins DD . Main features of vegetative incompatibility in Neurospora crassa . Fungal Genet Newsl. 1988;35:44–46.
- Mylyk OM . Heteromorphism for heterokaryon incompatibility genes in natural populations of Neurospora crassa . Genetics. 1976;83:275–284.
